Abstract
Qualitative expression of dissimilative sulfite reductase (dsrA), a key gene in sulfate reduction, and sulfide:quinone oxidoreductase (sqr), a key gene in sulfide oxidation was investigated. Neither of the two could be amplified from mRNA retrieved with Niskin bottles but were amplified from mRNA retrieved by the Deep SID. The sqr and sqr-like genes retrieved from the Cariaco Basin were related to the sqr genes from a Bradyrhizobium sp., Methylomicrobium alcaliphilum, Sulfurovum sp. NBC37-1, Sulfurimonas autotrophica, Thiorhodospira sibirica and Chlorobium tepidum. The dsrA gene sequences obtained from the redoxcline of the Cariaco Basin belonged to chemoorganotrophic and chemoautotrophic sulfate and sulfur reducers belonging to the class Deltaproteobacteria (phylum Proteobacteria) and the order Clostridiales (phylum Firmicutes).
Keywords: Cariaco Basin, cDNA, Gene expression, Sulfide:quinone oxidoreductase, Sulfite reductase, Sulfur cycle
INTRODUCTION
The Cariaco Basin, off the coast of Venezuela, is the largest truly marine permanently anoxic basin in the world [1]. Due to a shallow sill that separates the Basin from the Caribbean Sea and a strong salinity maximum at about 100 m the vertical and horizontal mixing of the waters is limited, generating complete anoxic conditions below 250-350 m [2]. The oxic-anoxic interface has been shown to vary in depth from 220 to 350 m and the redoxcline spans tens of meters, allowing for excellent sampling resolution of the redox gradient [3]. A prominent feature observed in Cariaco is dark inorganic carbon (DIC) assimilation, a large proportion of which takes place in zones where no O2 or light is detected. This implies that the terminal electron acceptor used by the bacterial population is not O2 but some other chemical species [3]. Recently, a localized sulfur cycle has been postulated for the redoxcline/anoxic zones of the Cariaco Basin that would explain the high DIC assimilation rates observed: a portion of the carbon fixed by chemoautotrophic microorganisms could be oxidized by sulfate-reducing microorganisms, which in turn could produce reduced sulfide compounds that fuel chemoautotrophic fixation of carbon [4]. Of course, this internal sulfur cycling in the redoxcline cannot be sustained without external sources of reductant and oxidant to provide chemical momentum. A similar phenomenon has been described for OMZs [5-7]. Support for this localized sulfur cycle has been provided by PCR-DGGE and pyrosequencing libraries of the 16S rRNA gene,whichshowed that themostprominent members ofthe bacterial community in the redox transition and anoxic zones of the Cariaco Basin putatively belong to sulfide oxidizing bacterial clades [8]. Microbial sulfide oxidation and sulfate reduction, therefore, appear to be important metabolic pathways in the water column of the Cariaco Basin.
Microbial sulfide oxidation involves a series of steps, the first of which is the oxidation of H2S to S 0, mediated by a membrane-bound electron transport system. Two enzymatic sulfide oxidizing systems have been described to catalyze this step, the flavocytochrome c, a sulfide:cytochrome c oxydoreductase (FCC), and the sulfide:quinone oxidoreductase (SQR) [9]. The sqr gene has been described in the majority of sulfide-oxidizing prokaryotes belonging to Proteobacteria, Chlorobi, Cyanobacteria, Aquificales and Archaea [9, 10]. It appears that sqr is the key enzyme for sulfide oxidation: a mutation in the sqr gene of Rhodobacter capsulatus leads to a loss of the ability to utilize sulfide [11] whereas a mutation of fcc in Chromatium vinosum did not affect its ability to utilize sulfide (C. vinosum also has a copy of sqr [12]). Phylogenetic analyses revealed that sqr gene sequences form six clades, each comprised of representatives from multiple bacterial phyla and, thus, showing incongruence with 16S rRNA gene-based phylogeny (i.e., likely lateral gene transfer [10]).
The dissimilatory sulfite reductase, encoded by the dsrAB genes [13], is ubiquitous to all known sulfate-reducing prokaryotes. Its highly conserved nucleotide sequence [13] has been successfully targeted to determine the diversity and distribution of SRB in several environments [e.g., 14-19]. DsrA is the primary enzyme in the dissimilatory sulfate reduction pathway of SRB and it catalyzes the six-electron reduction of bisulfite to sulfide in the last step of sulfate respiration [20, 21].
The diversity of dsrAB has been investigated for the Black Sea [16]. The diversity of either sqr or dsrA has not yet been studied in the Cariaco Basin. Moreover, simple detection of deoxyribonucleic acids (DNA) in an environmental sample does not necessarily mean that detected genes are actually being expressed [18]. On the other hand, preservation of intact mRNA during sample collection remains a difficult feat. Therefore, in order to build a cloning library for both DNA and mRNA of the sqr and dsrAB genes, a novel sampling devise “Deep-SID” was used and, as a result, the diversity and expression of the two sulfur metabolism genes in the Cariaco Basin redox transition zone were ascertained.
EXPERIMENTAL PROCEDURES
Sample Collection
Bacterioplankton samples were collected during the Car153 cruise on January 13th 2009 at station A (10.30°N, 64.40°W), the time series station for the CARIACO program (Carbon Retention In A Colored Ocean; http://www.imars.usf.edu/CAR/) situated in the eastern sub-basin, with a depth of 1400 m. Two different sampling methods were employed for collection of bacterial cells for nucleic acids isolation. In the first method, water samples were obtained with a Deep-SID (Submersible Incubation Device [22]). The devise takes several aliquots of ambient seawater at preprogrammed depths. Following sample collection, an aliquot of ProtectRNATM RNase inhibitor (500x, Sigma-Aldrich, St. Louis MO) was injected into the sample to a final concentration of 1x, according to manufacturer instructions. The mixture was filtered through polyvinylidene fluoride membranes (Durapore®, 0.22 μm pore size, 47 mm diameter; Millipore, Billerica, MA) and the filters were immersed in RNAlater (QIAGEN, Valencia CA), immediately frozen and stored at -20°C until delivered to the lab. In the second method water samples were obtained from 8 and 12-liter Teflon coated Niskin bottles under N2 atmosphere, filtered through polyvinylidene fluoride membranes (Durapore®, 0.22 μm pore size, 47 mm diameter; Millipore, Billerica, MA), immersed in RNAlater stabilization agent, immediately frozen and kept at -20°C until delivered to the lab, where they were stored at -80oC.
Total Nucleic Acids Extraction, mRNA Isolation, Poly(A)-tailing and cDNA Synthesis
For comparison of sampling methods and gene library construction, two samples were used, S2 (collected with method 1) and R6 (collected with method 2). Both samples were collected at 280 m, where the oxygen concentration was approximately 2 μmol kg-1, (CTD data), and the sulfide concentration was between 3.1 and 5.8 μM (http://www.imars.usf.edu/CAR/car_nodc/NODC-CAR153.txt).
All material and reagents used for nucleic acids extraction were nuclease free and all procedures were performed in a nuclease free environment. Samples consisted of a filter submerged in RNAlater stabilization reagent. The filters were removed from the RNAlater reagent and placed in a mortar to await extraction. The RNAlater reagent solution was pelleted down at 5500 rpm for 10 min in an Eppendorf tabletop centrifuge to separate the cells from the reagent. The pelleted cells were resuspended in 200 μL of β-mercaptoethanol/RLT plus (β-ME/RLT) buffer (10 μL of β-mercaptoethanol in 1 mL of RLT plus buffer, QIAGEN, Valencia, CA). The resuspended cells and 0.2 g of nuclease free sand were added to the mortar with the filters, covered with liquid nitrogen and grounded to a fine powder as recommended by the manufacturer’s instructions for the TruRNA MiniKit (Atom Sciences, Oak Ridge, TN). The slurry was then transferred to a 2 mL screw-capped centrifuge tube and covered with liquid nitrogen. After the nitrogen evaporated, 600 μL of β-ME/RLT buffer was added to the tube and the sample was vortexed at maximum speed for 10 min followed by 5 min incubation at room temperature. The extraction was then finished using an AllPrep DNA/RNA Mini kit, according to manufacturer’s instructions (QIAGEN, Valencia, CA). Integrity of the nucleic acids obtained was checked by gel electrophoresis and their concentration was determined spectrophotometrically with a NanoDrop ND1000 (NanoDrop Technologies, Inc., Wilmington, DE). The lack of DNA contamination in the total RNA extracted was verified by PCR amplification with eubacterial universal primers from Weisburg et al. [23]. The DNA extracted was stored at -20°C, while the total RNA was immediately used for mRNA isolation with an mRNA-ONLYTM Prokaryotic mRNA Isolation kit with Poly(A)-Tailing following manufacturer’s instructions (Epicentre, Madison, WI). The poly(A)-tailed mRNA obtained was immediately reverse transcribed using a ProtoScript® AMV LongAmpTM Taq RT-PCR kit, according to manufacterer’s instructions (New England BioLabs Inc., Ipswich, MA). Synthesized cDNA was stored at -20°C.
Functional Gene Amplification and Cloning
The sqr gene was amplified using two of the primer sets designed by Pham et al. [10] for Bacteria, with modifications to encompass a wider diversity of the sqr genes from Gamma- and Epsilonproteobacteria . To target the Gammaproteobacteria, the primer set SQR-G1-475FM (5’- TGY TWY GGB CCV GCB TAY GA -3’) and SQR-G1-964R (5’- GTS ACC ATS SWT TCR ATC AT -3’) was used. To target the Epsilonproteobacteria, the primer set SQR-G4-140FB (5’- TGG ATY CCM TCA AAY ATW TGG GT -3’) and SQR-G4-840RM (5’- AAT WAG CAT DGC RAA RTC RAA CTC -3’) was used. Each amplification reaction mixture (50 μL final volume) consisted of 25 μL of 2x GoTaq® Green Master Mix (Promega, Madison, WI), 5 μL of nucleic acid (concentration, 5 to 15 ng/μL), and each primer at a final concentration of 500 nM. The amplification cycling protocol had the following profile: initial denaturation at 95°C for 3 min, followed by 40 cycles of 30 seconds at 95°C, 30 seconds at XX°C, and 45 seconds at 72°C, followed by 5 min final extension at 72°C, where XX was 58 °C for the SQR-G1 primer set and 52 °C for the SQR-G4 primer set.
The dsrA gene from sulfate-reducing microorganisms was amplified using primers designed to target conserved regions of the gene, based on multiple alignments of cultured and environmental SRB sequences retrieved from GenBank. dsrA1FM (5’- ACS CAY TGG AAR CAY GG -3’; modified from Wagner et al. [13] to widen a range of amplified the dsrA genes) and dsr225R1 (5’- TCD CCD GTD GMR CCR TGS AWR TT -3’). Each amplification reaction mixture was as described above for the sqr gene. The amplification conditions were as follows: initial denaturation at 95°C for 3 min, followedby 35cycles of30 secondsat 95°C, 30 secondsat 55°C, and 45 seconds at 72°C, followed by 5 min final extension at 72°C. A gel with amplification products is shown in Fig (S1 (598.5KB, pdf) ).
The amplified fragments were ligated using the pGEM®-T easy vector system I kit and used to transform JM109 High Efficiency competent cells according to manufacturer’s instructions (Promega, Madison, WI). At least 50 white clones were picked and plasmids were extracted with the ZyppyTM Plasmid Miniprep kit as suggested by the manufacturer (Zymo Research, Irvine, CA). An enzymatic digestion with EcoRI (New England BioLabs) followed by gel electrophoresis was performed to check the presence of an insert. Plasmid insets were directly sequenced using a BigDye Terminator v3.1 Cycle sequencing kit (Applied Biosystems, Foster City, CA) with the primer T7 (5’- TAA TAC GAC TCA CTA TAG GG -3’) and a 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA).
Phylogenetic Analysis
Potential homologs of the generated sequences were identified in the GenBank database using the BLASTn program. To perform phylogenetic analyses of the cloned genes, alignments were built that included sequences of respective genes from several representatives of known cultured bacteria as well as sequences from the closest cultured and non-cultured relatives identified by BLASTn. Sequences were aligned using the ClustalW v2.0 aligner [24] through the EMBL-EBI framework [25] and the MAFFT aligner [26]. Phylogenetic trees were calculated with the neighbor joining and maximum likelihood algorithms using MEGA [27]. Bootstrap analysis (1000 replicates) was used to obtain confidence estimates for the phylogenetic tree topologies.
Clones Sequence Accession Numbers
Cloned sequences obtained from the sqr and dsrA gene libraries have been deposited in GenBank under accession numbers JX020805 through JX020940.
RESULTS
Nucleic Acids Isolation
Samples from the redoxcline of the Cariaco Basin were obtained using two different sampling methods: traditional Niskin bottles and the Deep-SID. It takes on average 2 hours for a sample from a Niskin bottle to be collected from the redox-transition zone and processed. The Deep-SID allows for immediate fixation of a sample following its collection. Both sets of samples were treated with the same mRNA/DNA extraction and isolation techniques. The yield of every nucleic acid fraction was higher for sample S2 and, in addition, the DNA isolated from sample S2 was of a higher molecular mass. mRNA comprised approximately 25% of total RNA for both samples.
cDNA obtained from sample S2 was amplifiable with dsrA and the two sqr primer sets. On the other hand, cDNA obtained with method 2 could not be amplified with either dsrA or sqr primer sets. Both cDNA preparations (i.e., S2 and R6) were amplifiable with universal bacterial 16S rRNA primers, albeit indicating incomplete removal of 16S rRNA (data not shown). DNA obtained by either method could be amplified with the universal bacterial 16S rRNA and dsrA primers. The SQR-G1 primers but not SQR-G4 successfully amplified the sqr gene from S2 DNA. With the R6 sample the situation was opposite, sqr was amplifiable with the SQR-G4 but not SQR-G1 primer sets.
The sqr Gene in the Cariaco Basin
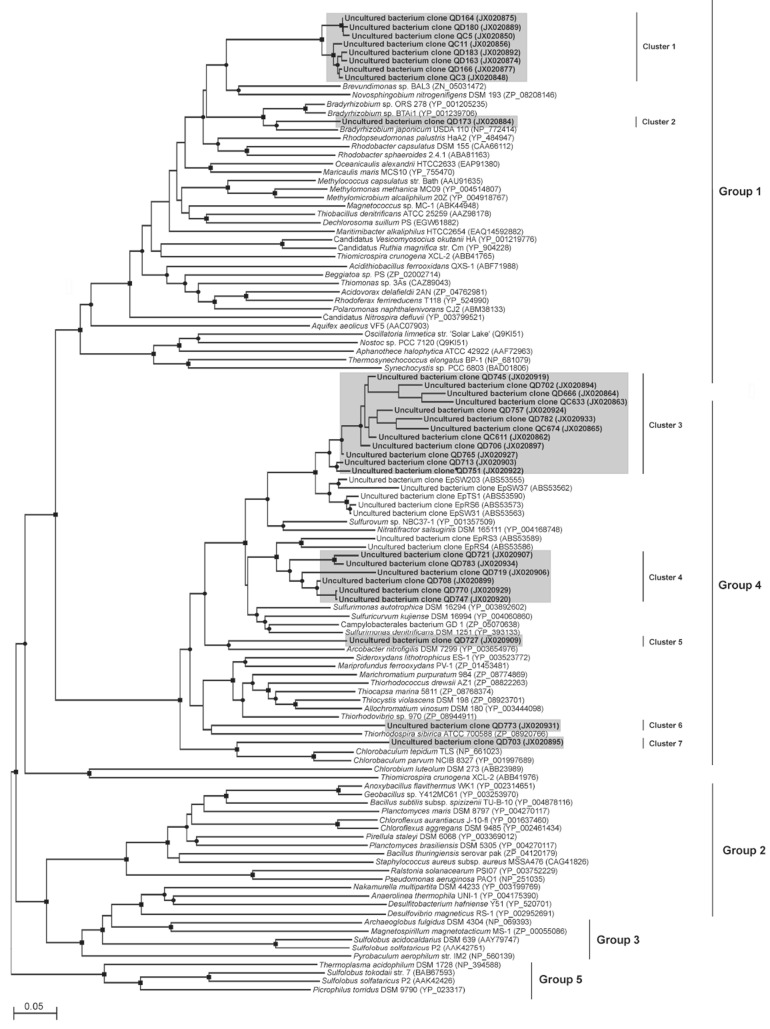
PCR products amplified with the dsrA and two sqr primers sets from S2 DNA and cDNA and R6 DNA were cloned and sequenced. The sqr sequencesexpected tobe foundin the CariacoBasin belongto groups 1 and 4as per Pham et al. [10] (Fig. 1 and Table 1). Other sqr groups, groups 2, 3 and 5 of Pham et al. [10], are not present in the Cariaco Basin. The SQR-G1 primer set amplified from both S2 cDNA and DNA almost exclusively sqr sequences related to those of gammaproteobacterial genera Methylomonas methanica and Methylomicrobium alcaliphilum. The only other sqr sequence amplified by this primer set is closely related to the sqr gene of Bradyrhizobium japonicum.
Fig. (1).
Neighbor joining phylogenetic analysis of sqr sequences. Names in bold correspond to sequences generated in this study. Clone names beginning with QC refer to sequences obtained from cDNA. Clone names beginning with QD refer to sequences obtained from DNA Groups according to Pham et al. (2008) are indicated to the right of the sequences. ■, indicates bootstrap values >75%; ●, indicates values between 50 and 75%. The scale bar indicates 0.05 substitutions per site.
Table 1.
sqr sequences identified in the redoxcline of the Cariaco Basin water column.
| Phylogenetic affiliation | Representative clone | No. of clones in library | Closest relative based on amino acid sequence, Accession No. (amino acid % identity, % similarity to the closest relative) | DNA based identity (%) | ||
|---|---|---|---|---|---|---|
|
S2
cDNA |
S2
DNA |
R6
DNA |
||||
| Proteobacteria | ||||||
| Alphaproteobacteria | QD713 | 1 | Bradyrhizobium japonicum, NP_772414 (86, 89) | 83 | ||
| Epsilonproteobacteria | QC611, QD745 | 11 | 24 | Sulfurovum sp. NBC37-1, YP_001357509 (86, 91) | 85 | |
| QD713 | 3 | Sulfurovum sp. NBC37-1, YP_001357509 (92, 93) | 83 | |||
| QD721 | 16 | Sulfurimonas autotrophica, YP_003892602 (87, 90) | 80 | |||
| QD727 | 3 | Arcobacter nitrofigilis, YP_003654976 (74, 73) | 72 | |||
| Gammaproteobacteria | QC3, QD183 | 16 | 27 | Methylomicrobium alcaliphilum 20Z YP_004918767(74, 81) | 71 | |
| QD773 | 1 | Thiorhodospira sibirica, ZP_08920766 (63, 79) | 70 | |||
| Chlorobi | QC703 | 1 | Chlorobium tepidum, NP_661023 (76, 72) | 68 | ||
The SQR-G4 primer set amplified a wider variety of sqr genes from S2 cDNA and R6 DNA, mostly those belonging to sulfur oxidizing epsilonproteobacteria (Sulfurovum, Arcobacter and Sulfurimonas spp.). One of the sqr sequences obtained from R6 DNA with this primer set was closely related to sqr of a gammaproteobacterium, Thiorhodospira sibirica, in family Ectothiorhodospiraceae. Interestingly, one of the sequenced sqr clones found in the S2 cDNA library was closely related to the sqr gene of Chlorobium tepidum and/or Chlorobaculum parvum, green sulfur bacteria.
The dsrA Gene in the Cariaco Basin
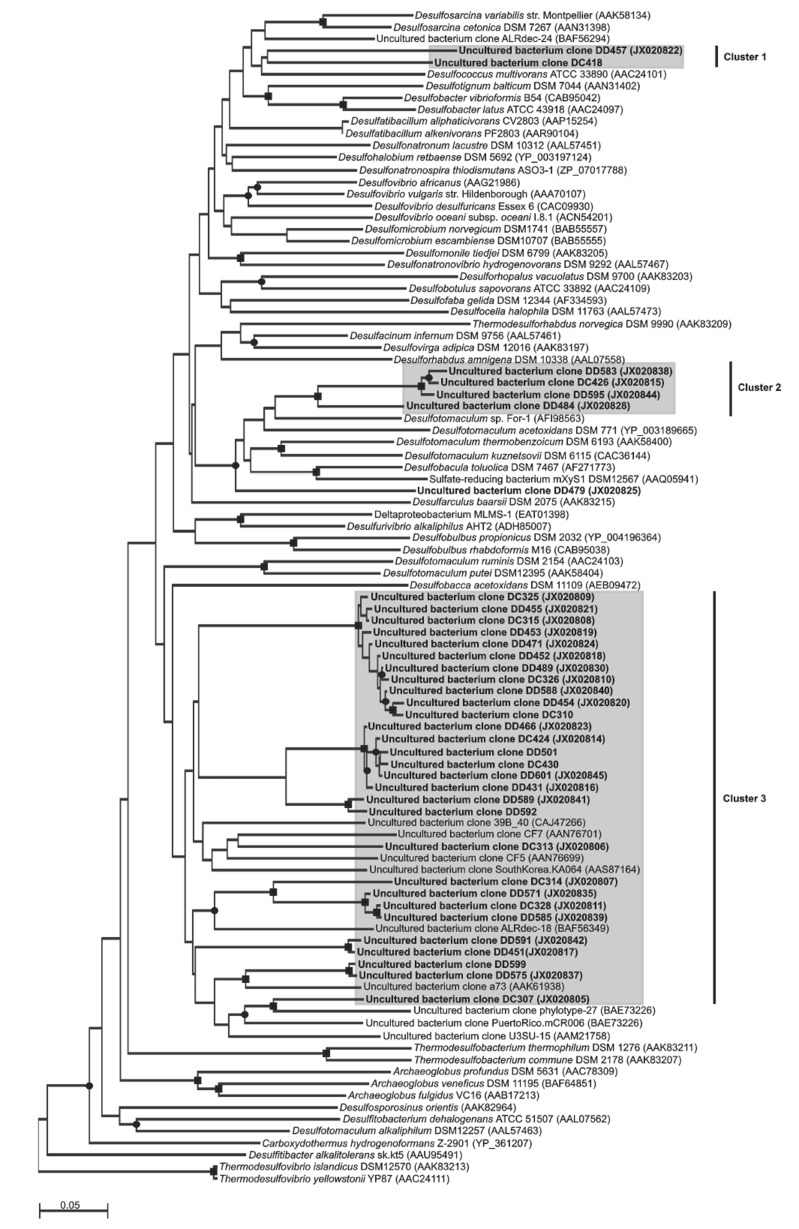
dsrA libraries were generated and analyzed for S2 and R6 DNA and S2 cDNA (Table 2). All dsrA sequences fell into three different phylogenetic clusters (Fig. 2). Cluster 1 was the smallest and contained dsrA sequences found only in S2 DNA and cDNA. dsrA cluster 1 sequences were closely related to dsrA from the uncultured sulfate reducing bacterium clone ALRdec-24 and more distantly to dsrA from Desulfococcus multivorans. Cluster 2 dsrA sequences were closely related to each other and were present in all three libraries. They were closely related to Desulfotomaculum spp., based on phylogenetic and BLAST analyses.
Table 2.
dsrA sequences identified in the redoxcline of the Cariaco Basin water column.
| Phylogenetic affiliation | Representative clone | No. of clones in library | Closest relative based on amino acid sequence, Accession No. (% identity; % similarity) | DNA based identity (%) | ||
|---|---|---|---|---|---|---|
|
S2
cDNA |
S2
DNA |
R6
DNA |
||||
| Deltaproteobacteria | ||||||
| Syntrophobacterales | DC307 | 1 | Phylotype 27, BAE73226 (84, 91) | 83 | ||
| DC313 | 1 | 3 | Clone 39b_40, CAJ47266 (80, 86) | 79 | ||
| DC314 | 1 | Clone ALRdec-18, BAF56349 (80, 92) | 78 | |||
| DC315, DD455, DD489 | 10 | 8 | 8 | Desulfobacca acetoxidans, AEB9473 (72, 83) | 70 | |
| DC328, DD585 | 2 | 4 | Clone ALRdec-18, BAF56349 (86, 93) | 79 | ||
| DC430, DD466, DD501 | 2 | 5 | 2 | Desulfobacca acetoxidans, AEB9473 (63, 74) | 72 | |
| DD451, DD591 | 1 | 2 | Desulfobacca acetoxidans, AEB9473 (69, 81) | 74 | ||
| DD575, DD599 | 1 | 1 | Clone a73, AAK61938 (82, 90) | 83 | ||
| DD589 | 2 | Clone 39b_40, CAJ47266 (77, 85) | 77 | |||
| Desulfobacterales | DC418, DD457 | 3 | 1 | Clone ALRdec-24, AB271553 (91, 94) | 81 | |
| Firmicutes | DC426, DD583 | 1 | 3 | Desulfotomaculum sp. For-1, AFI98563 (84, 88) | 77 | |
| DD479 | 1 | Desulfotomaculum sp. For-1, AFI98563 (81, 85) | 72 | |||
| DD484 | 1 | Desulfotomaculum sp. For-1, AFI98563 (83, 92) | 82 | |||
Fig. (2).
Neighbor joining phylogenetic analysis of dsrA sequences. Names in bold correspond to sequences generated in this study. Clone names beginning with DC refer to sequences obtained from cDNA. Clone names beginning with DD refer to sequences obtained from DNA. ■, indicates bootstrap values >75%; ●, indicates values between 50 and 75%. The scale bar indicates 0.05 substitutions per site.
Representatives of cluster 3 comprised 84% of all deltaproteobacterial clones sequenced and were found in all three libararies. Their dsrA genes were related to uncultured sulfate reducing bacteria and more distantly related to Desulfobacca acetoxidans.
DISCUSSION
Messenger RNA stabilization is a major issue when sampling bacterial communities in the environment. The average half-life time for mRNA in E. coli is below 6.8 minutes but can be as low as less than a 1 minute [28]. Thus, during sample collection, water samples need to be kept in conditions as similar to the natural environment as possible until fixed or frozen. Intact mRNA preservation is also critical for post-sampling transportation. Instant freezing in liquid nitrogen is generally recognized as the method of choice to stabilize and preserve RNA samples [29]. However, the logistics of transportation for international travel, for example, makes liquid nitrogen use a really complicated strategy. Here we tested a method based on the usage of the Deep-SID, which appears to be effective in stabilizing RNA and DNA for downstream molecular analysis.
Previous work [3, 4, 8, 30] strongly suggests the presence of an internal sulfur cycle in the Cariaco Basin, in which dominant sulfur Gammaproteobacteria and less prominent sulfur epsilonproteobacteria oxidize sulfide to sulfate and Deltaproteobacteria then reduce sulfate to sulfide. This suggested a choice of genes for investigation of expression: dissimilative sulfite reductase (dsrA gene as a proxy) is a key gene in sulfate reduction whereas sulfide:quinone oxidoreductase (sqr) is deemed to be a key gene in sulfide oxidation.
Sequences of the sqr gene have been shown to be present throughout a wide range of prokaryotic phyla [10]. Given the biogeochemical data available from previous studies involving the redoxcline of the Cariaco Basin [3, 4, 8, 31, 32], the phylogenetic groups of interest in the present study were the gamma- and epsilonproteobacteria whose sqr genes fall in the sqr groups 1 and 4 of Pham et al. [10].
Within the sqr group 1, retrieved sequences were related to sqr sequences from the gamma- (cluster 1, Fig. 2) and alphaproteobacteria (cluster 2). Representatives of alphaproteobacteria from the redoxcline of the Cariaco Basin found to harbor the sqr gene include genus Bradyrhizobium. Species of this genus can grow chemolithotrophically with molecular hydrogen, carbon dioxide and low levels of oxygen but cannot oxidize sulfur species [33]. The sqr-like genes from gammaproteobacterial genera Methylomonas methanica and Methylomicrobium alcaliphilum were related to the Cariaco sqr clones in cluster 1 (Fig. 2). Species of these genera are obligate methanotrophs that utilize methane and methanol for carbon and energy with oxygen as electron acceptor [34, 35]. Sulfide oxidation has not been reported for either of these two particular Alpha or Gammaproteobacterial genera. However, SQR belongs to a family of similar proteins, which can carry out various reactions unrelated to sulfur metabolism. Thus, Methylomicrobium alcaliphilum “sqr” is more likely a membrane bound formaldehyde dehydrogenase [36]. Sulfide-quinone reductase-like proteins are believed to help regulate cellular sulfide concentrations in humans [37] and in yeasts they are involved in heavy metal tolerance [38].
Group 4 sqr genes retrieved from the Cariaco Basin are closely related to sqr genes from sulfur bacteria and, thus, are most likely involved in in situ sulfide oxidation. One of these sulfur bacteria includes an epsilonproteobacterium, which is phylogenetically related to Sulfurovum sp. NBC37-1. 16S rDNA sequences closely related to Sulfurovum spp. have been previously found in the redoxcline of Cariaco [8]. Isolates of this genus have been demonstrated to grow chemolithoautotrophically with S 0 or thiosulfate as electron donors and oxygen or nitrate as electron acceptors [39]. The genome of Sulfurovum sp. NBC37-1 also includes genes related to utilization of H2 and S2-, in addition to S 0 and thiosulfate, as energy sources [40]. The sqr gene from Sulfurimonas autotrophica is related to sqr cluster 4 sequences found in this study (Fig. 1). S. autotrophica utilizes S 0, thiosulfate or sulfide as the sole electron donors for chemolithoautotrophic growth and so far only oxygen has been observed as an electron acceptor for this bacterium [41]. The sqr gene has been described in the genome of S. autotrophica [42]. Only one gammaproteobacterial sqr (from Thiorhodospira sibirica) was retrieved from the Cariaco Basin (Fig. 2). Sulfur oxidizing Gammaproteobacteria is believed to sometimes comprise up to 90% of all bacteria present in the Cariaco redox transition zone [8] and it is enigmatic that only one gammaproteobacterial sqr sequence was retrieved. The SQR-G1 and SQR-G4 primer sets used in this work should amplify all gammaproteobacterial sqr genes so far been deposited in GenBank. Perhaps, sulfur oxidizing Gammaproteobacteria use for sulfide oxidation an as yet unknown enzyme, which is distinct from sqr. T. sibirica has been described as a strictly anaerobic, obligately phototrophic, purple sulfur bacterium, which utilizes hydrogen sulfide and elemental sulfur as electron donors during photosynthesis [43]. One sqr gene sequence in the S2 cDNA library was closely related to sqr from the green-sulfur bacteria, phylum Chlorobi (Chlorobium tepidum and Chlorobaculum parvum). Chlorobia are obligate anaerobic photolithoautotrophs [44]. Reduced sulfur compounds oxidized by representatives of this phylum include sulfide, S 0, polysulfides, thiosulfates and tetrathionate [45, 46]. Given that there is no light present in the redoxcline of the Cariaco Basin, the importance of photosynthetic Chlorobi for the Cariaco biogeochemistry is questionable, even despite the fact that chlorobial sqr is being expressed in the Cariaco redox transition zone.
The dsrA gene sequences obtained from the redoxcline of the Cariaco Basin belonged to chemoorganotrophic and chemoautotrophic sulfate and sulfur reducers from the class Deltaprotebacteria (phylum Proteobacteria) and the order Clostridiales (phylum Firmicutes) (Fig. 2). The dsrA gene from Desulfococcus spp., bacteria which completely oxidize formate, lactate, pyruvate, alcohols, monocarboxylic acids, acetone and phenyl-substituted organic acids to CO2 with sulfate, sulfite and thiosulfate as electron acceptors, was related to two sequences retrieved from the Cariaco Basin. Fermentation by bacteria from this genus has also been observed in the absence of an external electron acceptor [47]. The majority of the Cariaco dsrA sequences were related to dsrA gene of Desulfobacca acetoxidans (order Syntrophobacterales). This bacterium utilizes sulfate, sulfite and thiosulfate as electron acceptors with acetate as electron donor and carbon source, oxidizing acetate completely to carbon dioxide [48]. Desulfotomaculum spp. detected in the Cariaco Basin include chemoorganotrophic strains that oxidize simple organic compounds either completely to carbon dioxide or incompletely to acetate. Several species can grow autotrophically with molecular hydrogen and carbon dioxide as sole carbon source. Species of this genus utilize sulfate, sulfite and thiosulfate as electron acceptors, and fermentation has been observed in the absence of sulfate [49].
Thus, we successfully amplified the genes encoding for SQR and DsrA from environmental DNA and RNA recovered from the Cariaco Basin. Further metagenome and metatranscriptom comparison for the Cariaco Basin microbial community is required to elucidate the role of abundant and rare bacteria in the Cariaco biogeochemistry.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publishers Website along with the published article.
ACKNOWLEDGEMENTS
Support for this work came from NSF grant MCB03-47811 to AYC, MIS, and GTT and NSF grant OCE-1061774 to VPE and CT. We gratefully acknowledge the captain and crew of the B/O Hermano Gines and the staff of the Estación de Investigaciones Marinas de Margarita (Fundación La Salle de Ciencias Naturales, Punta de Piedras, Edo. Nueva Esparta, Venezuela), specially Prof. Ramon Varela and Yrene Astor.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Richards F.A. The Cariaco Basin (trench). Oceanogr. Mar. Biol. Annu. Rev. 1975;13:11–67. [Google Scholar]
- 2.Scranton M.I., Astor Y., Bohrer R., Ho T.H., Muller-Karger F. Controls on temporal variability of the geochemistry of the deep Cariaco Basin. Deep Sea Res. Part I Oceanogr. Res. Pap. 2001;48:1605–1625. doi: 10.1016/S0967-0637(00)00087-X. [DOI] [Google Scholar]
- 3.Taylor G.T., Iabichella M., Ho T.Y., Scranton M.I., Thunnell R.C., Muller-Karger F., Varela R. Chemoautotrophy in the redox transition zone of the Cariaco Basin: a significant midwater source of organic carbon production. Limnol. Oceanogr. 2001;46:146–163. doi: 10.4319/lo.2001.46.1.0148. [DOI] [Google Scholar]
- 4.Li X.N., Taylor G.T., Astor Y., Varela R., Scranton M.I. The conundrum between chemoautotrophic production and reductant and oxidant supply: a case study from the Cariaco Basin. Deep Sea Res. Part I Oceanogr. Res. Pap. 2012;61:1–10. doi: 10.1016/j.dsr.2011.11.001. [DOI] [Google Scholar]
- 5.Stevens H., Ulloa O. Bacterial diversity in the oxygen minimum zone of the eastern tropical South Pacific. Environ. Microbiol. 2008;10(5):1244–1259. doi: 10.1111/j.1462-2920.2007.01539.x. [DOI] [PubMed] [Google Scholar]
- 6.Lavik G., Stührmann T., Brüchert V., Van der Plas A., Mohrholz V., Lam P., Mussmann M., Fuchs B.M., Amann R., Lass U., Kuypers M.M. Detoxification of sulphidic African shelf waters by blooming chemolithotrophs. Nature. 2009;457(7229):581–584. doi: 10.1038/nature07588. [DOI] [PubMed] [Google Scholar]
- 7.Canfield D.E., Stewart F.J., Thamdrup B., De Brabandere L., Dalsgaard T., Delong E.F., Revsbech N.P., Ulloa O. A cryptic sulfur cycle in oxygen-minimum-zone waters off the Chilean coast. Science. 2010;330(6009):1375–1378. doi: 10.1126/science.1196889. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Mora M.J., Scranton M.I., Taylor G.T., Chistoserdov A.Y. Bacterial community composition in a large marine anoxic basin: a Cariaco Basin time-series survey. FEMS Microbiol. Ecol. 2013;84(3):625–639. doi: 10.1111/1574-6941.12094. [DOI] [PubMed] [Google Scholar]
- 9.Griesbeck C., Günter H., Schütz M. Biological sulfide-oxidation: sulfide-quinone reductase (SQR), the primary reaction, In: Pandalai SG, editor. Recent Research Developments in Microbiology. India: Research Signpost.; 2000. pp. 129–203. [Google Scholar]
- 10.Pham V.H., Yong J-J., Park S-J., Yoon D-N., Chung W-H., Rhee S-K. Molecular analysis of the diversity of the sulfide : quinone reductase (sqr) gene in sediment environments. Microbiology. 2008;154(Pt 10):3112–3121. doi: 10.1099/mic.0.2008/018580-0. [DOI] [PubMed] [Google Scholar]
- 11.Schütz M., Maldener I., Griesbeck C., Hauska G. Sulfide-quinone reductase from Rhodobacter capsulatus: requirement for growth, periplasmic localization, and extension of gene sequence analysis. J. Bacteriol. 1999;181(20):6516–6523. doi: 10.1128/jb.181.20.6516-6523.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reinartz M., Tschäpe J., Brüser T., Trüper H.G., Dahl C. Sulfide oxidation in the phototrophic sulfur bacterium Chromatium vinosum. Arch. Microbiol. 1998;170(1):59–68. doi: 10.1007/s002030050615. [DOI] [PubMed] [Google Scholar]
- 13.Wagner M., Roger A.J., Flax J.L., Brusseau G.A., Stahl D.A. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 1998;180(11):2975–2982. doi: 10.1128/jb.180.11.2975-2982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minz D., Flax J.L., Green S.J., Muyzer G., Cohen Y., Wagner M., Rittmann B.E., Stahl D.A. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl. Environ. Microbiol. 1999;65(10):4666–4671. doi: 10.1128/aem.65.10.4666-4671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loy A., Lehner A., Lee N., Adamczyk J., Meier H., Ernst J., Schleifer K-H., Wagner M. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 2002;68(10):5064–5081. doi: 10.1128/AEM.68.10.5064-5081.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neretin L.N., Schippers A., Pernthaler A., Hamann K., Amann R., Jørgensen B.B. Quantification of dissimilatory (bi)sulphite reductase gene expression in Desulfobacterium autotrophicum using real-time RT-PCR. Environ. Microbiol. 2003;5(8):660–671. doi: 10.1046/j.1462-2920.2003.00452.x. [DOI] [PubMed] [Google Scholar]
- 17.Ben-Dov E., Brenner A., Kushmaro A. Quantification of sulfate-reducing bacteria in industrial wastewater, by real-time polymerase chain reaction (PCR) using dsrA and apsA genes. Microb. Ecol. 2007;54(3):439–451. doi: 10.1007/s00248-007-9233-2. [DOI] [PubMed] [Google Scholar]
- 18.Dar S.A., Yao L., van Dongen U., Kuenen J.G., Muyzer G. Analysis of diversity and activity of sulfate-reducing bacterial communities in sulfidogenic bioreactors using 16S rRNA and dsrB genes as molecular markers. Appl. Environ. Microbiol. 2007;73(2):594–604. doi: 10.1128/AEM.01875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blazejak A., Schippers A. Real-time PCR quantification and diversity analysis of the functional genes aprA and dsrA of sulfate-reducing prokaryotes in marine sediments of the Peru continental margin and the Black Sea. Front. Microbiol. 2011;2:253. doi: 10.3389/fmicb.2011.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo R., Shigematsu K., Butani J. Rapid enumeration of sulfate-reducing bacteria from aquatic environments using real-time PCR. Plankton Benthos Res. 2008;3:180–183. doi: 10.3800/pbr.3.180. [DOI] [Google Scholar]
- 21.Bradley A.S., Leavitt W.D., Johnston D.T. Revisiting the dissimilatory sulfate reduction pathway. Geobiology. 2011;9(5):446–457. doi: 10.1111/j.1472-4669.2011.00292.x. [DOI] [PubMed] [Google Scholar]
- 22.Taylor C.D., Doherty K.W. Submersible Incubation Device (SID), autonomous instrumentation for the in situ measurement of primary production and other microbial rate processes. Deep Sea Res A. 1990;37:343–358. doi: 10.1016/0198-0149(90)90132-F. [DOI] [Google Scholar]
- 23.Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 25.Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38(Web Server issue) Suppl. 2:W695-9. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selinger D.W., Saxena R.M., Cheung K.J., Church G.M., Rosenow C. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 2003;13(2):216–223. doi: 10.1101/gr.912603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mutter G.L., Zahrieh D., Liu C., Neuberg D., Finkelstein D., Baker H.E., Warrington J.A. Comparison of frozen and RNALater solid tissue storage methods for use in RNA expression microarrays. BMC Genomics. 2004;5:88. doi: 10.1186/1471-2164-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madrid V., Taylor G., Scranton M., Chistoserdov A. Characterization of bacterial communities in the anoxic zone of the Cariaco Basin. Appl. Environ. Microbiol. 2001;67:1663–1674. doi: 10.1128/AEM.67.4.1663-1674.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wakeham S.G., Turich C., Taylor G.T., Podlaska A., Scranton M.I., Li X.N., Varela R., Astor Y. Mid-chain mehtoxylated fatty acids within the chemocline of the Cariaco Basin: a chemoautotrophic source? Org. Geochem. 2010;41:498–512. doi: 10.1016/j.orggeochem.2010.01.005. [DOI] [Google Scholar]
- 32.Wakeham S.G., Turich C., Schubotz F., et al. Biomarkers, chemistry and microbiology show chemoautotrophy in a multilayer chemocline in the Cariaco Basin. Deep Sea Res. Part I Oceanogr. Res. Pap. 2012;63:133–156. doi: 10.1016/j.dsr.2012.01.005. [DOI] [Google Scholar]
- 33.Kuykendall L.D. Genus I. Bradyrhizobium. In: Brenner D.J., Krieg N.R., Staley J.T., editors. Bergey’s Manual of Systematic Bacteriology. 2nd ed. Vol. 2. New York: Springer-Verlag; 2005. pp. 438–443. [DOI] [Google Scholar]
- 34.Kalyuzhnaya M.G., Khmelenina V., Eshinimaev B., Sorokin D., Fuse H., Lidstrom M., Trotsenko Y. Classification of halo(alkali)philic and halo(alkali)tolerant methanotrophs provisionally assigned to the genera Methylomicrobium and Methylobacter and emended description of the genus Methylomicrobium. Int. J. Syst. Evol. Microbiol. 2008;58(Pt 3):591–596. doi: 10.1099/ijs.0.65317-0. [DOI] [PubMed] [Google Scholar]
- 35.Boden R., Cunliffe M., Scanlan J., Moussard H., Kits K.D., Klotz M.G., Jetten M.S., Vuilleumier S., Han J., Peters L., Mikhailova N., Teshima H., Tapia R., Kyrpides N., Ivanova N., Pagani I., Cheng J.F., Goodwin L., Han C., Hauser L., Land M.L., Lapidus A., Lucas S., Pitluck S., Woyke T., Stein L., Murrell J.C. Complete genome sequence of the aerobic marine methanotroph Methylomonas methanica MC09. J. Bacteriol. 2011;193(24):7001–7002. doi: 10.1128/JB.06267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vuilleumier S., Khmelenina V.N., Bringel F., Reshetnikov A.S., Lajus A., Mangenot S., Rouy Z., Op den Camp H.J., Jetten M.S., Dispirito A.A., Dunfield P., Klotz M.G., Semrau J.D., Stein L.Y., Barbe V., Médigue C., Trotsenko Y.A., Kalyuzhnaya M.G. Genome sequence of the haloalkaliphilic methanotrophic bacterium Methylomicrobium alcaliphilum 20Z. J. Bacteriol. 2012;194(2):551–552. doi: 10.1128/JB.06392-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shahak Y., Hauska G. In: Sulfide oxidation from cyanobacteria to humans: sulfide-quinone oxidoreductase (SQR). In Hell R, Dahl C, Knaff DB , Leustek T, editors. Netherland: Springer; 2008. [Google Scholar]
- 38.Vande Weghe J.G., Ow D.W. A fission yeast gene for mitochondrial sulfide oxidation. J. Biol. Chem. 1999;274(19):13250–13257. doi: 10.1074/jbc.274.19.13250. [DOI] [PubMed] [Google Scholar]
- 39.Inagaki F., Takai K., Nealson K.H., Horikoshi K. Sulfurovum lithotrophicum gen. nov., sp. nov., a novel sulfur-oxidizing chemolithoautotroph within the ε-Proteobacteria isolated from Okinawa Trough hydrothermal sediments. Int. J. Syst. Evol. Microbiol. 2004;54(Pt 5):1477–1482. doi: 10.1099/ijs.0.03042-0. [DOI] [PubMed] [Google Scholar]
- 40.Nakagawa S., Takaki Y., Shimamura S., Reysenbach A.L., Takai K., Horikoshi K. Deep-sea vent epsilon-proteobacterial genomes provide insights into emergence of pathogens. Proc. Natl. Acad. Sci. USA. 2007;104(29):12146–12150. doi: 10.1073/pnas.0700687104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inagaki F., Takai K., Kobayashi H., Nealson K.H., Horikoshi K. Sulfurimonas autotrophica gen. nov., sp. nov., a novel sulfur-oxidizing ε-proteobacterium isolated from hydrothermal sediments in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 2003;53(Pt 6):1801–1805. doi: 10.1099/ijs.0.02682-0. [DOI] [PubMed] [Google Scholar]
- 42.Sikorski J., Munk C., Lapidus A., Ngatchou Djao O.D., Lucas S., Glavina Del Rio T., Nolan M., Tice H., Han C., Cheng J.F., Tapia R., Goodwin L., Pitluck S., Liolios K., Ivanova N., Mavromatis K., Mikhailova N., Pati A., Sims D., Meincke L., Brettin T., Detter J.C., Chen A., Palaniappan K., Land M., Hauser L., Chang Y.J., Jeffries C.D., Rohde M., Lang E., Spring S., Göker M., Woyke T., Bristow J., Eisen J.A., Markowitz V., Hugenholtz P., Kyrpides N.C., Klenk H.P. Complete genome sequence of Sulfurimonas autotrophica type strain (OK10). Stand. Genomic Sci. 2010;3(2):194–202. doi: 10.4056/sigs.1173118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bryantseva I., Gorlenko V.M., Kompantseva E.I., Imhoff J.F., Süling J., Mityushina L. Thiorhodospira sibirica gen. nov., sp. nov., a new alkaliphilic purple sulfur bacterium from a Siberian soda lake. Int. J. Syst. Bacteriol. 1999;49(Pt 2):697–703. doi: 10.1099/00207713-49-2-697. [DOI] [PubMed] [Google Scholar]
- 44.Eisen J.A., Nelson K.E., Paulsen I.T., Heidelberg J.F., Wu M., Dodson R.J., Deboy R., Gwinn M.L., Nelson W.C., Haft D.H., Hickey E.K., Peterson J.D., Durkin A.S., Kolonay J.L., Yang F., Holt I., Umayam L.A., Mason T., Brenner M., Shea T.P., Parksey D., Nierman W.C., Feldblyum T.V., Hansen C.L., Craven M.B., Radune D., Vamathevan J., Khouri H., White O., Gruber T.M., Ketchum K.A., Venter J.C., Tettelin H., Bryant D.A., Fraser C.M. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. USA. 2002;99(14):9509–9514. doi: 10.1073/pnas.132181499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wahlund T.M., Woese C.R., Castenholz R.W., Madigan M.T. A thermophilic green sulfur bacterium from New Zealand hot springs, Chlorobium tepidum sp. nov. Arch. Microbiol. 1991;156:81–90. doi: 10.1007/BF00290978. [DOI] [Google Scholar]
- 46.Frigaard N.U., Bryant D.A. Genomic and evolutionary perspectives on sulfur metabolism in green-sulfur bacteria. In: Dahl C., Friedrich C.G., editors. Microbial Sulfur Metabolism. New York: Springer-Verlag; 2002. pp. 60–76. [Google Scholar]
- 47.Kuever J., Rainey F.A., Widdel F. Genus VI. Desulfococcus. In: Brenner D.J., Krieg N.R., Staley J.T., editors. Bergey’s Manual of Systematic Bacteriology. 2nd ed. Vol. 2. New York: Springer-Verlag; 2005. pp. 972–974. [DOI] [Google Scholar]
- 48.Oude Elferink S.J., Akkermans-van Vliet W.M., Bogte J.J., Stams A.J. Desulfobacca acetoxidans gen. nov., sp. nov., a novel acetate-degrading sulfate reducer isolated from sulfidogenic granular sludge. Int. J. Syst. Bacteriol. 1999;49(Pt 2):345–350. doi: 10.1099/00207713-49-2-345. [DOI] [PubMed] [Google Scholar]
- 49.Kuever J., Rainey F.A. Genus VII. Desulfotomaculum. In: De Vos P., Garrity G.M., Jones D., Krieg N.R., Ludwig W., Rainey F.A., et al., editors. Bergey’s Manual of Systematic Bacteriology. 2nd ed. vol. 3. New York: Springer-Verlag; 2009. pp. 989–996. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publishers Website along with the published article.