Abstract
AIMS--To study the geographical variation of the prevalence of hepatitis B virus (HBV) DNA in hepatitis B surface antigen (HBsAg) negative subjects. METHODS--A nested polymerase chain reaction (PCR) assay was used to amplify the core region of HBV. The assay was able to detect 10 molecules of a full length HBV plasmid. RESULTS--When applied to HBsAg negative paraffin wax embedded liver samples from Italy, Hong Kong, and the United Kingdom, a geographical variation in the prevalence of HBV-DNA positivity was noted. Two of 18 (11%) of Italian samples and 2/29 (6.9%) of Hong Kong samples were positive for HBV-DNA while none of the 70 cases from the United Kingdom was positive by nested PCR. Contamination by plasmid DNA was excluded using a novel method based on heteroduplex formation. One HBV-DNA positive case had idiopathic chronic active hepatitis, but the diagnoses in the other three HBV-DNA positive cases did not suggest any aetiological connection between HBV-DNA positivity and liver pathology. CONCLUSIONS--HBV-DNA could be detected in the liver tissues of a proportion of HBsAg negative subjects. The prevalence of such cases is related to the endemic rate of a geographical region. The use of HBV PCR on paraffin wax embedded tissues will be valuable for future studies on the molecular epidemiology of HBV.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- An S. F., Fleming K. A. Removal of inhibitor(s) of the polymerase chain reaction from formalin fixed, paraffin wax embedded tissues. J Clin Pathol. 1991 Nov;44(11):924–927. doi: 10.1136/jcp.44.11.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook M. G., Lever A. M., Kelly D., Rutter D., Trompeter R. S., Griffiths P., Thomas H. C. Antenatal screening for hepatitis B is medically and economically effective in the prevention of vertical transmission: three years experience in a London hospital. Q J Med. 1989 Apr;71(264):313–317. [PubMed] [Google Scholar]
- Bréchot C., Degos F., Lugassy C., Thiers V., Zafrani S., Franco D., Bismuth H., Trépo C., Benhamou J. P., Wands J. Hepatitis B virus DNA in patients with chronic liver disease and negative tests for hepatitis B surface antigen. N Engl J Med. 1985 Jan 31;312(5):270–276. doi: 10.1056/NEJM198501313120503. [DOI] [PubMed] [Google Scholar]
- Bréchot C. Polymerase chain reaction. A new tool for the study of viral infections in hepatology. J Hepatol. 1990 Jul;11(1):124–129. doi: 10.1016/0168-8278(90)90282-v. [DOI] [PubMed] [Google Scholar]
- Figus A., Blum H. E., Vyas G. N., De Virgilis S., Cao A., Lippi M., Lai E., Balestrieri A. Hepatitis B viral nucleotide sequences in non-A, non-B or hepatitis B virus-related chronic liver disease. Hepatology. 1984 May-Jun;4(3):364–368. doi: 10.1002/hep.1840040303. [DOI] [PubMed] [Google Scholar]
- Fowler M. J., Monjardino J., Weller I. V., Bamber M., Karayiannis P., Zuckerman A. J., Thomas H. C. Failure to detect nucleic acid homology between some non-A, non-B viruses and hepatitis B virus DNA. J Med Virol. 1983;12(3):205–213. doi: 10.1002/jmv.1890120306. [DOI] [PubMed] [Google Scholar]
- Gough N. M., Murray K. Expression of the hepatitis B virus surface, core and E antigen genes by stable rat and mouse cell lines. J Mol Biol. 1982 Nov 25;162(1):43–67. doi: 10.1016/0022-2836(82)90161-9. [DOI] [PubMed] [Google Scholar]
- Kaneko S., Feinstone S. M., Miller R. H. Rapid and sensitive method for the detection of serum hepatitis B virus DNA using the polymerase chain reaction technique. J Clin Microbiol. 1989 Sep;27(9):1930–1933. doi: 10.1128/jcm.27.9.1930-1933.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S., Miller R. H., Feinstone S. M., Unoura M., Kobayashi K., Hattori N., Purcell R. H. Detection of serum hepatitis B virus DNA in patients with chronic hepatitis using the polymerase chain reaction assay. Proc Natl Acad Sci U S A. 1989 Jan;86(1):312–316. doi: 10.1073/pnas.86.1.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok S., Higuchi R. Avoiding false positives with PCR. Nature. 1989 May 18;339(6221):237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- Lampertico P., Malter J. S., Colombo M., Gerber M. A. Detection of hepatitis B virus DNA in formalin-fixed, paraffin-embedded liver tissue by the polymerase chain reaction. Am J Pathol. 1990 Aug;137(2):253–258. [PMC free article] [PubMed] [Google Scholar]
- Liang T. J., Blum H. E., Wands J. R. Characterization and biological properties of a hepatitis B virus isolated from a patient without hepatitis B virus serologic markers. Hepatology. 1990 Aug;12(2):204–212. doi: 10.1002/hep.1840120205. [DOI] [PubMed] [Google Scholar]
- Lo Y. M., Lo E. S., Patel P., Tse C. H., Fleming K. A. Heteroduplex formation as a means to exclude contamination in virus detection using PCR. Nucleic Acids Res. 1991 Dec 11;19(23):6653–6653. doi: 10.1093/nar/19.23.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
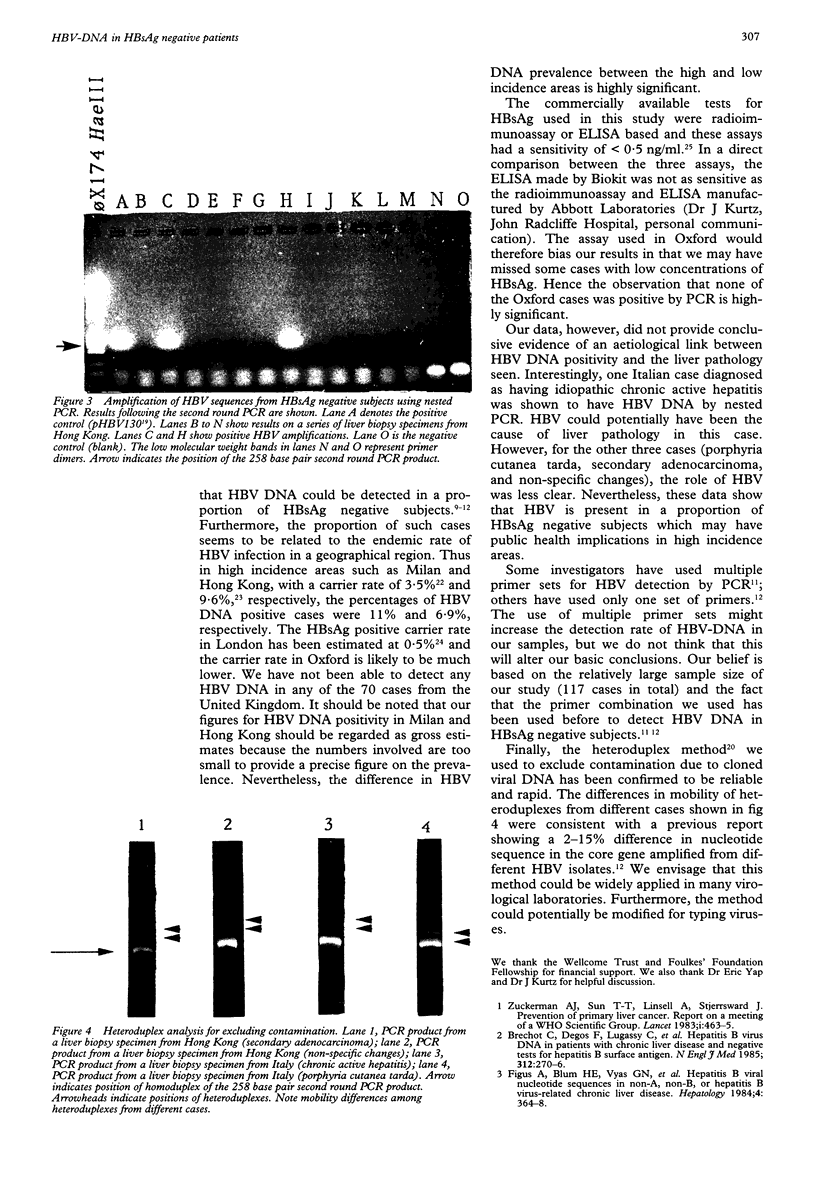
- Lo Y. M., Mehal W. Z., Fleming K. A. False-positive results and the polymerase chain reaction. Lancet. 1988 Sep 17;2(8612):679–679. doi: 10.1016/s0140-6736(88)90487-4. [DOI] [PubMed] [Google Scholar]
- Lo Y. M., Mehal W. Z., Fleming K. A. In vitro amplification of hepatitis B virus sequences from liver tumour DNA and from paraffin wax embedded tissues using the polymerase chain reaction. J Clin Pathol. 1989 Aug;42(8):840–846. doi: 10.1136/jcp.42.8.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevention of primary liver cancer. Report on a meeting of a W.H.O. Scientific Group. Lancet. 1983 Feb 26;1(8322):463–465. [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Slusarczyk J., Hess G., Hansson B. G., Meyer Zum Büschenfelde K. H. Lack of hepatitis B virus DNA sequences in sera from patients with acute and chronic liver diseases diagnosed as non-A, non-B-hepatitis. Liver. 1986 Dec;6(6):337–340. doi: 10.1111/j.1600-0676.1986.tb00301.x. [DOI] [PubMed] [Google Scholar]
- Stroffolini T., Pasquini P., Mele A. HBsAg carriers among pregnant women in Italy: results from the screening during a vaccination campaign against hepatitis B. Public Health. 1988 Jul;102(4):329–333. doi: 10.1016/s0033-3506(88)80102-1. [DOI] [PubMed] [Google Scholar]
- Thiers V., Nakajima E., Kremsdorf D., Mack D., Schellekens H., Driss F., Goudeau A., Wands J., Sninsky J., Tiollais P. Transmission of hepatitis B from hepatitis-B-seronegative subjects. Lancet. 1988 Dec 3;2(8623):1273–1276. doi: 10.1016/s0140-6736(88)92891-7. [DOI] [PubMed] [Google Scholar]
- Yokosuka O., Omata M., Hosoda K., Tada M., Ehata T., Ohto M. Detection and direct sequencing of hepatitis B virus genome by DNA amplification method. Gastroenterology. 1991 Jan;100(1):175–181. doi: 10.1016/0016-5085(91)90598-f. [DOI] [PubMed] [Google Scholar]
- Yokosuka O., Omata M., Imazeki F., Ito Y., Okuda K. Hepatitis B virus RNA transcripts and DNA in chronic liver disease. N Engl J Med. 1986 Nov 6;315(19):1187–1192. doi: 10.1056/NEJM198611063151903. [DOI] [PubMed] [Google Scholar]