The Vaccinia virus D4/A20 complex is the DNA polymerase cofactor. The heterodimer interface has been analysed using three new crystal structures of the complex in which key residues forming a cation–π interaction have been mutated.
Keywords: Vaccinia virus, DNA replication, X-ray structure, cation–π interaction, protein–protein interface
Abstract
The Vaccinia virus polymerase holoenzyme is composed of three subunits: E9, the catalytic DNA polymerase subunit; D4, a uracil-DNA glycosylase; and A20, a protein with no known enzymatic activity. The D4/A20 heterodimer is the DNA polymerase cofactor, the function of which is essential for processive DNA synthesis. The recent crystal structure of D4 bound to the first 50 amino acids of A20 (D4/A201–50) revealed the importance of three residues, forming a cation–π interaction at the dimerization interface, for complex formation. These are Arg167 and Pro173 of D4 and Trp43 of A20. Here, the crystal structures of the three mutants D4-R167A/A201–50, D4-P173G/A201–50 and D4/A201–50-W43A are presented. The D4/A20 interface of the three structures has been analysed for atomic solvation parameters and cation–π interactions. This study confirms previous biochemical data and also points out the importance for stability of the restrained conformational space of Pro173. Moreover, these new structures will be useful for the design and rational improvement of known molecules targeting the D4/A20 interface.
1. Introduction
Vaccinia virus (VACV) is the prototype of the Orthopoxvirus genus of the family Poxviridae. The Orthopoxvirus genus contains a number of pathogens infecting humans, such as Monkeypox virus, Cowpox virus and the best-known member, Variola virus. Viral genome replication takes place in the cytoplasm of the infected host cell and is thought to depend almost exclusively on virally encoded proteins. Four of these proteins presumably located at the replication fork are essential for DNA synthesis (Moss, 2013 ▸). These are E9, the DNA polymerase catalytic subunit; D5, an NTPase/primase with putative helicase activity (Boyle et al., 2007 ▸; De Silva et al., 2007 ▸; Evans et al., 1995 ▸); D4, a uracil-DNA glycosylase (Upton et al., 1993 ▸); and A20, a central component interacting with E9, D5 and D4 (McCraith et al., 2000 ▸; Ishii & Moss, 2002 ▸). The function of A20 remains unclear, but it may be involved in the recruitment of the other factors at the replication fork. Under physiological conditions, E9 alone is an inherently distributive enzyme (McDonald & Traktman, 1994 ▸). However, it becomes highly processive when bound to its heterodimeric cofactor D4/A20, forming the DNA polymerase holoenzyme (Stanitsa et al., 2006 ▸).
We have recently reported the high-resolution structure of VACV D4 bound to the first 50 residues of A20 (D4/A201–50; Contesto-Richefeu et al., 2014 ▸). Although a large contact surface is formed between the proteins (∼1840 Å2 of buried surface), the interface is strikingly flat. The D4/A20 contacts involve residues 167–180 and 191–206 of D4 and a leucine-rich α-helix found at the very N-terminal end of A20, together with a second contact involving Trp43 of A20 sandwiched between Pro173 and Arg167 of D4, simultaneously forming a prominent cation–π interaction. Site-directed mutagenesis highlighted the key roles of these three residues in the D4/A20 interaction. This peculiar structure may represent a hotspot at the D4/A20 interface that can potentially be targeted by small molecules, disrupting the interaction and inhibiting viral replication. In order to further investigate the role of each of these residues in the interaction, we solved the crystal structures of the three individual mutants D4-R167A/A201–50, D4-P173G/A201–50 and D4/A201–50-W43A. These new structures together with the wild-type (WT) structure will be helpful for the optimization of existing molecules (Schormann et al., 2011 ▸) or the design of new molecules inhibiting the D4/A20 interaction.
2. Materials and methods
2.1. Macromolecule production
The experimental procedures for generation of the vectors pETDuet-D4R-R167A-A20R1–50WT, pETDuet-D4R-P173G-A20R1–50WT and pETDuet-D4R-A20R1–50W43A have been described previously (Contesto-Richefeu et al., 2014 ▸). These constructs allow the production of the three different mutants (D4, Arg167Ala and Pro173Gly; A20, Trp43Ala). Protein complexes were produced as described in Contesto-Richefeu et al. (2014 ▸). Dimers isolated from the second size-exclusion chromatography step (Superdex 75 10/300 GL, GE Healthcare) equilibrated in 50 mM Tris–HCl pH 7.5, 100 mM NaCl were concentrated to 9 mg ml−1 prior to crystallization.
2.2. Crystallization
The D4-R167A/A201–50 complex crystallized in 0.1 M Bicine pH 7.8, 1.6 M ammonium sulfate. D4-P173G/A201–50 crystallized under the same conditions at pH 8.9 and D4/A201–50-W43A in 0.1 M Bicine pH 8.9, 1.5 M ammonium sulfate. All crystallization experiments were performed using the hanging-drop vapour-diffusion method at 293 K with a 1.5 µl:1.5 µl protein:reservoir ratio.
2.3. Data collection and processing
Prior to data collection, crystals were successively transferred into 10 and 20%(v/v) glycerol in the reservoir solution as a cryosolution before flash-cooling in liquid nitrogen. Data sets were collected at a wavelength of 0.978 Å at 107 K on beamline ID23-1 at the European Synchrotron Radiation Facility (ESRF), Grenoble, France using a Pilatus 6M detector. Data were processed with XDS (Kabsch, 2010 ▸) and scaled with AIMLESS (Evans & Murshudov, 2013 ▸) from the CCP4 suite (Winn et al., 2011 ▸). Data-collection statistics are given in Table 1 ▸.
Table 1. Data collection and processing.
| D4/A201–50-W43A | D4-R167A/A201–50 | D4-P173G/A201–50 | |
|---|---|---|---|
| Resolution range (Å) | 48.51–2.59 (2.71–2.59) | 28.84–2.80 (2.95–2.80) | 48.61–2.49 (2.59–2.49) |
| No. of observed reflections | 297885 (34796) | 181073 (25880) | 248402 (26386) |
| No. of unique reflections | 23077 (2648) | 18696 (2618) | 26149 (2811) |
| Completeness (%) | 99.6 (95.5) | 99.6 (97.5) | 99.5 (96.0) |
| Multiplicity | 12.9 (13.6) | 9.7 (9.9) | 9.5 (9.4) |
| Mean I/σ(I) | 16.1 (3.2) | 15.4 (3.5) | 19.7 (3.8) |
| R r.i.m. † | 0.099 (0.779) | 0.113 (0.695) | 0.073 (0.573) |
| Mosaicity (°) | 0.21 | 0.22 | 0.26 |
| Overall B factor from Wilson plot (Å2) | 56.8 | 52.1 | 47.4 |
Estimated R r.i.m. = R merge × [N/(N − 1)]1/2, where N is the data multiplicity.
2.4. Structure solution, refinement and analysis
Structures were solved by molecular replacement using the previously determined His-D4/A201–50 structure from PDB entry 4oda (Contesto-Richefeu et al., 2014 ▸) and MOLREP (Vagin & Teplyakov, 2010 ▸) from the CCP4 suite (Winn et al., 2011 ▸). Initial structures were improved by model building with Coot (Emsley et al., 2010 ▸) and were refined with REFMAC5 (Murshudov et al., 2011 ▸). The final model quality was analyzed using the PDB validation server. Interacting surfaces were analyzed with PISA (Krissinel & Henrick, 2007 ▸) and cation–π interactions were analyzed with CAPTURE (Gallivan & Dougherty, 1999 ▸). Crystallographic coordinates and structure factors have been deposited in the PDB with accession codes 5jkr, 5jks and 5jkt for D4/A201–50-W43A, D4-R167A/A201–50 and D4-P173G/A201–50, respectively. Structure-refinement statistics are given in Table 2 ▸.
Table 2. Structure refinement and model composition.
| D4/A201–50-W43A | D4-R167A/A201–50 | D4-P173G/A201–50 | |
|---|---|---|---|
| Resolution range (Å) | 48.51–2.59 | 28.84–2.80 | 48.61–2.49 |
| Completeness (%) | 99.6 | 99.6 | 99.5 |
| σ Cutoff | None | None | None |
| No. of reflections, working set | 23039 | 18665 | 26112 |
| No. of reflections, test set | 1162 | 940 | 1306 |
| Final R cryst | 0.195 | 0.176 | 0.183 |
| Final R free | 0.250 | 0.233 | 0.238 |
| No. of non-H atoms | |||
| Protein | 4329 | 4323 | 4351 |
| Ligand | 30 | 30 | 33 |
| Water | 63 | 25 | 105 |
| Total | 4428 | 4378 | 4489 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.015 | 0.015 | 0.017 |
| Bond angles (°) | 1.85 | 1.83 | 1.99 |
| Average B factor (Å2) | |||
| Protein | 64.9 | 54.9 | 58.3 |
| Ramachandran plot (%) | |||
| Favoured regions | 93 | 93 | 96 |
| Additionally allowed regions | 6 | 6 | 4 |
| Outliers | 1 | 1 | 0 |
3. Results and discussion
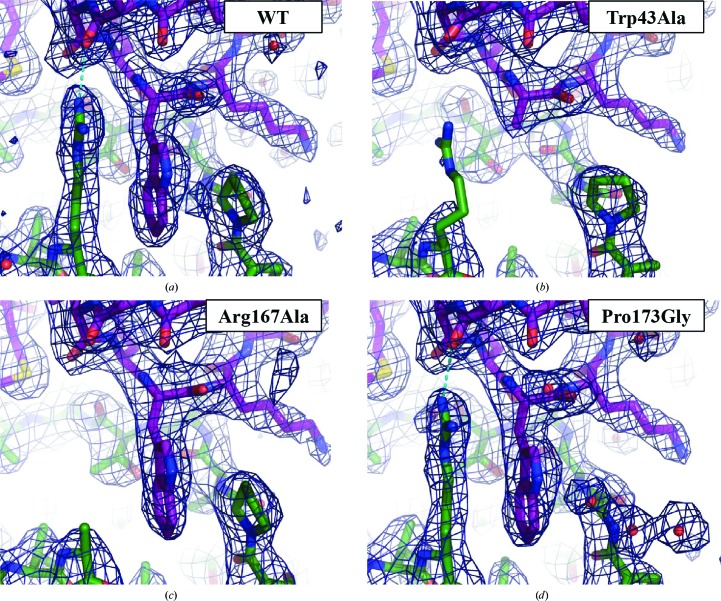
The D4-R167A/A201–50, D4-P173G/A201–50 and D4/A201–50-W43A mutant complexes crystallized under conditions similar to those for WT D4/A201–50 with two molecules in the asymmetric unit. The crystals were isomorphous to those of the WT complex (space group P3121, unit-cell parameters a = b = 93.0, c = 145.7 Å). Diffraction data were collected to 2.8, 2.5 and 2.6 Å resolution, respectively (Table 1 ▸). The overall mutant structures are identical to that of WT D4/A201–50 (PDB entry 4oda); the effect of each point mutation is strictly local and only affects the mutated residue (Figs. 1 ▸ and 2 ▸ and Table 3 ▸). However, the A20 Trp43Ala mutant affecting the central residue of the interaction also induced disorder of the previously well positioned Arg167 residue of D4 (Fig. 2 ▸ b). The Arg167Ala mutant of D4 results in loss of the cation–π interaction with Trp43 and the hydrogen bond to Thr41 of A20 (Fig. 2 ▸ c and Table 3 ▸). Finally, the Pro173Gly mutant has the least apparent effect (Fig. 2 ▸ d).
Figure 1.
Surface representations of the WT D4 (green)/A201–50 (violet) complex and the individual partner molecules (top row) from PDB entry 4oda. The three mutant structures are presented at the bottom.
Figure 2.
Crystal structure of the WT His-D4/A201–50 complex (a) and of the Trp43Ala (b), Arg167Ala (c) and Pro173Gly (d) mutants. σA-Weighted 2F o − F c electron-density maps of the refined models contoured at 1σ are also shown. A cyan dotted line indicates the hydrogen bond between Arg167 and Thr41. Colouring and orientation are as in Fig. 1 ▸.
Table 3. Analysis of the A20/D4 interface.
| Reference value | Difference from the reference value | |||
|---|---|---|---|---|
| Construct | WT D4/A201–50 | D4/A201–50-W43A | D4-R167A/A201–50 | D4-P173G/A201–50 |
| R.m.s.d. of Cα positions compared with WT (Å) | 0.29 ± 0.03 | 0.32 ± 0.02 | 0.20 ± 0.01 | |
| Buried surface (from PISA)† (Å2) | 1837 ± 21 | −196 ± 9 | −106 ± 7 | −24 ± 11 |
| Hydrogen bonds | 10 | −1‡ | −1‡ | 0 |
| Solvation free-energy gain (from PISA)† (kcal mol−1) | −13.0 ± 0.5 | 2.22 ± 0.16 | −1.00 ± 0.10 | 0.50 ± 0.06 |
| Energy of the cation–π interaction of Trp43 (from CAPTURE)§ (kcal mol−1) | −4.2 ± 0.1 | 4.2 ± 0.1 | 4.2 ± 0.1 | 0.4 ± 0.3 |
| Total of interaction energies¶ (kcal mol−1) | −17.2 ± 0.6 | 6.4 ± 0.3 | 3.2 ± 0.2 | 0.9 ± 0.4 |
| Melting-temperature change (from Contesto-Richefeu et al., 2014 ▸) (°C) | 47.3 | −5.3 | −3.6 | −4.6 |
| Complex formation upon co-expression (from Contesto-Richefeu et al., 2014 ▸) | +++ | +/− | +++ | ++/− |
Only residues involved in the contact around Trp43 were used in order to calculate the change. Average value for the two copies in the asymmetric unit.
Loss of the hydrogen bond between the carbonyl of Thr41 and Arg167 Nη1 shown in Figs. 2 ▸(a) and 2 ▸(d).
Average value for the two copies in the asymmetric unit.
Sum of the solvation free-energy gain and the energy of the cation–π interaction.
The effect of these mutations at the interface was studied with the PISA and CAPTURE servers and was correlated with previous data on the stability of the complex (Table 3 ▸). PISA analyzes the interface using atomic solvation parameters (ASPs) but does not allow the examination of cation–π interactions (Krissinel & Henrick, 2007 ▸), which were analyzed with the CAPTURE server (Gallivan & Dougherty, 1999 ▸). The Arg167Ala and Trp43Ala mutations considerably reduce the surface of the interface and lead to loss of the cation–π interaction, whereas the Pro173Gly mutation only leads to a slightly reduced contact surface. The predicted strength of the interaction was compared with the stability of the different constructs observed in a thermal shift assay and with the amount of complex formed upon co-expression (Table 3 ▸; Contesto-Richefeu et al., 2014 ▸). All three mutant complexes displayed a melting temperature that is about 4 to 5°C lower (T m) compared with WT D4/A201–50 in thermal shift assays (Table 3 ▸). It was further observed that A20 Trp43 and D4 Pro173 are critical residues at the D4/A20 interface and that mutations of these amino acids interfere with complex formation in solution. In contrast, D4 Arg167 did not seem to be essential for D4/A20 complex formation (Contesto-Richefeu et al., 2014 ▸). In silico analysis of the interface properties using a combination of ASPs and cation–π interactions allowed the correct prediction of the measured stability for the WT complex and the Trp43Ala and Arg167Ala mutants (i.e. WT > Arg167Ala > Trp43Ala). In contrast, using only ASPs, an increase in stability of the Arg167Ala mutant is predicted compared with the WT. Thus, loss of the hydrogen bond (cyan dotted line in Figs. 2 ▸ a and 2 ▸ d) does not seem to be detrimental; its contribution is small in any case as it cannot stabilize the conformation of Arg167 without the contribution of Trp43 (see the Trp43Ala mutant in Fig. 2 ▸ b). Unexpectedly, the calculated free-energy change of 0.9 kcal mol−1 for the interaction of the Pro173Gly mutant underestimates by far the effects of the mutation observed in the thermal stability assay and in complex formation, which are similar to those of the Trp43Ala mutation (Table 3 ▸). This indicates a major contribution of the restrained conformational space of the proline residue to the strength of the interaction, a term which cannot be identified by the analysis of surface properties. This indirect role of proline residues in contact interfaces should be taken into account in mutagenesis studies.
Small molecules interfering with protein–protein interfaces within the poxvirus replication machinery and in particular with the D4/A20 interaction could represent an attractive new antiviral strategy (Flusin et al., 2012 ▸; Saccucci et al., 2009 ▸). A recent study identified compounds binding to D4 and interfering with the complex formed by VACV D4 and the first 100 residues of A20 (A201–100; Schormann et al., 2011 ▸). Using a molecular-docking approach, we further showed that some of these molecules could reproduce key contact features observed at the D4/A201–50 interface. In particular, they contain a hydrophobic phenyl group which was predicted to mimic the stacking interactions involving A20 Trp43 (Contesto-Richefeu et al., 2014 ▸).
Thus, the structure of the WT D4/A201–50 complex together with the new structures involving the mutation of critical residues at the interface will be useful for in silico drug-design experiments aiming to select compounds interfering with the D4/A20 interaction and also for improving hit compounds which were selected during high-throughput screening to specifically target this interaction (Schormann et al., 2011 ▸).
Supplementary Material
PDB reference: D4/A201–50-W43A, 5jkr
PDB reference: D4-R167A/A201–50, 5jks
PDB reference: D4-P173G/A201–50, 5jkt
Acknowledgments
We wish to thank the European Synchrotron Radiation Facility for access to the ID23-1 beamline and the local contacts for help. We received funding from the Service de Santé des Armées, the Délégation Générale pour l’Armement and grant ANR-13-BSV8-0014 (REPLIPOX).
References
- Boyle, K. A., Arps, L. & Traktman, P. (2007). J. Virol. 81, 844–859. [DOI] [PMC free article] [PubMed]
- Contesto-Richefeu, C., Tarbouriech, N., Brazzolotto, X., Betzi, S., Morelli, X., Burmeister, W. P. & Iseni, F. (2014). PLoS Pathog. 10, e1003978. [DOI] [PMC free article] [PubMed]
- De Silva, F. S., Lewis, W., Berglund, P., Koonin, E. V. & Moss, B. (2007). Proc. Natl Acad. Sci. USA, 104, 18724–18729. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, E., Klemperer, N., Ghosh, R. & Traktman, P. (1995). J. Virol. 69, 5353–5361. [DOI] [PMC free article] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Flusin, O., Saccucci, L., Contesto-Richefeu, C., Hamdi, A., Bardou, C., Poyot, T., Peinnequin, A., Crance, J.-M., Colas, P. & Iseni, F. (2012). Antiviral Res. 96, 187–195. [DOI] [PubMed]
- Gallivan, J. P. & Dougherty, D. A. (1999). Proc. Natl Acad. Sci. USA, 96, 9459–9464. [DOI] [PMC free article] [PubMed]
- Ishii, K. & Moss, B. (2002). Virology, 303, 232–239. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- McCraith, S., Holtzman, T., Moss, B. & Fields, S. (2000). Proc. Natl Acad. Sci. USA, 97, 4879–4884. [DOI] [PMC free article] [PubMed]
- McDonald, W. F. & Traktman, P. (1994). J. Biol. Chem. 269, 31190–31197. [PubMed]
- Moss, B. (2013). Cold Spring Harb. Perspect. Biol. 5, a010199. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Saccucci, L., Crance, J.-M., Colas, P., Bickle, M., Garin, D. & Iseni, F. (2009). Antiviral Res. 82, 134–140. [DOI] [PubMed]
- Schormann, N., Sommers, C. I., Prichard, M. N., Keith, K. A., Noah, J. W., Nuth, M., Ricciardi, R. P. & Chattopadhyay, D. (2011). Antimicrob. Agents Chemother. 55, 5054–5062. [DOI] [PMC free article] [PubMed]
- Stanitsa, E. S., Arps, L. & Traktman, P. (2006). J. Biol. Chem. 281, 3439–3451. [DOI] [PubMed]
- Upton, C., Stuart, D. T. & McFadden, G. (1993). Proc. Natl Acad. Sci. USA, 90, 4518–4522. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: D4/A201–50-W43A, 5jkr
PDB reference: D4-R167A/A201–50, 5jks
PDB reference: D4-P173G/A201–50, 5jkt