Abstract
Aim
Massive digitalization of natural history collections is now leading to a steep accumulation of publicly available species distribution data. However, taxonomic errors and geographical uncertainty of species occurrence records are now acknowledged by the scientific community – putting into question to what extent such data can be used to unveil correct patterns of biodiversity and distribution. We explore this question through quantitative and qualitative analyses of uncleaned versus manually verified datasets of species distribution records across different spatial scales.
Location
The American tropics.
Methods
As test case we used the plant tribe Cinchoneae (Rubiaceae). We compiled four datasets of species occurrences: one created manually and verified through classical taxonomic work, and the rest derived from GBIF under different cleaning and filling schemes. We used new bioinformatic tools to code species into grids, ecoregions, and biomes following WWF's classification. We analysed species richness and altitudinal ranges of the species.
Results
Altitudinal ranges for species and genera were correctly inferred even without manual data cleaning and filling. However, erroneous records affected spatial patterns of species richness. They led to an overestimation of species richness in certain areas outside the centres of diversity in the clade. The location of many of these areas comprised the geographical midpoint of countries and political subdivisions, assigned long after the specimens had been collected.
Main conclusion
Open databases and integrative bioinformatic tools allow a rapid approximation of large‐scale patterns of biodiversity across space and altitudinal ranges. We found that geographic inaccuracy affects diversity patterns more than taxonomic uncertainties, often leading to false positives, i.e. overestimating species richness in relatively species poor regions. Public databases for species distribution are valuable and should be more explored, but under scrutiny and validation by taxonomic experts. We suggest that database managers implement easy ways of community feedback on data quality.
Keywords: Cinchoneae, data quality, GBIF, occurrence data, Rubiaceae, species richness, SpeciesGeoCoder
Introduction
Museums and herbaria with natural history collections are invaluable sources of knowledge for science and society. These repositories contain a sample of the world's biodiversity collected over centuries of field exploration (Smith & Blagoderov, 2012). This information is becoming increasingly available over the Internet through interactive digital databases, thanks to the rapid emergence of new computational platforms and bioinformatic tools (Soberón & Peterson, 2004; Newbold, 2010). Numerous databases provided by different institutions are now publicly available, covering the majority of taxonomic groups and geographical regions. As a consequence of the growing number of data providers and amount of information, it has become necessary to assemble species occurrence data under standardized formats to enable the easy exchange and use of this information around the world (Graham et al., 2004). The Global Biodiversity Information Facility (GBIF, http://www.gbif.org/) is at the moment one of the largest and most widely used biodiversity databases (Beck et al., 2012, 2014; Jetz et al., 2012), with the objective to ‘make the world's primary data on biodiversity freely and universally available via the Internet’ (Chapman, 2005a; Yesson et al., 2007; GBIF, 2008; Newbold, 2010). Currently, GBIF provides a single portal to access more than 500 million records.
The massive availability of biodiversity data, together with the rapid emergence of new techniques and tools to analyse such information (e.g. Geographic Information Systems, and statistical analysis packages), has facilitated large‐scale analyses and interpretation of biodiversity and distribution data. Such data thus provide an invaluable resource to document biodiversity and its distribution through time and space for research, education and policy making (Williams et al., 1996; Winker, 2004). Biodiversity data, integrated with environmental spatial data, allow many uses extending from aspects of ecological and evolutionary theory to applications in conservation, biogeography, agriculture and human health, among others (Peterson et al., 1998; Chapman, 1999; Faith et al., 2001; Graham et al., 2004; Selama et al., 2013; Ficetola et al., 2014).
In response to the growing number of users and different requirements for the use of data, GBIF regularly improves its data portal adding more details for each data entry and allowing new types of extraction and analyses (GBIF, 2014). Nevertheless, there are still important limitations in data quality, which may influence the results and conclusions of biodiversity studies using GBIF if the data are used uncritically. For example, attention should be paid to potentially poor quality in terms of geographic position of locations, erroneous taxonomic identifications, and when dealing with groups with no recent taxonomic revision or for which no taxonomic expert is available (Graham et al., 2004; Wieczorek et al., 2004; Chapman, 2005b; Newbold, 2010; Hjarding et al., 2014). These limitations have called into question the usefulness of public databases, even if all available data could be gathered exhaustively (Hortal et al., 2007).
Ideally, careful quality evaluation of the primary information in a dataset downloaded from GBIF should be conducted, before the data is used for further analyses – in particular checking, where possible, the species identification and locality data, including their georeferences (coordinates for latitude and longitude). Otherwise, results could be flawed and research money unnecessarily spent (Dov, 2007). However, ecologists, biogeographers, conservationists, policy makers and stakeholders worldwide are using the information available in GBIF and other public databases to rapidly assess patterns of diversity, often without much attention being paid to the quality and reliability of the underlying data. Given the rapid development of computational tools for handling, cleaning and analysing biodiversity data, to what extent can we use public databases without the intervention of taxonomists?
Here we address this question by analysing and comparing diversity patterns inferred from datasets compiled manually and automatically, comprising species distribution data for the plant tribe Cinchoneae (Rubiceae). We use these data as a test case to investigate to what extent currently available data in GBIF are sufficient to correctly infer distribution and biodiversity patterns, across multiple spatial and altitudinal scales.
Methods
Study group and distribution
The tribe Cinchoneae comprises nine genera: Ciliosemina, Cinchona, Cinchonopsis, Joosia, Ladenbergia, Remijia, Stilpnophyllum, Maguireocharis, and Pimentelia totalling 121 accepted names of species so far (Andersson & Antonelli, 2005). The tribe occurs exclusively in the Neotropics, primarily in the foothills of the Andes from Bolivia to Colombia, but extending north to Venezuela, Costa Rica, the Guianas and southeastern Brazil. The tribe Cinchoneae forms a well‐defined clade characterized by morphology and DNA sequence data (Andersson, 1995; Andersson & Antonelli, 2005; Manns & Bremer, 2010). The tribe is historically and economically important as a source of Cinchona bark and quinine, the only treatment for malaria for c. 400 years (Kaufman & Ruveda, 2005).
Compilation of datasets
Four datasets were compiled including all taxa identified at species level in the tribe Cinchoneae: one created manually and verified through classical taxonomic work, and the other three derived from GBIF under different cleaning and filling schemes: a non‐cleaned dataset, a cleaned dataset and a cleaned dataset with the manual addition of records. For all datasets, infraspecific taxa were treated at the species level, and only georeferenced records were included. Records with strikingly incorrect georeferences (e.g. points on the sea, inverted latitude/longitude, inverted signs) were excluded from all analyses.
Verified dataset (VD)
This dataset was manually compiled through classical taxonomic work. GBIF was not considered during compilation. We attempted to include all the main herbaria in South America (Appendix S1 in Supporting Information) as well as the TROPICOS database at the Missouri Botanical Garden (http://mobot.mobot.org/W3T/Search/vast.html), which is the most comprehensive information resource for the flora of the Andean region (Taylor, 1999). Additionally, new records were digitalized, including new collections made by us during fieldwork in the last three years. Three new species under description were also included (C.M. Taylor, pers. comm. 2015). Cultivated and hybrid specimens were consistently excluded.
Synonyms were checked in The Plant List portal (http://www.theplantlist.org, accessed 15 May 2014) to ensure that all records were assigned with an accepted name (Appendix S2). Numerous specimens with non‐reliable identification were detected, e.g. duplicates of the same collection that were identified with different names in different herbaria. To validate species identity, a taxonomic check was carried out at the Missouri Botanical Garden's Herbarium (which holds one of the largest curated collections for the group), by comparing specimens with their respective types and protologues (see Appendix S3 for a list of the main references used). Because not all specimens were readily available for validation, we decided to remove specimens not identified by taxonomists working on Cinchoneae. For specimens without georeferences but with an accurate description of the collection locality, a coordinate was assigned using Google Earth (V 7.1.2.2041, 2014).
GBIF non‐cleaned dataset (‘GBIF’)
All available species records for tribe Cinchoneae were downloaded from GBIF (on 1 July 2014). Only records without ‘known georeference issues’ (an alternative in the data portal) were selected. The search was made for each genus separately.
GBIF cleaned from records with presumably wrong georeferences (‘GBIF cleaned’)
We removed from the GBIF dataset all those records without information in the fields Locality and/or State/Province, since we considered that their georeferences were more likely to be imprecise, often by assignments a posteriori.
GBIF cleaned and increased with additional records (‘GBIF cleaned_increased’)
This dataset was created by adding records to ‘GBIF cleaned’. These comprised records not available in GBIF, such as those from herbaria that are not linked to GBIF but were important for our study group. We also added the new records and species we found during fieldwork. In contrast to VD, we did not correct taxonomy and misidentifications in this dataset.
Analyses
We analyzed species distribution and species‐richness based on all four datasets to test the impact of dataset quality on the results. Since the spatial resolution of richness maps is affected by data volume and quality, these analyses were performed at three spatial scales: one‐degree grids, ecoregions, and biomes (Olson et al., 2001). We also computed the altitudinal distribution of all species in both datasets.
The analyses were conducted using SpeciesGeoCoder (Töpel et al., 2014), a software package written in Python and R (Zizka et al., in prep.). The program combines geographic polygons with occurrence points from multiple species, tests which species occurs in which polygon and computes summary statistics on species distribution and diversity. QuantumGIS (QGIS V 2.2.0‐Valmier) was then used to edit and export some of the summary maps to other formats. The Mollweide map projection was applied for all maps.
Analytical scales
Grids: The finest spatial level of our analyses is represented by one‐degree cells covering the entire range of the tribe. Ecoregions: The second spatial level was the terrestrial ecoregions used by WWF (Olson et al., 2001; Burgess et al., 2004); (http://maps.tnc.org/gis_data.html, accessed 1 March 2014). Biomes: They are polygons also defined under WWF's classification, the next hierarchical level above ecoregions. Altitudinal distribution: An altitude was assigned to every occurrence based on the Shuttle Radar Topography Mission global elevation model at 90 meters resolution (http://srtm.csi.cgiar.org). Altitudinal patterns of distribution were analysed per genus and per species.
For the altitudinal analysis, the VD and GBIF datasets were compared using a non‐parametric Mann–Whitney U‐test. For each analytical unit, species richness was calculated and mapped. We then created additional maps and diagrams to show the qualitative and quantitative differences found between both datasets. The statistical tests were performed using the R statistical software package (R_Core_Team, 2013).
Results
For the VD we obtained 4192 records in total, but after removing entries without georeferences the dataset was reduced to 2670 records. The complete query on GBIF returned 9592 records for the tribe Cinchoneae. Of these 8680 (90%) were identified at the species level and only 3720 (43%) were georeferenced. GBIF cleaned reduced this number to 3572, and GBIF cleaned_increased reached a total of 3756 records. Occurrence maps with both the VD and GBIF datasets are provided in Fig. 1.
Figure 1.
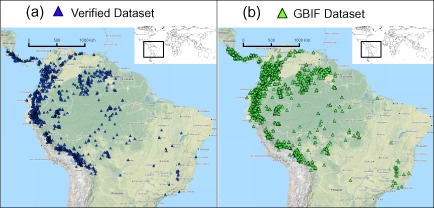
Plot of all species occurrences in the plant tribe Cinchoneae (Rubiaceae). (a) Verified dataset (i.e., manually compiled through classical taxonomic work including herbarium visits, fieldwork, and information from monographs); (b) unverified dataset downloaded from the Global Biodiversity Information Facility GBIF using minor automated cleaning functions (e.g. excluding points in the ocean).
A total of 114 species were included in this study (109 accepted names, two unresolved names, and three new species still undescribed. See Appendix S2).
Grid analysis
The maps obtained by the 1‐degree grid analyses are shown in Fig. 2. The map using VD (Fig. 2a) indicates that the most species‐rich areas for tribe Cinchoneae are located in the northern and central parts of Peru. The map using GBIF dataset (Fig. 2b) shows almost the same result, but additional areas appear to be particular species rich, namely the northern and central parts of Ecuador, central Venezuela and central Brazil. Differences between these two maps are highlighted in Fig. 2 (c).
Figure 2.
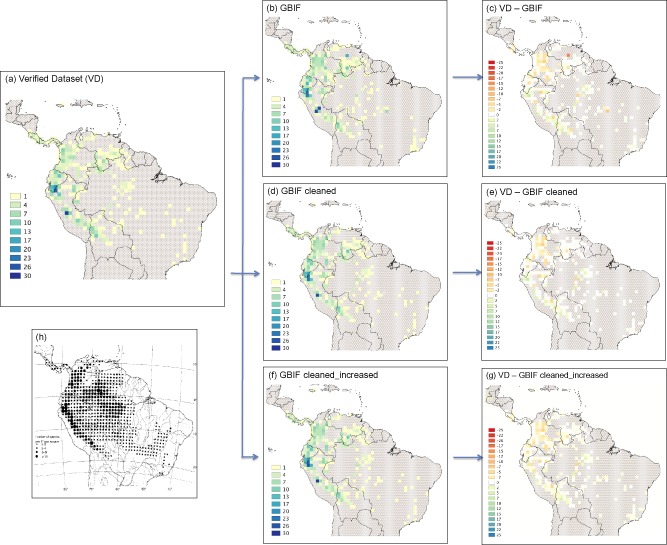
One‐degree grid maps showing species richness of tribe Cinchoneae. (a) Verified dataset (VD); (b) GBIF dataset; (c) Difference between VD and GBIF; (d) GBIF cleaned by the exclusion of uncertain georeferences; (e) Difference between VD and GBIF cleaned; (f) “GBIF cleaned” increased through the addition of records compiled manually; (g) Difference between VD and GBIF cleaned_increased; and (h) grid map showing a previous compilation of specimen records from the taxonomic literature, with dots proportional to species numbers (Antonelli et al., 2009).
As shown in Fig. 2 (b), using GBIF we identified some incorrect centres of diversity. Two cells stood out by a very high number of species. One is located exactly in the geographic midpoint of Peru. An assessment of the occurrence of species in this grid showed that 26 species were registered at exactly the same georeference (−74.14, −9.1839). Most of these entries were old specimens, collected between 1778–1900 by Ruiz & Pavón, Weddell, and Spruce among others, who did not provide coordinates for their collection sites. The available information in these labels only stated ‘Peru’ as the collection locality, and the georeference subsequently assigned to them when the data was entered into GBIF was simply the one corresponding to the central point of the country. The same issue was found in the second most species rich grid, which was located in the central part of Venezuela (−69.9119, 7.0758) and appeared to constitute a second centre of diversity with 16 species. To assess whether this was a recurrent problem, we therefore checked the central points of each Latin American country, and found the same problem for Ecuador (with nine species at −78.1869, −1.4628), Brazil (with 5 species at −52.8731, −10.8339), and Bolivia (with 5 species at −64.435, −16.7261).
GBIF cleaned (Fig. 2d) led to the inference of diversity patterns that were more similar to those obtained from VD (Fig. 2e), which means that most of the erroneous coordinates identified above were among the imprecise georeferences removed in GBIF cleaned. The addition of verified records not originally available through GBIF resulted in even more similar results (Figs. 2f & 2g), which means that some data are still missing on GBIF.
Ecoregion analysis
Species records included in either occurrence dataset were present in a total of 72 Terrestrial Ecoregions (Appendix S4a). Fig. 3 shows richness maps for each of those, using VD (Fig. 3a) and the three GBIF‐based datasets (Fig. 3b–d). The richness map using GBIF shows a higher number of areas with high diversity as compared to the one using VD.
Figure 3.
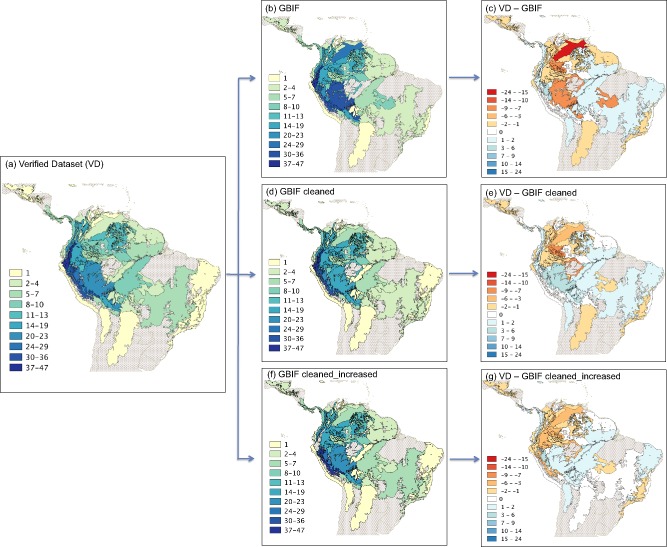
Species richness maps coded by ecoregions using the following datasets: (a) verified (VD), (b) GBIF, (c) VD‐GBIF, (d) GBIF cleaned, (e) VD‐GBIF cleaned, (f) GBIF cleaned_increased, and (g) VD‐GBIF cleaned_increased. The colour coding (see legend) refers to species numbers in tribe Cinchoneae. Only ecoregions containing at least one species are delimited.
We found high correspondence in the number of species between both datasets in the richest ecoregions: ‘Peruvian Yungas’ (with 36 species in both datasets) and the ‘Eastern Cordillera Real Mountain Forest’ (with 47 species registered in VD and 46 in GBIF). However, considerable differences in some ecoregions were identified (Fig. 3c). For instance, the ‘Llanos’, in the central part of Venezuela and in northeastern Colombia, included 27 species when GBIF was used, and only 3 when VD was used. ‘Southwest Amazon Moist Forest’, the ecoregion located between Peru and Brazil, had 33 species registered in GBIF but only 24 in VD. The ‘Mato Grosso Seasonal Forest’ ecoregion, located in central Brazil, had 10 species in GBIF but only 2 in VD. In those cases, a careful examination of the data points revealed a similar effect of poor georeferences as found for our grid level analyses: ecoregions comprising the geographical midpoints of some countries erroneously appear to constitute centres of diversity.
A related problem with inaccurate georeferences is exemplified by the ‘Southwest Amazon Moist Forest’ terrestrial ecoregion (Appendix S4a). Using GBIF, for this ecoregion we detected exactly the same coordinate (−12.00, −70.25) for six different species: Cinchona micrantha, Joosia umbellifera, Ladenbergia carua, L. graciliflora, L. oblongifolia and Remijia firmula. The records in all these cases state ‘Peru, Madre de Dios’. The coordinate was apparently assigned to the centre of the department Madre de Dios, which is very imprecise given the department's total area of over 85.000 km2 (equivalent to the size of Austria) and adds considerable noise to the final results. Additionally, in this case, most species are typical of montane forests (except for Remijia firmula, which is very common in the lowland). When we mapped the complete distribution of the six species, we found no overlap between the known ranges of these species with the point assigned to them in the GBIF dataset. Likewise, this repeatedly assigned but imprecise coordinate also increased the number of species in the ‘Southwest Amazon Moist Forest’ ecoregion. When we used GBIF cleaned (Fig. 3d) most of these erroneous records were eliminated and we obtained a better approximation to VD (Fig. 3a).
We also detected that the number of species in each ecoregion was not consistently higher when using GBIF; higher numbers of species were sometimes detected when using VD. Examples include the ‘Bolivian Yungas’, which hold 19 vs. 16 species, the ‘Iquitos Varzea’ with 12 vs. 9, and ‘Ucayali Moist Forest’ with 29 vs. 23 using VD and GBIF, respectively. These ecoregions are highly consistent with the places where we obtained extra records by digitizing new information from herbaria and through our own fieldwork, as well as by adding information from recent taxonomic work on the group. Consequently, these higher occurrences in the VD data are caused by improved data quality, not by noise or errors in either dataset. Using the GBIF cleaned_increased dataset, these records were included and the results (Fig. 3f & 3g) were even more consistent with the one obtained with VD.
Biome analysis
At the biome level, tribe Cinchoneae occurred in six of the world's fourteen biomes (Appendix S4b). Because the analytical units in this case are larger than the ecoregions, most species simply correspond to the largest biome included here, which is the ‘Tropical and Subtropical Moist Broadleaf Forests’ (containing 102 species in both datasets) (Fig. 4a & 4b). The largest difference between datasets was found in the ‘Tropical and Subtropical Grasslands and Savannas and Shrublands’ biome, which appears to comprise 32 species when VD was used and 10 when GBIF was used. This difference is depicted in Fig. 4c, and is again due to the inaccurate georeference given to several species in central Venezuela. In this case, the other inaccurate georeferences found (coordinates assigned for several species in the centre of other countries) are not visible here since they all are part of the larger biome considered. As in the previous cases, using GBIF cleaned (Fig. 4d & 4e) and GBIF cleaned_increased (Fig. 4f & 4g), the results show a better approximation to those obtained with VD (Fig. 4a).
Figure 4.
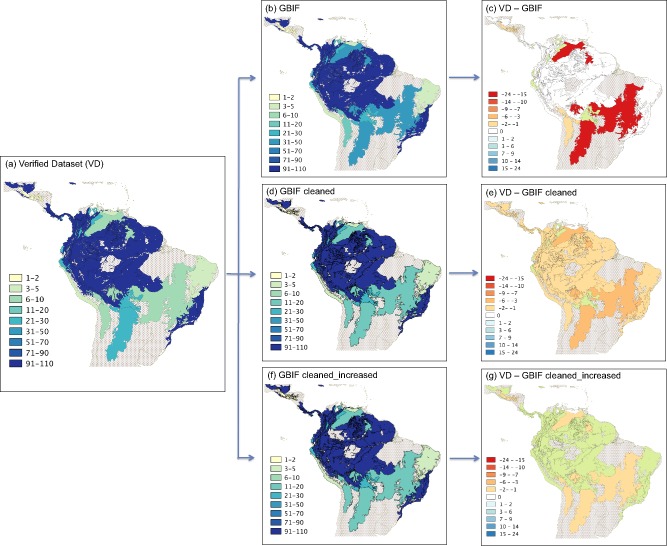
Maps of species richness coded at the biome level using the following datasets: (a) verified (VD), (b) GBIF, (c) VD‐GBIF, (d) GBIF cleaned, (e) VD‐GBIF cleaned, (f) GBIF cleaned_increased, and (g) VD‐GBIF cleaned_increased. The colour coding (see legend) refers to species numbers in tribe Cinchoneae. Only biomes containing at least one species are delimited.
Altitudinal analysis
Figures 5 and 6 show boxplots depicting the mean and interquartile ranges of altitude for genera and species, using VD and GBIF. At both levels, the altitudinal ranges are highly consistent between the two datasets.
Figure 5.
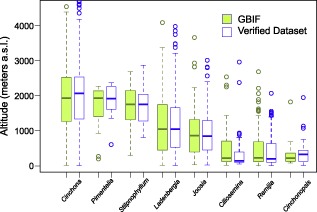
Altitudinal range for each analysed genus in tribe Cinchoneae, using both the Verified and the GBIF datasets. Boxes indicate the interquartile range (IQ) of all estimates, with the median shown as a horizontal line and the whiskers indicating data range outside the quartiles. There were no significant differences between the ranges of any genus (Mann–Whitney U‐test; P > 0.05).
Figure 6.
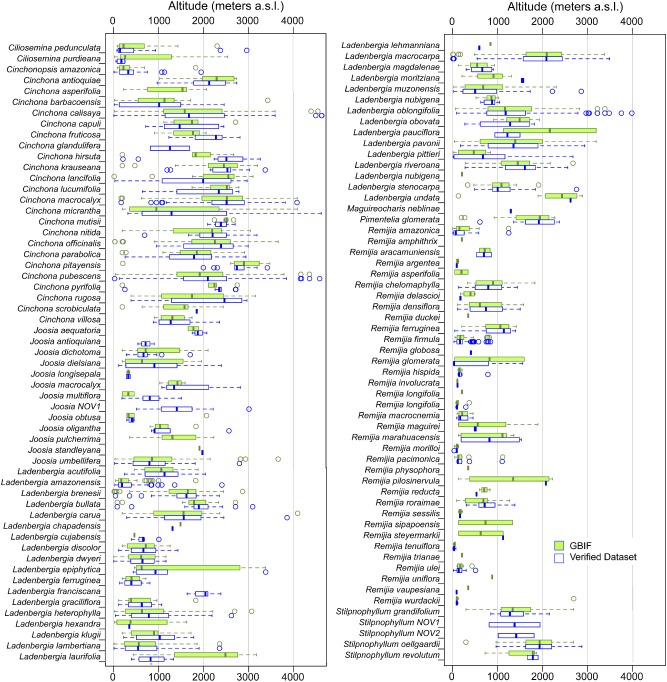
Altitudinal range for each analysed species in tribe Cinchoneae, using both the Verified and the GBIF datasets. Boxes indicate the interquartile range (IQ) of all estimates, with the median shown as a horizontal line and the whiskers indicating data range outside the quartiles. NOV means new species. There were no significant differences between the ranges of any species (Mann–Whitney U‐test; P > 0.05).
Discussion
The impact of spatial scales on the results
We found biases associated with erroneous georeferences at all spatial scales analysed (1‐degree grids, ecoregions, and biomes; Figs 2, 3 & 4). Contrary to our expectation, increasing the spatial scale did not reduce these problems. Below we discuss the common as well as particular issues of these separate analyses.
Grids are the most commonly used operational units for computing diversity metrics. Although they suffer from the fact that species records come from presence data only and therefore closely reflect sampling effort (Geri et al., 2013), they can refine species range maps when some parts of the region were not surveyed as rigorously as others (Franklin, 2010).
The gridded richness maps based on VD (Fig. 2a) are congruent with prior knowledge based on the manual compilation of species richness from specimen citations (Fig. 2h) (see Antonelli et al., 2009 and references therein) and are most likely an adequate representation of the distribution and diversity pattern of our tribe under study. Furthermore, these maps offer a detailed insight in the distribution of the clade and refine previous estimates. For instance, our study confirmed that some areas in Eastern and Southern Brazil previously thought to be part of the distribution range are void of this tribe. However, we acknowledge that part of these patterns might be a result of spatial biases in collecting activity or be specific to the area under study. Furthermore, the gridded maps have proven to be a valuable tool to identify major sources of geographic errors, in particular the false assignment of occurrences to the centroids of political units (Fig. 2b & 2c).
Comparing the VD and GBIF datasets using grids suggests that poor accuracy of georeferences can considerably affect the patterns obtained. We identified several centres of diversity for the tribe using GBIF (Fig. 2b) that are associated with incorrect coordinates assigned to the specimens, and we predict even less accuracy at finer scales.
The ecoregion analysis provided an arguably adequate estimate of the distribution and species richness of tribe Cinchoneae across the Neotropics. Ecoregions more closely reflect natural boundaries of plant and animal communities than arbitrarily defined political borders or grid cells (Olson et al., 2001; Wikramanayake, 2002; Burgess et al., 2004; Kier et al., 2005). This makes them useful analytical units in biodiversity, conservation and biogeography (Burgess et al., 2006; Giam et al., 2012; Jordon‐Thaden et al., 2013). We think that our ecoregion‐level analysis provided a fairly accurate estimate of the distribution of the tribe Cinchoneae, but only in the ecoregions along the Andean Mountains and around the Guiana Highlands. For the remaining ecoregions, which are generally larger, even a single occurrence led to a relatively large effect on the estimated biodiversity map. Indeed, by using GBIF, several large ecoregions appear to have false levels of species richness (e.g. the ‘Dry Chaco’, the ecoregion between Bolivia, Paraguay and Argentina with a single non‐verified record in Bolivia, but apparently affecting a large area; Fig. 3a). Minimum occurrence thresholds for coding species into polygons (an option already implemented in SpeciesGeoCoder) could be one automated way of reducing this bias when expert verification is not possible.
The biome level is larger than ecoregions and generally defined as major types of natural vegetation originating from a particular mix of climatic and edaphic conditions (Olson & Dinerstein, 1998; Riddle et al., 2011). We did not expect to find substantial differences at this level using VD or GBIF datasets; however, we did. As depicted in Fig. 4c, there are large areas in northern and central South America showing considerable differences. This is likely an effect of biomes not being continuous units, with several of them being composed of more than one polygon in different regions. For instance, the biome ‘Tropical and subtropical grasslands, savannas, and shrublands’, includes the ecoregion ‘Llanos’ (Appendix S4) for which a conspicuous bias was detected (Fig. 4c). The biome‐level comparison between datasets, as conducted here, thus also affects the same biome in central Brazil, despite the geographic distance. The same effect was found with the other two biomes in central South America: ‘Montane grasslands and shrublands’, and ‘Tropical and subtropical dry broadleaf forests’.
Due to the effect described above, using biomes shows less difference between the two datasets than using ecoregions. Moreover, the expected distribution of the tribe Cinchoneae is nearly fully consistent with the biome ‘Tropical and subtropical moist broadleaf forest’ and it has the same richness of species using both datasets (VD and GBIF). This suggests that at this level, the wrong coordinates or misidentifications found in GBIF do not affect the results considerably. In our study case, they do not reflect a real richness of the tribe Cinchoneae throughout the biome.
At the three levels, the additional datasets analysed (GBIF_cleaned and GBIF cleaned_increased) showed an improvement in the results (Figs 2, 3 & 4). Removing uncertain coordinates significantly reduced mistakes introduced using RD. Despite our expectation of a major improvement using GBIF_cleaned_increased, this was not very obvious, mainly because the new data added came from the Andes and did not cover the full distribution of the tribe.
Surprisingly, applying the Mann–Whitney U‐test, we did not find relevant differences between VD and GBIF concerning the altitudinal range of genera (Fig. 5) and species (Fig. 6). Accordingly, despite potentially erroneous references, the differences found using VD and GBIF at altitudinal range level were generally negligible.
Using unverified data from GBIF: a critical evaluation
Advantages
Despite the differences and errors detected in the GBIF dataset, there were also several advantages (Edwards, 2004; Chapman, 2005b; Guralnick & Hill, 2009). One clear advantage observed for this study is the fact that the number of institutions providing information to GBIF greatly exceeded what we could gather manually for the VD (Appendix S1) resulting in 76% more records in the GBIF dataset.
Saving time and money is also a clear advantage using GBIF. We were able to download a complete set of entries for tribe Cinchoneae in less than half an hour. By comparison, obtaining the clean list of our entries for the VD took us almost six months and necessitated visits to major collections in herbaria on several continents.
Another major benefit of GBIF was the uniformity of data. GBIF has compiled a very substantial database with data in the same format, which is ready to use for many analyses. Building the VD implied merging many small datasets into a single large file, which required uniformity of data based on a single data standard. To individual researchers, this is still largely a manual and time‐consuming process.
Disadvantages
The quality of unverified GBIF dataset depends mainly on the data providers, and therefore it varies substantially. Increasingly the GBIF network are adopting strong peer review processes for data publishers (T. Robertson, Head of Informatics‐GBIF, pers. comm.), after realizing the need for data validation before they become publicly available. This practice is however not yet common and consequently both inaccurate and incorrect information may be published in databases for public use.
Two critical points were detected in our study: mistakes in the taxonomic identification of specimens, and inaccurate georeferencing. Mistakes in taxonomic identification can often be corrected by a taxonomist checking the identifications, provided that it is possible to access the specimen record (or at least images of it). A correct species name should ideally be a minimum requirement for including the data in GBIF or any other public database. However, there are many cases of taxonomic disagreement regarding species delimitations, synonymisation and nomenclatural problems, making such a requirement not straightforward. Moreover, considering that there are relatively few people in the world able to correctly identify species of many understudied organism groups, and that there are millions of species yet to be described it would not be feasible to require a peer‐review of the accuracy of identifications before making occurrence data publicly available (Dov, 2007; Mora et al., 2011).
It has been suggested that a relatively large proportion of available georeferences are erroneous (Hijmans et al., 1999). Georeferencing errors can sometimes only be detected by very careful examination of all records by an expert (Graham et al., 2004). Wrong or inaccurate georeferencing is mainly caused by the lack of data associated with older collections. In these cases it becomes the responsibility of the digitizer to assign a best possible coordinate based on the information available on the labels. An accurate georeference assigned to the specimen is often crucial, since a small deviation here could imply wrong interpretation of the results. This would not only affect studies aiming at identifying general biodiversity patterns, but also others such as reconstructing the environmental niche of species and predicting their total distribution.
In this study we repeatedly found a lumping of specimens with imprecise collection information to the central point of the geographical reference (e.g. the country or province). When data quality cannot be improved, and the assignment of a country‐level coordinate is deemed necessary for some reason, it is crucial that data repositories at least clearly state how the coordinates were obtained, and what the level of accuracy is. We emphasize that it is crucial for the future use of the data that the institutions providing information to GBIF include information on whether the coordinate assignment was done manually or automatically, and if possible indicate their level of precision. For many studies, researchers may then consider removing records with uncertain geographical reference prior to data analysis.
Another weakness detected using GBIF data is that there are few requirements for data publishers beyond providing stable record identifiers wherever possible and some form of taxonomic identification. Implementing formal peer‐reviewing or control processes is beyond the scope of GBIF; instead, it is the ambition of GBIF to educate the scientific community and institutions to take responsibility for ensuring the best quality of the data for a large variety of uses (T. Robertson, pers. comm.). Consequently, the decision on which records to publish and how much detail to supply rests solely on the data owner, relying on good quality practice by both providers and users of data. However, in this study we show that such best practice needs improvement. For example, we found entries where the data providers did not even provide the collection number for the specimen (or this information was lost when entering GBIF), precluding subsequent validation of the specimen's identification and georeference.
Some additional minimum requirements for data providers, in particular the collection number, could significantly improve the quality of specimen data in GBIF, saving considerable time for users who wish to verify the data (and avoid the need for dismissing uncertain records). Review work of taxonomists always improves data, analyses and interpretations (Hjarding et al., 2014), but with the extended and multidisciplinary use of data, quality assurance must be a joint effort by all parties at all stages. Promoting best practice at all stages from recording data on the specimens at the time of collecting, providing comprehensive and accurate data to public databases, and evaluating downloaded datasets for further use, will contribute to improve data quality for many important applications in biodiversity research and policy making. Finally, an important issue must be highlighted: there is a clear need for a feedback system between data users and custodians. Data users should be able to assess data quality and inform about publications including results of quality checks directly, so that rectifications can be fed back into the original provider's database. This kind of feedback is still missing in most available systems at the moment.
Conclusion
In a time of rapidly increasing biological databases, using species distribution data and bioinformatic tools holds a large potential to inferring patterns of species diversity and distribution. However, they have to be used critically due to important concerns with data quality.
For our study case, the usefulness of the GBIF portal did not depend on the spatial scale at which the information was coded, but on the precision of the data. In contrast, our results suggest that analyses on altitudinal ranges at genus and species level are less sensitive to georeferencing mistakes. This could however become a problem for groups with fewer records or narrower distribution.
Our study demonstrates that the correct estimation of species distribution and species richness still requires occurrence data of good quality. In practice, this means applying substantial amounts of taxonomic knowledge, time and funding on verifying and cleaning up subsets of public databases. When this is not a viable option, automatically removing uncertain data (e.g. records without locality names) may be sufficient to reveal general diversity patterns and identify main centres of diversity for the focal group.
It is unfortunate that GBIF still do not allow their users to easily provide feedback on specific records, e.g. correcting misidentifications and erroneous georeferencing as outlined in this study. We therefore urge the managers of public biological databases to implement this sort of ‘crowd science’ evaluation – comparable to the open rating of hotels and restaurants on the Internet. It will then be up to individual researchers to decide to which extent they wish to rely on this community‐based assessment of biological records, e.g. whether they want to perform their analyses only based on unchallenged data. Some initiatives have started to emerge (i.e. http://www.idigbio.org, http://www.ispotnature.org and http://www.inaturalist.org), but a rapid change is required to advance our knowledge on biodiversity and distribution patterns in the era of Big Data.
Additional references to the data sources used in this study are found in Appendix S3.
The Antonelli group (http://antonelli‐lab.net) focuses on understanding how biological diversity has evolved and how it will be affected by on‐going climate and habitat changes, with a focus on the American tropics. The Rønsted group (http://snm.ku.dk/phylogenetic_prediction/) explores the evolution of plants and the correlation between phylogeny, biological interactions and natural products, to explain patterns and processes of diversity.
Author contributions: A.A., C.M. and N.R. conceived the ideas and supervised the research. C.M. assembled and analysed the data with contributions from C.I.M., A.Z. and A.A. C.M. wrote the manuscript with contributions from A.A. and N.R. All authors read and improved the manuscript.
Supporting information
Appendix S1 Acronyms of institutions providing information for the tribe Cinchonae.
Appendix S2 Genera and species in tribe Cinchoneae.
Appendix S3 References used for identification of specimens from the Cinchoneae tribe.
Appendix S4 Ecoregions and Biomes where tribe Cinchoneae was registered.
Acknowledgements
The research presented in this publication was supported by a grant from the Carlsberg foundation and a Freja stipend from the Faculty of Science, University of Copenhagen to Nina Rønsted. Alexandre Antonelli is supported by funding from the European Research Council under the European Union's Seventh Framework Programme (FP/2007–2013), the Swedish Research Council and the Wallenberg Foundation. Tim Robertson at GBIF is thanked for helpful discussions. Fernanda Antunes Carvalho, Henrik Nilsson and Rønsted's group made useful suggestions to improve the manuscript.
Editor: John‐Arvid Grytnes
References
- Andersson, L. (1995) Tribes and genera of the Cinchoneae complex (Rubiaceae). Annals of the Missouri Botanical Garden, 82, 409–427. [Google Scholar]
- Andersson, L. & Antonelli, A. (2005) Phylogeny of the tribe Cinchoneae (Rubiaceae), its position in Cinchonoideae, and description of a new genus, Ciliosemina. Taxon, 54, 17–28. [Google Scholar]
- Antonelli, A. , Nylander, J.A. , Persson, C. & Sanmartín, I. (2009) Tracing the impact of the Andean uplift on Neotropical plant evolution. Proceedings of the National Academy of Sciences USA, 106, 9749–9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, J. , Ballesteros‐Mejia, L. , Buchmann, C.M. , Dengler, J. , Fritz, S.A. , Gruber, B. , Hof, C. , Jansen, F. , Knapp, S. & Kreft, H. (2012) What's on the horizon for macroecology? Ecography, 35, 673–683. [Google Scholar]
- Beck, J. , Böller, M. , Erhardt, A. & Schwanghart, W. (2014) Spatial bias in the GBIF database and its effect on modeling species' geographic distributions. Ecological Informatics, 19, 10–15. [Google Scholar]
- Burgess, N. , Hales, J.D. , Underwood, E. , Dinerstein, E. , Olson, D. , Itoua, I. , Schipper, J. , Ricketts, T. , Newman, K. & Hales, J.A. (2004) Terrestrial ecoregions of Africa and Madagascar: a conservation assessment. Island Press, Washington, DC. [Google Scholar]
- Burgess, N.D. , Hales, J.D.A. , Ricketts, T.H. & Dinerstein, E. (2006) Factoring species, non‐species values and threats into biodiversity prioritisation across the ecoregions of Africa and its islands. Biological Conservation, 127, 383–401. [Google Scholar]
- Chapman, A.D. (1999) Quality control and validation of point‐sourced environmental resource data, version 1.0 Spatial accuracy assessment: land information uncertainty in natural resources (ed. by Lower K. and Jaton A.), pp. 409–418, Chelsea, Michigan. [Google Scholar]
- Chapman, A.D. (2005a) Principles of data quality, version 1.0. Global Biodiversity Information Facility, Copenhagen. [Google Scholar]
- Chapman, A.D. (2005b) Principles and Methods of Data Cleaning – Primary Species and Species‐Occurrence Data, version 1.0. Global Biodiversity Information Facility, Copenhagen. [Google Scholar]
- Dov, F. (2007) A ‘taxonomic affidavit’: why it is needed? Integrative Zoology, 2, 57–59. [DOI] [PubMed] [Google Scholar]
- Edwards, J.L. (2004) Research and societal benefits of the Global Biodiversity Information Facility. BioScience, 54, 485–486. [Google Scholar]
- Faith, D.P. , Walker, P.A. & Margules, C.R. (2001) Some future prospects for systematic biodiversity planning in Papua New Guinea‐and for biodiversity planning in general. Pacific Conservation Biology, 6, 325–343. [Google Scholar]
- Ficetola, G.F. , Rondinini, C. , Bonardi, A. , Katariya, V. , Padoa‐Schioppa, E. & Angulo, A. (2014) An evaluation of the robustness of global amphibian range maps. Journal of Biogeography, 41, 211–221. [Google Scholar]
- Franklin, J. (2010) Mapping species distributions: spatial inference and prediction. Cambridge University Press, Cambridge. [Google Scholar]
- GBIF (2008) Training manual 1: digitisation of natural history collections data. Global Biodiversity Information Facility, Copenhagen. [Google Scholar]
- GBIF (2014) New features introduced in GBIF Portal. In: GBits – the GBIF newsletter , p. 1 Global Biodiversity Information Facility, Copenhagen. [Google Scholar]
- Geri, F. , Lastrucci, L. , Viciani, D. , Foggi, B. , Ferretti, G. , Maccherini, S. , Bonini, I. , Amici, V. & Chiarucci, A. (2013) Mapping patterns of ferns species richness through the use of herbarium data. Biodiversity and Conservation, 22, 1679–1690. [Google Scholar]
- Giam, X. , Scheffers, B.R. , Sodhi, N.S. , Wilcove, D.S. , Ceballos, G. & Ehrlich, P.R. (2012) Reservoirs of richness: least disturbed tropical forests are centres of undescribed species diversity. Proceedings of the Royal Society B: Biological Sciences, 279, 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, C.H. , Ferrier, S. , Huettman, F. , Moritz, C. & Peterson, A.T. (2004) New developments in museum‐based informatics and applications in biodiversity analysis. Trends in Ecology and Evolution, 19, 497–503. [DOI] [PubMed] [Google Scholar]
- Guralnick, R. & Hill, A. (2009) Biodiversity informatics: automated approaches for documenting global biodiversity patterns and processes. Bioinformatics, 25, 421–428. [DOI] [PubMed] [Google Scholar]
- Hijmans, R. , Schreuder, M. , De la Cruz, J. & Guarino, L. (1999) Using GIS to check co‐ordinates of genebank accessions. Genetic Resources and Crop Evolution, 46, 291–296. [Google Scholar]
- Hjarding, A. , Tolley, K.A. & Burgess, N.D. (2014) Red List assessments of East African chameleons: a case study of why we need experts. Oryx, FirstView, 1–7. [Google Scholar]
- Hortal, J. , Lobo, J.M. & Jimenez‐Valverde, A. (2007) Limitations of biodiversity databases: case study on seed‐plant diversity in Tenerife, Canary Islands. Conservation Biology, 21, 853–863. [DOI] [PubMed] [Google Scholar]
- Jetz, W. , McPherson, J.M. & Guralnick, R.P. (2012) Integrating biodiversity distribution knowledge: toward a global map of life. Trends in Ecology and Evolution, 27, 151–159. [DOI] [PubMed] [Google Scholar]
- Jordon‐Thaden, I.E. , Al‐Shehbaz, I.A. & Koch, M.A. (2013) Species richness of the globally distributed, arctic–alpine genus Draba L.(Brassicaceae). Alpine Botany, 123, 97–106. [Google Scholar]
- Kaufman, T.S. & Ruveda, E.A. (2005) The quest for quinine: those who won the battles and those who won the war. Angewandte Chemie International Edition, 44, 854–885. [DOI] [PubMed] [Google Scholar]
- Kier, G. , Mutke, J. , Dinerstein, E. , Ricketts, T.H. , Küper, W. , Kreft, H. & Barthlott, W. (2005) Global patterns of plant diversity and floristic knowledge. Journal of Biogeography, 32, 1107–1116. [Google Scholar]
- Manns, U. & Bremer, B. (2010) Towards a better understanding of intertribal relationships and stable tribal delimitations within Cinchonoideae s.s. (Rubiaceae). Molecular Phylogenetics and Evolution, 56, 21–39. [DOI] [PubMed] [Google Scholar]
- Mora, C. , Tittensor, D.P. , Adl, S. , Simpson, A.G.B. & Worm, B. (2011) How many species are there on Earth and in the ocean? PLoS Biology, 9, e1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbold, T. (2010) Applications and limitations of museum data for conservation and ecology, with particular attention to species distribution models. Progress in Physical . Geography, 34, 3–22. [Google Scholar]
- Olson, D.M. & Dinerstein, E. (1998) The Global 200: a representation approach to conserving the Earth's most biologically valuable ecoregions. Conservation Biology, 12, 502–515. [Google Scholar]
- Olson, D.M. , Dinerstein, E. , Wikramanayake, E.D. , Burgess, N.D. , Powell, G.V.N. , Underwood, E.C. , D'Amico, J.A. , Itoua, I. , Strand, H.E. & Morrison, J.C. (2001) Terrestrial ecoregions of the world: a new map of life on earth a new global map of terrestrial ecoregions provides an innovative tool for conserving biodiversity. BioScience, 51, 933–938. [Google Scholar]
- Peterson, A.T. , Navarro‐Sigüenza, A.G. & Benítez‐Díaz, H. (1998) The need for continued scientific collecting; a geographic analysis of Mexican bird specimens. Ibis, 140, 288–294. [Google Scholar]
- R_Core_Team (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: ISBN 3‐900051‐07‐0. Available at: http://www.R‐project.org [Google Scholar]
- Riddle, B.R. , Ladle, R.J. , Lourie, S.A. & Whittaker, R.J. (2011) Basic biogeography: estimating biodiversity and mapping nature Conservation biogeography (ed. by Ladle R.J. and Whittaker R.J.), pp. 45–92. John Wiley & Sons, Ltd, Chichester. [Google Scholar]
- Selama, O. , James, P. , Nateche, F. , Wellington, E.M. & Hacène, H. (2013) The world bacterial biogeography and biodiversity through databases: a case study of NCBI Nucleotide Database and GBIF Database. BioMed Research International, 2013, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, V.S. & Blagoderov, V. (2012) Bringing collections out of the dark. ZooKeys, 209, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberón, J. & Peterson, T. (2004) Biodiversity informatics: managing and applying primary biodiversity data. Philosophical Transactions of the Royal Society B: Biological Sciences, 359, 689–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, C.M. (1999) Rubiaceae Catálogo de las plantas vasculares del Ecuador. Monographs in Systematic Botany from the Missouri Botanical Garden (ed. by Jørgensen P.M. and León‐Yánez S.), pp. 855–878. Missouri Botanical Garden, Missouri. [Google Scholar]
- Töpel, M. , Calió, M.F. , Zizka, A. , Scharn, R. , Silvestro, D. & Antonelli, A. (2014) SpeciesGeoCoder: fast categorisation of species occurrences for analyses of biodiversity, biogeography, ecology and evolution. BioRxiv, 009274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek, J. , Guo, Q. & Hijmans, R. (2004) The point‐radius method for georeferencing locality descriptions and calculating associated uncertainty. International Journal of Geographical Information Science, 18, 745–767. [Google Scholar]
- Wikramanayake, E. , Dinerstein, E. & Loucks, C. (2002) Terrestrial ecoregions of the Indo‐Pacific: a conservation assessment. Island Press, Washington DC. [Google Scholar]
- Williams, P. , Gibbons, D. , Margules, C. , Rebelo, A. , Humphries, C. & Pressey, R. (1996) A comparison of richness hotspots, rarity hotspots, and complementary areas for conserving diversity of British birds. Conservation Biology, 10, 155–174. [Google Scholar]
- Winker, K. (2004) Natural history museums in a postbiodiversity era. BioScience, 54, 455–459. [Google Scholar]
- Yesson, C. , Brewer, P.W. , Sutton, T. , Caithness, N. , Pahwa, J.S. , Burgess, M. , Gray, W.A. , White, R.J. , Jones, A.C. & Bisby, F.A. (2007) How global is the global biodiversity information facility? PLoS ONE, 2, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Acronyms of institutions providing information for the tribe Cinchonae.
Appendix S2 Genera and species in tribe Cinchoneae.
Appendix S3 References used for identification of specimens from the Cinchoneae tribe.
Appendix S4 Ecoregions and Biomes where tribe Cinchoneae was registered.


