Abstract
Biosensor development has been a highly dynamic field of research and has progressed rapidly over the past two decades. The advances have accompanied the breakthroughs in molecular biology, nanomaterial sciences, and most importantly computers and electronics. The subfield of evanescent wave fluorescence biosensors has also matured dramatically during this time. Fundamentally, this review builds on our earlier 2005 review. While a brief mention of seminal early work will be included, this current review will focus on new technological developments as well as technology commercialized in just the last decade. Evanescent wave biosensors have found a wide array applications ranging from clinical diagnostics to biodefense to food testing; advances in those applications and more are described herein.
Keywords: optical biosensor, evanescent wave, fluorescence, applications
1.0 Introduction
Since the theory and practice of fluorescence-based biosensors that exploit the surface sensitive nature of the evanescent wave first appeared in the mid-1970’s, the types of evanescent wave biosensors and the applications for which those biosensors have been utilized has exploded. In our first review (Taitt et al. 2005), we covered the history, theory and practice of evanescent wave biosensors up to that date. Now, some ten years later, the time has come to provide an update on the field and describe the highlights of the last decade.
Biosensors encompass the entire range of instruments which utilize a biological component for the target recognition / capture step. For this review we are limiting ourselves to only those instruments where that biological recognition step and subsequent signal transduction of the binding event occurs within the confines of an evanescent wave. The evanescent wave is remarkable phenomena, the details of which are often poorly understood – even by those responsible for assay development on the biosensor systems.
The evanescent wave, first described by (Hirschfeld 1966) arises from the manner in which light behaves when confined in an optical waveguide. Guided light is totally internally reflected when it meets the interface of the waveguide and a surrounding medium with a lower index of refraction. For total internal reflectance, the Fresnel transmission coefficients for the transverse electric wave and the transverse magnetic wave are non-zero. This means that, although the light energy is totally reflected, an electromagnetic field extends out from the interface into the lower index medium. This field, the evanescent wave, decays exponentially with distance from the surface, generally over the distance of 100 nm to approximately a wavelength. For multimode waveguides, the penetration depth dp, the distance from the surface at which the strength of this field is 1/e of its value at the surface, is a function of the two refractive indices (nwaveguide and, nmedium), the angle of incidence of the light, and the wavelength (λ). Looking at these parameters, one can appreciate the key to generating a strong evanescent field is the angle of incidence of the light at the interface. One can easily show that nearly all the power in the evanescent wave comes from light that contacts the interface at an angle just above that required to be become leaky, an important consideration during the instrument design phase (Anderson et al. 1994).
Since the evanescent wave is such a near-surface phenomena, biosensors which employ evanescent wave excitation to generate the fluorescent signal are by their nature surface-sensitive measurements, meaning that only fluorescent molecules near the surface are excited. This geometric limitation can help minimize unwanted background signal from a bulk sample while enhancing just the signal from fluorophores captured on the surface. What has been most remarkable is the profuse variety of biosensors that have been developed based on the same fundamental scientific principle.
2.0 Fast Forward to the Present
The first biosensors to take advantage of technology evolving from the optical communications community relied on optical fibers and relatively large lasers. Since then, lasers have gotten much smaller, cheaper, and so easy to use that they are found in consumer goods. In many cases, they have been replaced by light emitting diodes (LEDs) and even polymer LEDs. Concurrent advances in waveguide materials have ushered in a transition from silica fibers to planar waveguides. The expansion from silicon and silica materials to polymers has not only decreased the cost and facilitated manufacture, but also opened the doors to a much wider variety of geometries for waveguiding and signal detection. This geometric flexibility is leading to integrated, automated biosensors where the sample processing, excitation and fluorescence signal collection can all occur on a single substrate — which could even be disposable.
The fiber optic biosensors of today have evolved significantly from the initial devices that relied on long, partially clad, tapered silica fibers (Thompson and Villarruel 1991) or unclad rods of glass with diameters large enough for coupling light easily into the end (Hirschfeld and Block 1985). The molecular recognition elements are still attached to the fiber surface and the fluorescence is excited with evanescent light--and the advantages of tapering the fiber to improve the delivery of light to the surface are well appreciated. However, fibers used by most groups have evolved into small, molded polystyrene disposable probes with the in-coupling lens integrated with the fiber. Research International in particular (www.resrchintl.com, Monroe, WA USA) currently sells two fully automated systems with the same fiber probes: the RAPTOR, which requires manual sample introduction, and the BioHawk, which is integrated with an air sampler in a back pack for use by first responders and military (Anderson and McCrae 2007; Saaski 2009). The RAPTOR has four probes per cassette with an off-chip reservoir of fluorescent reagents (Jung et al. 2003), while the BioHawk has eight probes contained with reagents in a reusable cassette to test for 8 different targets simultaneously. Separating the reagents according to the specific assay on the probe has the benefit of eliminating the problem of crossreacting antibodies as cocktails of tracer reagents become more complex in more highly multiplexed assays. The other difference is that the signal in the RAPTOR is collected back up the fiber and through a ball lens into the detector while the BioHawk collects the signal normal to the fiber probe (Figure 1). In a recent modification of the Research International probes, Ton et al. demonstrated a “reagentless” assay format, coating the probes with a fluorophore-containing molecularly imprinted polymer (Ton et al. 2015); the researchers detected a small molecule target in the low nM range without the addition of fluorescent reagents during the assay.
Figure 1.
Fiber probe waveguide and biosensing scheme used in the BioHawk (Saaski 2009). Reprinted with permission from SPIE and the author.
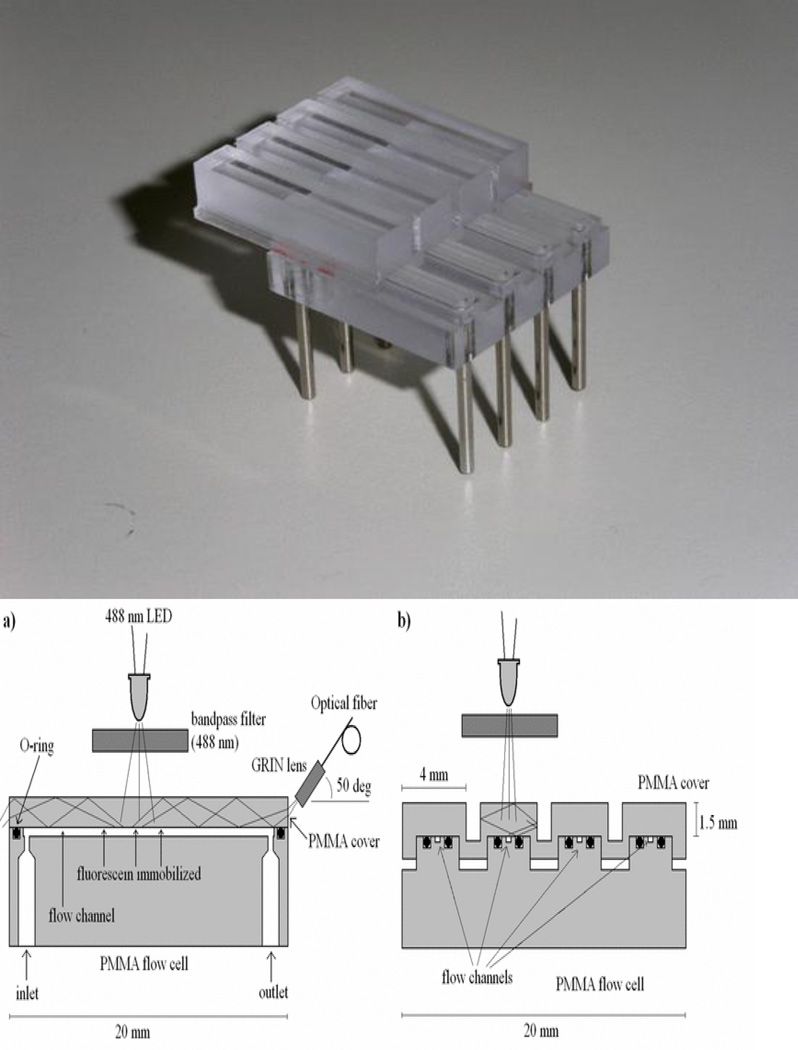
The use of short glass rods as waveguides took a very different evolutionary turn. Ligler, Feldstein and MacCraith realized that a glass capillary could serve as a waveguide while the sensing molecules were attached to the inner wall and the lumen used to deliver sample and reagents (Feldstein et al. 2000). This system had three other advantages: 1. The excitation could be provided easily from the side, either at a critical angle or normal to the capillary. 2. The emission signal could be integrated as it guided down the capillary/waveguide into a simple photodiode. The geometric integration of the signal without concomitant increase in stray excitation light enhanced the signal-to-noise ratio significantly. 3. The capillaries could be bundled and easily used in parallel with a single light source. This unique geometry has been used by two other groups to develop biosensor products, Creatv Microtech (www.creatvmicrotec.com, Potomac, MD, USA) and an Italian consortium funded by the European Union CAREMAN project. Creatv’s Signalyte™-II uses a single fused-silica capillary waveguide to harvest target from relatively large volumes and achieve sensitivities 1000 times that of a fluorescent plate reader. Samples tested include clinical fluids, food, water and environmental samples. In concert with immunomagnetic separation and antibody-coated capillaries, the Signalyte™-II system achieved a detection level of 10 Escherichia coli cells per mL in water and beef homogenate (Zhu et al. 2011). The Italian consortium led by Francesco Baldini has produced a polymethylmethacrylate (PMMA) chip with parallel channels and multiple waveguides in the roof (Figure 2). Flood illumination from an LED provides the excitation light normal to the waveguides and an optical fiber with a grin lens collects the fluorescence traveling to the end of the waveguide (Baldini et al. 2008). The configuration for the parallel waveguiding channels appears to be much more manufacturable than the original Ligler biosensor (Ligler et al. 2002), but the collection optics could be improved for a more robust device.
Figure 2.
Integrating waveguide capillary sensor (Baldini et al. 2008). Schematics a and b diagram the waveguide and channel from the side and end, respectively, while a photo of a 4-channel device is shown on the right. Reprinted with permission from IEEE.
Prototype experiments have been recently published describing using the signal integrating capability of capillary waveguides to enhance fluorescence signals to the level they could be detected using a cell phone camera (Balsam et al. 2013). However, they still need to demonstrate in a real assay that the signal they are measuring is generated by the target at the wall of the capillary and not in the liquid core. Liquid core waveguides are of significant interest for integration into microfluidic systems, but have not yet been demonstrated for evanescent signal generation or collection. In many sensor applications, the distinction between fluorophores bound to a waveguide surface vs free in solution is critical for sensitivity.
Total internal reflectance (TIRF) microscopy evolved from the same understanding of how light behaves at interfaces that produce fiber optic communication systems. The first planar waveguide biosensor essentially extracted the planar substrates from TIRF microscopes and used a camera to generate a 2D image. Most of the commercial planar waveguide biosensors available today evolved from the work started in the 80’s by either the group at the Naval Research Laboratory (NRL) or the one at Ciba-Corning. Hansen Technologies and mBio Diagnostics (ww.mBioDx.com, Boulder, CO, USA) licensed the NRL patents while the group at Ciba-Geigy transitioned through Novartis to become the startup company Zeptosens, which became a subsidiary of Bayer (http://www.bayertechnology.com/solutions/technology-development/proteomic-profiling/technology.html; Leverkusen, Germany). The NRL technology includes a slightly rough (leaky) surface of the planar waveguide for an extended penetration depth multimode waveguide while the Zeptosens technology has a very thin, higher refractive index, light guiding layer on the sensing side of the substrate to extend the penetration depth by creating a single mode waveguide. Light is injected into the NRL waveguides through a cylindrical lens while the light is coupled into the Zeptosens waveguide via a diffractive grating. Hanson Technologies (Carlisle, PA, USA) has married the NRL automated biosensor (Golden et al. 2005) to a large scale filtration system for harvesting and identifying bacteria and toxins in during food production (http://www.prnewswire.com/news-releases/hanson-technologies-inc-introduces-ultrarapidtm-food-and-beverage-safety-solutions-at-food-safety-and-security-summit-expo--conference-52127082.html) while mBio has eliminated active fluidic elements (Ligler et al. 2007) to produce an easy-to-use, inexpensive point-of-care device for detection of infection (Lochhead et al. 2011; Logan et al. 2013) and toxins in lake water (Murphy et al. 2015) (Figure 3). While initially developed as an antibody-capture array (Pawlak et al. 2002), the Zeptosens system is now primarily used for protein profiling in which samples are spotted onto the waveguide in microarrays which are subsequently interrogated with antibodies, enzymes, or ligands (Assadi et al. 2013).
Figure 3.
Planar waveguide biosensors for fluorescence detection in the evanescent region: NRL system (left) and mBio (right).
Combining the concepts pioneered by fiber optic and planar waveguides yielded a construct known as ridge waveguides. These devices are essentially composed of a ridge etched out of a silicon (most often) or polymer substrate such that the light is focused primarily into the ridge as it travels across the planar surface. Due to the fabrication methods utilized and design flexibility, ridge waveguides are especially attractive for integration into microfluidic systems including automated biosensors. Ryan Bailey has combined the concepts of ridge waveguides and microring resonators to create multiplexed chips on silicon for analysis of complex matrices (Figure 4). Off-chip excitation light is coupled into a silicon waveguide using a grating, the waveguide couples the light into a liquid core ring resonator, and the light propagates around the cavity by TIRF with a path length that is much larger than the circumference of the cavity. Target analytes binding to the surface of the ring resonator shift the ring resonance. Initially, one of the most attractive aspects of this system was the ability to perform a wide variety of label-free interaction analyses (Kindt and Bailey 2013), but the need for higher sensitivity for clinical diagnostics has led to the use of fluorescent tags, nanoparticles or enzymatically generated precipitates to increase the signal to noise. Bailey’s version of microring resonators is now commercially available from Genaltye (http://genalyte.com; San Diego, CA, USA) in an automated system called the Maverick. An interesting future direction for the microring resonance technology may include the integration of a circular organic photodiode into the middle of the microring for on-chip detection (Lamprecht et al. 2011). While their system most commonly is used in fully ‘reagentless’ mode (measuring resonance shifts) or with nanoparticles or beads for signal amplification, fluorophores have also been used to boost signals (Lamprecht et al. 2011). Furthermore, horseradish peroxidase (HRP)-based catalyzed reporter deposition of a chromogenic precipitate has been incorporated to achieve sub pg/mL detection (Kindt et al. 2013); there is no reason why fluorescent substrates could not also be used.
Figure 4.
Biosensing with ring resonators (Kindt and Bailey 2013). Reprinted with permission from Elsevier.
Companies like Research International and mBio Diagnostics have demonstrated the practicality of using molded polymers to make waveguides in relatively conventional formats. New fabrication methods and materials for waveguides offer even more flexibility for waveguide design and integration into biosensor systems. Polystyrene waveguides have been screen printed onto both polymer and glass substrates, offering the potential for scale up using roll-to-roll manufacturing technology (Sagmeister et al. 2013). Agnarsson and colleagues (Agnarsson et al. 2010) describe the production of waveguides composed of a fluoropolymer Cytop™ that can be fabricated on a wide variety of substrates. They demonstrate that it is closely index-matched to water and thus provides a tunable penetration depth for evanescent wave sensing. It can even be used for fluorescence imaging of cultured cells, which makes it perfect for continuous monitoring of tissue-on-chip systems.
Pushing the idea of biocompatible polymer waveguides even further, Manocchi’s group created planar waveguides made of gelatin clad between agarose sheets (Manocchi et al. 2009). Though relatively lossy, the biocompatibility of the waveguides as well as the easy of embedding fluorophore or other signal generating molecules, the lost cost, and the ease of fabrication make them attractive for use in wet or in vivo applications.
Metal-clad optical waveguides are probably at the opposite end of the fabrication spectrum from polymer waveguides in terms of materials and fabrication technologies. Thin metal films have been used for decades to increase the power at the surface in surface plasmon resonance sensors. While the original appeal of these devices was for label-free sensors that measured a change in refractive index at the surface, many investigators discovered they needed fluorescence tags to achieve the desired sensitivity and the increase in photonic mode density in surface plasmon biosensors could enhance fluorescence intensity (Sato et al. 2009). Dostalek and Knoll recently reviewed the progress in biosensors based on surface plasmon-enhanced fluorescence spectroscopy (Dostalek and Knoll 2008). They describe the detection of 100 fM DNA, receptor binding in lipid membranes, and immunoassay sensitivities as low as 0.6 pg/mL. The addition of time resolution fluorescence measurement further reduced the sensitivity to ~10 molecules mm−2 min−1 (Yu et al. 2004).
In a comparable waveguide conformation, a very thin metal layer (~20 nm) minimizes the reflectivity for optical coupling and guides zero-order TE and TM modes with the lowest possible losses (Margheri et al. 2014). The underlying SiO2 layer thickness was selected to propagate the same modes. The utility of this waveguide was demonstrated by attaching a fluorescent molecule to the surface that quenches in the presence of Hg2+. In this case the fluorophore was part of the recognition complex rather than an exogenously added label.
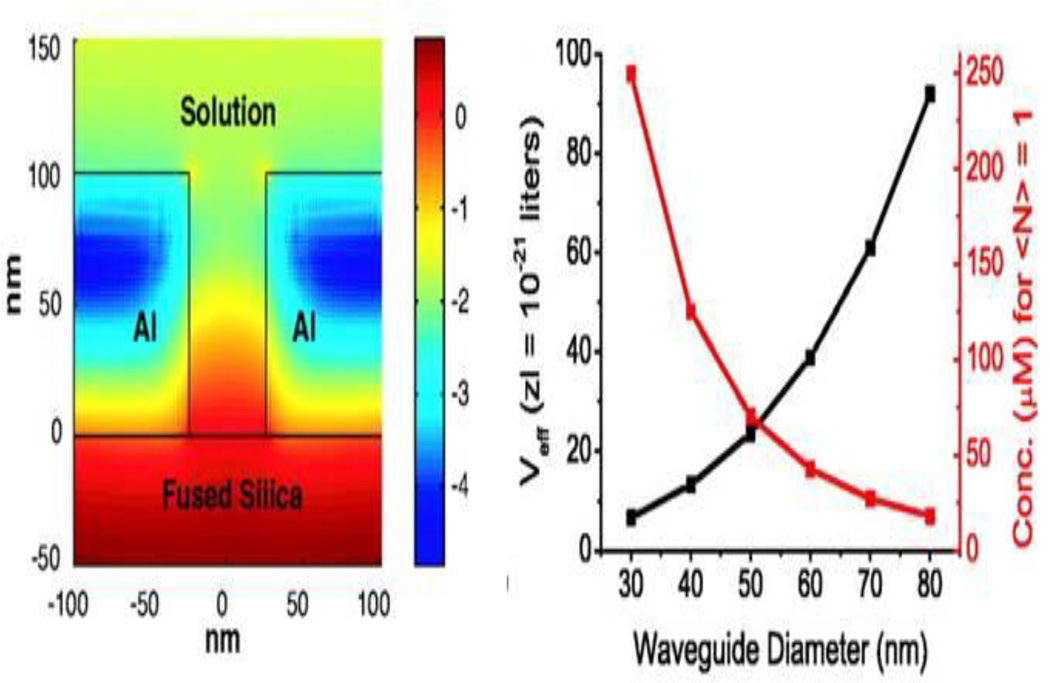
One of the most exciting examples of zero-mode waveguides for biosensor applications was promulgated by Harold Craighead and colleagues. The devices consisted of a zero-mode waveguide with arrays of small holes with well-defined volumes (Levene et al. 2005, 2007). The light from the waveguide is confined at the bottom of the hole, effectively separating bound from free molecules for fluorescence excitation. This technology has been used for single molecule analysis (Levene et al. 2003) (Figure 5), sequencing of individual DNA molecules (Korlach et al. 2006), and even analysis of protrusions from the plasma membranes of living cells (Moran-Mirabal et al. 2007). Pacific Biosciences (http://www.pacificbiosciences.com, Menlo Park, CA, USA) has commercialized the zero-mode waveguide technology for single molecule DNA sequencing with a focus on the ability to sequence very long chains of oligonucleotides in highly parallel analyses.
Figure 5.
Zero mode waveguides (Levene et al. 2003) Left: Simulation of the log of the intensity distribution for a zero-mode waveguide 50 nm in diameter and 100 nm long. Right: The effective volume Veff and the corresponding concentration for which an average of one molecule enters the volume. Reprinted with permission from Science.
The relatively new field of plasmonics exploits metallic nanostructures on surfaces to route and manipulate light (Schuller et al. 2010). Just as continuous metal thin films can modify the waveguiding of light, so can nano-sculptured thin films (Abdulhalim 2014). The nanosculptured films can enhance fluorescence at a sensor surface partially due to the existence of surface plasmon waves. This enhancement has been documented at factors greater than 70 with surface films made of silver and aluminum. Yet another configuration of metal plasmonic films composed of nanostructured gold has created “trapped rainbows” in the visible range, offering the potential to perform spectroscopy on a chip as well as fluorescence enhancement (Smolyaninova et al. 2012).
3.0 Applications of Evanescent Wave Fluorescence Biosensors
The field of evanescent wave biosensors using fluorescence for detection has matured greatly since its inception (Hirschfeld and Block 1985). Improvements to the technology during the 1990s and early 2000s have resulted in a push towards commercial production and application in the areas of environmental monitoring, biosecurity and law enforcement, food safety, and medicine and disease diagnosis. It should be noted that, for the most part, many of these fields of potential application overlap. For example, food safety monitoring also takes into account the need for security to prevent and detect intentional acts of contamination or adulteration; the susceptibility of the public food supply to nefarious contamination has been recognized as a significant challenge by the Centers for Disease Control and the World Health Organization (Sobel et al. 2002; World Health Organization 2008).
3.1 Environmental and drinking water analysis
Environmental contaminants, including pollutants, industrial byproducts, endocrine effectors, and natural toxins and contaminants, can adversely affect human health and welfare both directly (e.g., through consumption or direct contact) or indirectly (e.g., via alterations to local flora/fauna). Environmental monitoring through groundwater and soil surveillance, stream sampling, or aerosolized particulate testing can provide early warning of hazardous conditions or contamination events and can also be used to monitor the progress of remediation efforts.
In spite of the importance of environmental applications, surprisingly few detection systems have achieved the capability of on-site, continuous monitoring in recent years. In addition to requirements for robust, weather-proof operation, biosensors intended for environmental surveillance must also contend with complex matrices comprising many chemical and biological components, wide ranges of pH and salinity, and a broad variety of targets interest. Therefore, there is often not sufficient economic motivation for development, manufacture, and widespread deployment of commercial sensors. That being said, several systems – both commercially produced and laboratory prototypes – have been successfully applied to monitoring of real-world environmental samples (Table 1).
Table 1.
Sensors applied to analysis of environmental samples (publications since 2005)
| System | Target | Notes | Reference |
|---|---|---|---|
| River Analyzer, Automated Water Analyser Computer Supported System (AWACSS) | Multiplexed: atrazine, isoproturon, propanil; sulphamethizole, estrone | Automated, unattended, and networked surveillance of river water at: Spain: 3 sites; Germany: 3 sites; Slovakia: 4 sites; Validate in multisite study. | (Le Blanc et al. 2009; Rodriguez-Mozaz et al. 2009; Tschmelak et al. 2006; Tschmelak et al. 2004) |
| mBio | Multiplexed: domoic acid, okadaic acid, saxitoxin, cylindrosperopsin, microcystin | Multiplexed analysis in cultures, seawater, tap water; automated sample prep including cell lysis improves sensitivity | (Devlin et al. 2013; McNamee et al. 2014; Murphy et al. 2015) |
| Automated online optical biosensing system (AOBS) | Microcystin LR | Long-term continuous monitoring of Lake Tai (China); validated with gold standard method | (Shi et al. 2013) |
| Leopard Array Biosensor (Hanson) | Microcystin-LR | Reservoir water (from 4 sites in Spain); regenerated 15 times; nodularin used as internal control | (Herranz et al. 2012) |
| RAPTOR | Enterococcus faecalis | Tampa Bay municipal beach water (samples were pre-filtered to concentrate) | (Leskinen and Lim 2008) |
| Laboratory prototype evanescent wave immunosensor | Bisphenol A; Multiplexed: 2,4-D, microcystin-LR, bisphenol A, melamine | Bottled water, lake water, tap water; regenerated 300 times | (Xiao-Hong et al. 2014; Xiao-hong et al. 2014) |
| Fiber optic biosensor | Trinitrotoluene (TNT) | Soil extracts; specificity tested with tetryl, 4-nitrophenol; regenerated 10 times | (Ehrentreich-Forster et al. 2008) |
| Fiber optic biosensor | Mercury | Wastewater, tap water, bottled water; Sludge water; specificity-tested with other metals; regenerated 100 times | (Long et al. 2011; Long et al. 2013) |
| Fiber optic biosensor | Atrazine | Soil extracts; correlated with gold standard method (R2=0.9968) | (Long et al. 2015) |
3.2 Defense and security
Over the past 20 years, concern about terrorist and criminal activities has increased interest in development and integration of biosensors for drugs of abuse, explosives, and chemical and biological weapons. As indicated earlier, security and defense-related applications overlap considerably with other fields of use, as chemical and biological agents can be spread by multiple modes of dissemination (food, air, water), and therefore myriad sample types may be encountered; similarly, sample types used for drug testing can also vary from purified stocks to environmental samples to clinical fluids, depending on the intended purpose. Furthermore, the effects of other interfering materials – potentially used to mimic (Crighton et al. 2012) or mask (Krebs et al. 2000) the presence of target – must be accounted for.
Table 2 describes biosensor technologies using total internal reflectance fluorescence (TIRF) for detection of explosives, toxins and pathogenic threat agents, drugs of abuse, and other banned substances. In most cases, detection of a specific target is desired. However, the presence of closely related compounds and metabolites may also be indicative of the targets of interest when testing for explosives, drugs of abuse, and other banned substances. For this reason, specificity studies may be important (Ehrentreich-Forster et al. 2008; Shriver-Lake et al. 1997). At the opposite end of the spectrum, interest has been growing in use of semi-selective recognition reagents for “anomaly” detection, such that unknown (or unanticipated) threat agents might be detected; these reagents include peptides (Kulagina et al. 2014), enzymes (Ramanathan and Simonian 2007; Viveros et al. 2006), and carbohydrates (Ngundi et al. 2006c; Ngundi et al. 2007; Ngundi et al. 2006d; Taitt et al. 2008). Broadly selective recognition reagents have also been used in evanescent wave fluorescence sensors for detection of other targets such as viruses (Kale et al. 2008), foodborne pathogens and toxins (Kulagina et al. 2014; Kulagina et al. 2007; Kulagina et al. 2006; Wang et al. 2015), and markers of infection (Sakamuri et al. 2014).
Table 2.
Biosensor platforms amenable to application to defense, security, and law enforcement (publications since 2005)
| Target | Platform | Notes | Reference |
|---|---|---|---|
| Biothreat organisms | |||
| Bacillus anthracis cells, protective antigen | Planar waveguide array | Sandwich immunoassay on avidin-decorated supported membranes; | (Martinez et al. 2005; Mukundan et al. 2009b) |
| B. anthracis protective antigen | planar waveguide | Sandwich immunoassay in serum; PEG-decorated surfaces improved signal-to-noise | (Anderson et al. 2008) |
| B. anthracis | Integrating waveguide (capillary) | Immunoassay detection, followed by growth and PCR | (Hang et al. 2008) |
| Yersinia pestis cells, F1 antigen | Fiber optic immunosensor (multi-specific) | Detected in liver, spleen, lung of infected mice in blind study; specificity tested against other Yersiniae, B. anthracis, Escherichia coli | (Wei et al. 2007a; Wei et al. 2007b) |
| B. anthracis DNA and cellular extracts | Planar waveguide array | Simultaneous detection of DNA and proteins using molecular beacons and antibodies | (Asanov et al. 2012) |
| Brucella abortus, Coxiella burnettii (multi-specific) | Planar waveguide array | Semi-selective detection, classification using array of antimicrobial peptides | (Kulagina et al. 2014) |
| B. anthracis spores | BioHawk/TacBioHawk (fiber optic biosensor) | Aerosolized Bacillus spores from air samples after 325 lpm aerosol collection | (Schaudies 2013) |
| B. anthracis spores | Capillary waveguide (Creatv Microtech) | Environmental swabs (+ dust); growth following capture/detection; | (Hang et al. 2008) |
| Chemical threats, toxins | |||
| Organophosphates | Fiber optic biosensor (multi-specific) | Enzyme-based assay | (Viveros et al. 2006) |
| Organophosphates | Planar waveguide array (multi-specific) | Enzyme-based assay using pH sensitive dye; kinetic measurements, specificity-tested | (Ramanathan and Simonian 2007) |
| Botulinum toxoids A, B, and E | Planar waveguide array | Semi-selective detection, classification using array of antimicrobial peptides; binding kinetics determined | (Kulagina et al. 2007) |
| Botulinum toxoid A, staphylococcal enterotoxin B (SEB) | Planar waveguide array | Multiplexed detection of two toxins in canned foods | (Sapsford et al. 2005) |
| Ricin | RAPTOR (fiber optic biosensor) | immunoassay; detected in deionized and tap water | (Yue et al. 2009) |
| Cholera toxin, ricin | NRL array biosensor | semi-selective toxin detection using array of carbohydrates | (Ngundi et al. 2006c; Ngundi et al. 2007; Taitt et al. 2008) |
| Explosives, drugs of abuse, banned substances | |||
| TNT | Fiber optic biosensor | Aptamer-based assay; used with soil extracts; specificity tested with tetryl, 4-nitrophenol; regenerated 10 times | (Ehrentreich-Forster et al. 2008) |
| Methyl-boldenone (anabolic steroid) | Planar grating waveguide | Competitive assay; 2-photon fluorescence excitation improved signal-to-noise, signal-to-backgound | (Muriano et al. 2011; Muriano et al. 2012) |
3.3 Medicine/Infectious disease diagnosis
In spite of the tremendous success that glucose sensors have had in the commercial market for diagnostics since the 1960s (Updike and Hicks 1967), evanescent wave fluorescence biosensors have not yet made as significant a contribution to the medical field. However, with the recent commercialization of a number of waveguide-based sensors (e.g., nGimat/Los Alamos Optical Biosensor, mBio, Zeptosens, Fraunhofer ivD platform), we anticipate that these sensors will make a major impact in healthcare-related fields. Table 3 lists a number of recent studies using commercial and lab-engineered platforms for analysis of clinical samples; publications describing assay development for physiologically relevant targets, but which lack data in clinical fluids, are too numerous to include here.
Table 3.
EW fluorescence sensors for medical applications that have been demonstrated in physiological matrices (publications since 2005)
| Target | System | Matrix | LOD | Reference |
|---|---|---|---|---|
| Anti-Mycoplasma antibodies (serology) | Fiber optic biosensor (RAPTOR) | Plasma | Blind trial: 92% samples correctly classified (positive/negative) | (Brown et al. 2008) |
| Mycobacterium tuberculosis | Fiber optic biosensor (RAPTOR) | Lung tissue homogenates | 106 cells/mL (80% detection) | (Denton et al. 2009) |
| M. tuberculosis mycobactin T (tuberculosis marker); M. leprae phenolic glycolipid-I (leprosy marker) | Planar waveguide with lipid bilayer (specificity conferred by “tracer” antibody | Serum | 500 nM (serum) | (Sakamuri et al. 2014) |
| M. tuberculosis lipoarabinomannan (tuberculosis marker) | Planar waveguide with lipid bilayer (specificity conferred by “tracer” antibody | Serum | 10 fM (serum) | (Mukundan et al. 2012) |
| Carcinoembryonc antigen (breast cancer marker) | Planar array | Serum, nipple aspirate | 0.5 pM | (Mukundan et al. 2009a) |
| Human platelet antigen DNA | Planar waveguide | DNA extracts from plasma (following concentration and “bar coding”) | 2 pM | (Trévisan et al. 2010) |
| Interleukin-6 (IL-6) (lupus marker) | Fiber optic biosensor | Serum, lymphoma sample | 5 pM (serum) not detected in lymphoma | (Wang et al. 2010) |
| B. anthracis protective antigen (anthrax marker) | Planar array | Serum | 83 ng/ml (1 nM) | (Anderson et al. 2008) |
| Procalcitonin (sepsis marker) | Planar waveguide | Whole blood (septic/non-septic) | Correlated with sepsis in patient samples (R2=0.988) | (Rascher et al. 2014) |
| Urokinase-type plasminogen activator (uPA), plasminogen activator inhibitor-1 (PAI-1), vascular endothelial growth factor (VEGF) (Breast cancer markers) | Planar grating array (Zeptosens) | Breast cancer tissue extracts | Multiplexed: 1 pg/mL (uPA), 33 pg/mL (PAI-1), 1 pg/mL VEGF) | (Weissenstein et al. 2006) |
| RNA transcripts (14 genes, incl. controls) (Multiplexed) | Planar grating array (Zeptosens) | RNA extracted from breast tissues | 0.01 pM (control transcripts) | |
| C-reactive protein (cardiac marker) | RIANA | Serum (100× diluted) | 0.13 ug/mL (undiluted) | (Albrecht et al. 2008) |
| Nerve growth factor (NGF) (cardiac marker) | Fiber optic biosensor (Analyte 2000) | Plasma | 1 ng/mL (plasma) | (Tang et al. 2007) |
| HIV, Treponema pallidum, hepatitis C antigens (multiplexed) | mBio Dx planar waveguide array | Serum, plasma | 100%, 98%, 98% positives identified correctly, respectively; no false-positives | (Lochhead et al. 2011) |
| CD4 T cell counts | mBio Dx planar waveguide array | Whole blood | 99.8% accurate counts (flow cytometry as gold standard) | (Logan et al. 2013) |
For point-of-care diagnostics — especially in resource- or personnel-limited clinics — one of the key requirements is to minimize the number of manipulations required to process and analyze the samples. This is critical in situations where resources and trained personnel are limited, as well as when highly virulent or infectious materials may be present in the samples (Leski et al. 2015). For this reason, lab-on-chip-type systems are being rapidly developed (Lochhead et al. 2011; Parks et al. 2014; Schumacher et al. 2012), although to date, only a handful have been fully validated in real clinical samples. mBio Diagnostics recently published a study assessing performance of their platform using 251 clinical samples from subjects comprising healthy individuals and those positive for HIV, syphilis, and hepatitis C (Lochhead et al. 2011); sample preparation — performed independently from microarray analysis — was limited to loading a preparative cartridge, adding a chase buffer, and then adding the tracer mix. Excellent specificity was demonstrated. The Fraunhofer ivD platform (Fraunhofer IZI-BB, Potsdam, Germany) appears to have similar capabilities, with additional modules for DNA amplification/hybridization and electrochemical readout (Schumacher et al. 2013; Schumacher et al. 2012), but to our knowledge, data from fully integrated analyses of clinical samples have not yet been published.
3.4 Food testing
Food safety is of major concern for consumers, regulatory agencies, food processors, retailers, and the medical community. Globally, approximately 1.8 million deaths occur each year due to contaminated food and water (Newell et al. 2010). While a core set of bacterial, viral, and parasitic pathogens and toxins have been identified as significant causes of food-related morbidity and mortality within the United States, additional sources of concern include the presence of allergens, pesticides, and antimicrobial compounds in tainted foodstuffs. As the analytical techniques used to identify the causative agents of food-related illnesses in general are often lengthy (requiring up to several days), labor intensive, and limited to one or two agents, biosensors capable of detecting multiple targets in a wide variety of food matrices have been developed (Table 4).
Table 4.
TIRF-based biosensors used for detection in spiked and naturally contaminated foodstuffs (publications since 2005)
| Target | Multiplex | Matrix | Detection limit | Reference |
|---|---|---|---|---|
| Antibiotics, hormones | ||||
| Chloramphenicol Streptomycin Desfuroylceftiofur |
3-plex | raw milk | 0.1 ng/mL 11 ng/mL 2 ng/mL |
(McGrath et al. 2015) |
| Progesterone | - | milk | 45–56 pg/mL | (Tschmelak et al. 2005)(Tschmelak et al. 2005)(Tschmelak et al. 2005)(Tschmelak et al. 2005)(Tschmelak et al. 2005)(Tschmelak et al. 2005) |
| Progesterone | - | milk | 0.04 ng/mL (could be used to determine estrus cycle) | (Kappel et al. 2007) |
| Bacteria, viruses, parasites | ||||
| Escherichia coli O157:H7 | - | Ground beef, turkey sausage, chicken rinse, apple juice | 1–5 ×104 cell/mL | (Shriver-Lake et al. 2007b)(Shriver-Lake et al. 2007b)(Shriver-Lake et al. 2007b)(Shriver-Lake et al. 2007b)(Shriver-Lake et al. 2007b)(Shriver-Lake et al. 2007b)(Shriver-Lake et al. 2007b) |
| SEB Salmonella typhimurium |
2-plex | Apple juice, milk (blind trial) | 1 ng/mL 5×105 colony-forming units (cfu)/mL | (Shriver-Lake et al. 2007a) |
| E. coli O157:H7 | - | Produce wash | 0.01 cfu/g produce | (Dyer et al. 2009) |
| E. coli O157:H7 | Ground beef | 900 cfu/g beef | (Zhu et al. 2011) | |
| Listeria monocytogenes | - | Read-to-eat meats (beef, chicken, turkey) | 1×103 cfu/mL* | (Ohk et al. 2010) |
| Toxins | ||||
| SEB botulinum toxoid |
2-plex | Apple juice, tomato products, mushroom, canned corn, canned green beans, tuna | 0.1–0.5 ng/ml 20–500 ng/mL |
(Sapsford et al. 2005) |
| Ochratoxin (OTA) | - | Barley, wheat pasta, corn, coffee, wine | 3.8–10 ng/g | (Ngundi et al. 2005) |
| Deoxynivalenol (DON) | - | Corn, wheat, barley oats | 1–10 ng/g | (Ngundi et al. 2006a) |
| Aflatoxin B1 | - | Corn, peanuts, pecans, peanut butter | 0.6–5 ng/g | (Sapsford et al. 2006) |
| OTA, DON | 2-plex | Corn, barley, wheat | 1–85 ng/g | (Ngundi et al. 2006b) |
| OTA | Oat samples | 1 ng/mL (buffer) | ||
100 cfu/25 g meat detected after 18h enrichment
Noticeably, TIRF-based sensors have generally yielded poor sensitivities for bacterial and viral contaminants, with detection limits clearly above FDA and international regulatory limits. Most operators, however, have incorporated enrichment steps – either by culture or by filtration and/or immunomagnetic separation). Although any procedure to enrich these targets will necessarily increase both the number of manual manipulations and time-to-result, results may still be available in a shorter time than otherwise obtained using standard microbiological methods (up to several days). Sensitivity for proteins and small molecule targets is typically better, with detection limits in many cases well below regulatory limits. A key benefit of TIRF-based sensors is the ability to detect these targets without significant sample cleanup. Mycotoxins and phycotoxins are often simply extracted with organic solvent and diluted before analysis. For proteinaceous targets, simple filtration or centrifugation is often sufficient for sample prep.
While detection of genetically modified organisms in foods and feed by SPR and electrochemical methods has been widely reported (Arugula et al. 2014), to our knowledge, TIRF-based sensors have not been applied to this application.
3.5 Basic research
The ability to simultaneously measure multiple analytes on planar waveguides has encouraged the use of fluorescence-based multiplexed analyses in research. The capacity to separate signal from background eliminates the need to pre-purify many of the samples, and the analyses can be accomplished in culture media, body fluids, or environmental samples. The array biosensors have proved useful for looking at cell viability and gene expression (Johnson-White et al., 2007) and even commercialized for interrogating cells and tissues for protein expression and drug sensitivities.
While not producing microarrays specific for an individual disease or syndrome, Zeptosens/Bayer manufactures waveguides and instrumentation for individual research teams to pattern and interrogate cells and tissues as “reverse phase protein arrays” (RPPAs). RPPAs are distinct from standard antibody- or DNA-type arrays in that instead of using an array of different “capture” molecules immobilized in a planar array to interrogate a sample mixture of different proteins, RPPAs comprise spots of immobilized protein lysates, which are then subsequently probed with different antibodies, enzymes, or ligands. Zeptosens’ waveguide-based microarrays have been used for biomarker discovery (Assadi et al. 2013; Weissenstein et al. 2006), following protein expression and modification over time (Pirnia et al. 2009), profiling enzymatic activities and signaling pathways (Van Oostrum et al. 2009; Voshol et al. 2009), and drug discovery and testing (Saturno et al. 2007; Tegnebratt et al. 2014) using tissues and lysates supplied by each research group and appropriate for each application.
4.0 Conclusions
This review updates the reader up with the progress in the field of evanescent wave biosensors since our previously published review (Taitt et al. 2005). While we have endeavored to include the most noteworthy progress that has occurred during the last decade, undoubtedly and regretfully some pertinent contributions will have been overlooked. Nonetheless, we hope this update will serve to reinvigorate the research community to make even greater strides in the coming decade, as we work towards developing ever more sensitive, versatile, and user-friendly biosensors. Unquestionably, this will be accomplished by exploiting advances in photonics technology and the ever expanding technology behind mobile phones. Advances in nanophotonics, metamaterials, integrated optics and fluidics elements, and advanced manufacturing will provide these new opportunities. Future bioanalytical systems integrating waveguides and fluorescence can be smaller, cheaper, faster, and more flexible in terms of the type of analyses that can be conducted. The driving force for these advances has been and continues to be the desire for reliable biosensors that are easy to use and provide rapid and sensitive results for anywhere from one to one hundred analytes at a time. The challenge as the field of biosensor development has matured is to continue to move the ball forward. Many highly effective and efficient instruments have been and are being manufactured, and the utility of these systems will spark the creation of even more remarkable instrumentation in the future.
Acknowlegements
Preparation of this manuscript was supported in part by the Office of Naval Research and NRL 6.2 funding (work units 6547 and 6703) and a grant from the North Carolina Translational and Clinical Sciences Institute (NIH 1UL1TR001111).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdulhalim I. Small. 2014;10(17):3499–3514. doi: 10.1002/smll.201303181. [DOI] [PubMed] [Google Scholar]
- Agnarsson B, Halldorsson J, Arnfinnsdottir N, Ingthorsson S, Gudjonsson T, Leosson K. Microelectronic Eng. 2010;87(1):56–61. [Google Scholar]
- Albrecht C, Kaeppel N, Gauglitz G. Anal. Bioanal. Chem. 2008;391(5):1845–1852. doi: 10.1007/s00216-008-2093-x. [DOI] [PubMed] [Google Scholar]
- Anderson AS, Dattelbaum AM, Montaño GA, Price DN, Schmidt JG, Martinez JS, Grace WK, Grace KM, Swanson BI. Langmuir. 2008;24(5):2240–2247. doi: 10.1021/la7033438. [DOI] [PubMed] [Google Scholar]
- Anderson GP, Golden JP, Ligler FS. IEEE Trans. Biomed. Eng. 1994;41(6):578–584. doi: 10.1109/10.293245. [DOI] [PubMed] [Google Scholar]
- Anderson GP, McCrae DA. RAPTOR: Development of a fiber-optic biosensor. In: Marks RS, Cullen DC, Karube I, Lowe CR, Weetail HH, editors. Handbook of Biosensors and Biochips. London: John Wiley & Sons; 2007. pp. 1281–1287. [Google Scholar]
- Arugula MA, Zhang Y, Simonian AL. Anal. Chem. 2014;86(1):119–129. doi: 10.1021/ac402898j. [DOI] [PubMed] [Google Scholar]
- Asanov A, Zepeda A, Vaca L. Sensors. 2012;12:1800–1815. doi: 10.3390/s120201800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assadi M, Lamerz J, Jarutat T, Farfsing A, Paul H, Gierke B, Breitinger E, Templin MF, Essioux L, Arbogast S, Venturi M, Pawlak M, Langen H, Schindler T. Mol. Cell Proteomics. 2013;12(9):2615–2622. doi: 10.1074/mcp.M112.023051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldini F, Carloni A, Giannetti A, Mencaglia A, Porro G, Tedeschi L, Trono C. IEEE Sensors J. 2008;8(7–8):1305–1309. [Google Scholar]
- Balsam J, Bruck HA, Rasooly A. Sens. Act. B Chem. 2013;186:711–717. doi: 10.1016/j.snb.2013.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DR, Wendland LD, Ortiz GV, Kramer MF, Lim DV, Brown MB, Klein PA. Vet. Immunol. Immunopathol. 2008;124:322–331. doi: 10.1016/j.vetimm.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Crighton T, Hoile R, Coleman NV. Forensic Sci. Internat. l. 2012;219(1–3):88–95. doi: 10.1016/j.forsciint.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Denton KA, Kramer MF, Lim DV. J. Rapid Meth. Automation in Microbiol. 2009;17(1):17–31. [Google Scholar]
- Devlin S, Meneely JP, Greer B, Greef C, Lochhead MJ, Elliott CT. Anal. Chim. Acta. 2013;769(0):108–113. doi: 10.1016/j.aca.2013.01.033. [DOI] [PubMed] [Google Scholar]
- Dostalek J, Knoll W. Biointerphases. 2008;3(3):FD12–FD22. doi: 10.1116/1.2994688. [DOI] [PubMed] [Google Scholar]
- Dyer M, Oberholtzer J, Mulligan D, Hanson W. Proc. SPIE. 2009;7306:73060G. [Google Scholar]
- Ehrentreich-Forster E, Orgel D, Krause-Griep A, Cech B, Erdmann VA, Bier F, Scheller FW, Rimmele M. Anal. Bioanal. Chem. 2008;391:1793–1800. doi: 10.1007/s00216-008-2150-5. [DOI] [PubMed] [Google Scholar]
- Feldstein MJ, MacCraith BD, Ligler FS. Integrating multi-waveguide sensor. 6137117. US patent. 2000
- Golden JP, Taitt CR, Shriver-Lake LC, Shubin YS, Ligler FS. Talanta. 2005;65(5):1078–1085. doi: 10.1016/j.talanta.2004.03.072. [DOI] [PubMed] [Google Scholar]
- Hang J, Sundaram A, Zhu P, Shelton D, Karns J, Marktin P, Li S, Amstutz P, Tang C-M. J. Microbiol. Methods. 2008;73(3):242–246. doi: 10.1016/j.mimet.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz S, Marazuela MD, Moreno-Bondi MC. Biosensors and Bioelectronics. 2012;33(1):50–55. doi: 10.1016/j.bios.2011.12.016. [DOI] [PubMed] [Google Scholar]
- Hirschfeld TB, Block MJ. Assay apparatus and methods. 4558014. US patent. 1985
- Hirschfeld TE. Canadian J. Spectroscopy. 1966;10:128–130. [Google Scholar]
- Johnson-White B, Lin B, Ligler FS. Anal. Chem. 2007;79:140–146. doi: 10.1021/ac061229l. [DOI] [PubMed] [Google Scholar]
- Jung CC, Saaski EW, McCrae DA, Lingerfelt BM, Anderson GP. IEEE Sensors J. 2003;3(4):352–360. [Google Scholar]
- Kale RR, Mukundan H, Price DN, Harris JF, Lewallen DM, Swanson BI, Schmidt JG, Iyer SS. J. Am. Chem. Soc. 2008;130(26):8169–8171. doi: 10.1021/ja800842v. [DOI] [PubMed] [Google Scholar]
- Kappel N, Proll F, Gauglitz G. Biosens. Bioelectron. 2007;22:2295–2300. doi: 10.1016/j.bios.2006.11.030. [DOI] [PubMed] [Google Scholar]
- Kindt JT, Bailey RC. Current Opinion Chem. Biol. 2013;17(5):818–826. doi: 10.1016/j.cbpa.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindt JT, Luchansky MS, Qavi AJ, Lee SH, Bailey RC. Anal. Chem. 2013;85(22):10653–10657. doi: 10.1021/ac402972d. [DOI] [PubMed] [Google Scholar]
- Korlach J, Webb WW, Levene M, Turner S, Craighead HG, Foquet M. Method for sequencing nucleic acid molecules. 7056661. US patent. 2006
- Krebs CP, Costelloe MT, Jenks D. International Journal of Drug Policy. 2000;11(5):351–356. doi: 10.1016/s0955-3959(00)00052-9. [DOI] [PubMed] [Google Scholar]
- Kulagina N, Taitt C, Anderson GP, Ligler FS. Affinity-based detection of biological targets. 8965710. US patent 8658372 and US patent. 2014
- Kulagina NV, Anderson GP, Ligler FS, Shaffer KM, Taitt CR. Sensors. 2007;7(11):2808–2824. doi: 10.3390/s7112808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulagina NV, Shaffer KM, Anderson GP, Ligler FS, Taitt CR. Anal. Chim. Acta. 2006;575(1):9–15. doi: 10.1016/j.aca.2006.05.082. [DOI] [PubMed] [Google Scholar]
- Lamprecht B, Kraker E, Sagmeister M, Köstler S, Galler N, Ditlbacher H, Ungerböck B, Abel T, Mayr T. Physica Status Solidi (RRL) – Rapid Research Letters. 2011;5(9):344–346. [Google Scholar]
- Le Blanc A, Albrecht C, Bonn T, Fechner P, Proll G, Proll F, Carlquist M, Gauglitz G. Anal. Bioanal. Chem. 2009;395:1769–1776. doi: 10.1007/s00216-009-3038-8. [DOI] [PubMed] [Google Scholar]
- Leski TA, Ansumana R, Taitt CR, Lamin JM, Bangura U, Lahai J, Mbayo G, Kanneh MB, Bawo B, Bockarie AS, Scullion M, Phillips CL, Horner CP, Jacobsen KH, Stenger DA. J. Clin. Microbiol. 2015;15:00527. doi: 10.1128/JCM.00527-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskinen SD, Lim DV. Appl. Environ. Microbiol. 2008;74(15):4792–4798. doi: 10.1128/AEM.00052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene MJ, Korlach J, Turner SW, Craighead HG, Webb WW. Zero-mode clad waveguides for performing spectroscopy with confined effective observation volumes. 6917726. US patent. 2005
- Levene MJ, Korlach J, Turner SW, Craighead HG, Webb WW. Zero-mode waveguides. 7181122. US patent. 2007 doi: 10.1126/science.1079700. [DOI] [PubMed]
- Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- Ligler FS, Breimer M, Golden JP, Nivens DA, Dodson JP, Green TM, Haders DP, Sadik OA. Anal. Chem. 2002;74(3):713–719. doi: 10.1021/ac015607s. [DOI] [PubMed] [Google Scholar]
- Ligler FS, Sapsford KE, Golden JP, Shriver-Lake LC, Taitt CR, Dyer MA, Barone S, Myatt CJ. Anal. Sci. 2007;23(1):5–10. doi: 10.2116/analsci.23.5. [DOI] [PubMed] [Google Scholar]
- Lochhead MJ, Todorof K, Delaney M, Ives J, Greef C, Moll K, Rowley K, Vogel K, Myatt C, Zhang X-Q, Logan C, Benson C, Reed S, Schooley R. J. Clin. Microbiol. 2011;49(10):3584–3590. doi: 10.1128/JCM.00970-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan C, Givens M, Ives JT, Delaney M, Lochhead MJ, Schooley RT, Benson CA. J. Immunol. Methods. 2013;387(1–2):107–113. doi: 10.1016/j.jim.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, Gao C, Shi HC, He M, Zhu AN, Klibanov AM, Gu AZ. Biosens. Bioelectron. 2011;26:4018–4023. doi: 10.1016/j.bios.2011.03.022. [DOI] [PubMed] [Google Scholar]
- Long F, Zhu A, Shi H, Sheng J, Zhao Z. Chemosphere. 2015;120(0):615–620. doi: 10.1016/j.chemosphere.2014.09.072. [DOI] [PubMed] [Google Scholar]
- Long F, Zhu A, Shi H, Wang H, Liu J. Scientific Reports. 2013;3:2308. doi: 10.1038/srep02308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manocchi AK, Domachuk P, Omenetto FG, Yi H. Biotechnol. Bioeng. 2009;103(4):725–732. doi: 10.1002/bit.22306. [DOI] [PubMed] [Google Scholar]
- Margheri G, Giorgetti E, Marsili P, Zoppi A, Lascialfari L, Cicchi S. 2014;53(7):071816–071816. [Google Scholar]
- Martinez JS, Grace K, Grace K, Hartman N, Swanson BI. J. Mat. Chem. 2005;15:4639–4647. [Google Scholar]
- McGrath T, McClintock L, Dunn J, Husar G, Lochhead M, Sarver R, Klein F, Rice J, Campbell K, Elliott C. Anal. Bioanal. Chem. 2015:1–14. doi: 10.1007/s00216-015-8526-4. [DOI] [PubMed] [Google Scholar]
- McNamee SE, Elliott CT, Greer B, Lochhead M, Campbell K. Environ. Sci. Tech. 2014;48(22):13340–13349. doi: 10.1021/es504172j. [DOI] [PubMed] [Google Scholar]
- Moran-Mirabal J, Torres A, Samiee K, Baird B, Craighead H. Nanotechnology. 2007;18(19):195101. [Google Scholar]
- Mukundan H, Kubicek JZ, Holt A, Shively JE, Martinez JS, Grace K, Grace WK, Swanson BI. Sens. Act. B: Chem. 2009a;138(2):453–460. [Google Scholar]
- Mukundan H, Price DN, Goertz M, Parthasarathi R, Montaño GA, Kumar S, Scholfield MR, Anderson AS, Gnanakaran S, Iyer S, Schmidt J, Swanson BI. Tuberculosis. 2012;92(1):38–47. doi: 10.1016/j.tube.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Mukundan H, Xie H, Anderson A, Grace K, Martinez JS, Swanson BI. Proc. SPIE. 2009b;7167:71670A-71671–71670A-71610. [Google Scholar]
- Muriano A, Salvador JP, Galve R, Marco MP, Thayil KNA, Loza-Alvarez P, Soria S. Proc. SPIE. 2011:794114–794117. [Google Scholar]
- Muriano A, Thayil KNA, Salvador JP, Loza-Alvarez P, Soria S, Galve R, Marco MP. Sens. Act. B: Chemical. 2012;174:394–401. [Google Scholar]
- Murphy C, Stack E, Krivelo S, McPartlin DA, Byrne B, Greef C, Lochhead MJ, Husar G, Devlin S, Elliott CT, O’Kennedy RJ. Biosens. Bioelectron. 2015;67:708–714. doi: 10.1016/j.bios.2014.10.039. [DOI] [PubMed] [Google Scholar]
- Newell DG, Koopmans M, Verhoef L, Duizer E, Aidara-Kane A, Sprong H. Internat. J. Food Microbiol. 2010;139(Supplement):S1–S15. doi: 10.1016/j.ijfoodmicro.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngundi MM, Qadri S, Wallace E, Moore MH, Lassman ME, Shriver-Lake LC, Ligler FS, Taitt CR. Environ. Sci. Tech. 2006a;40(7):2352–2356. doi: 10.1021/es052396q. [DOI] [PubMed] [Google Scholar]
- Ngundi MM, Shriver-Lake LC, Moore MH, Lassman ME, Ligler FS, Taitt CR. Anal. Chem. 2005;77(1):148–154. doi: 10.1021/ac048957y. [DOI] [PubMed] [Google Scholar]
- Ngundi MM, Shriver-Lake LC, Moore MH, Ligler FS, Taitt CR. J. Food Protect. 2006b;69:3047–3051. doi: 10.4315/0362-028x-69.12.3047. [DOI] [PubMed] [Google Scholar]
- Ngundi MM, Taitt CR, Ligler FS. Biosens. Bioelectron. 2006c;22(1):124–130. doi: 10.1016/j.bios.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Ngundi MM, Taitt CR, Ligler FS. Sensor Letts. 2007;5(3–4):621–624. [Google Scholar]
- Ngundi MM, Taitt CR, McMurry SA, Kahne D, Ligler FS. Biosens. Bioelectron. 2006d;21(7):1195–1201. doi: 10.1016/j.bios.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohk SH, Koo OK, Sen T, Yamamoto CM, Bhunia AK. J. Appl. Microbiol. 2010;109(3):808–817. doi: 10.1111/j.1365-2672.2010.04709.x. [DOI] [PubMed] [Google Scholar]
- Parks JW, Olson MA, Kim J, Ozcelik D, Cai H, Carrion R, Patterson JL, Mathies RA, Hawkins AR, Schmidt H. Biomicrofluidics. 2014;8(5):054111. doi: 10.1063/1.4897226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak M, Schick E, Bopp MA, Schneider MJ, Oroszlan P, Ehrat M. Proteomics. 2002;2(4):383–393. doi: 10.1002/1615-9861(200204)2:4<383::AID-PROT383>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Pirnia F, Pawlak M, Thallinger GG, Gierke B, Templin MF, Kappeler A, Betticher DC, Gloor B, Borner MM. Proteomics. 2009;9(13):3535–3548. doi: 10.1002/pmic.200800159. [DOI] [PubMed] [Google Scholar]
- Ramanathan M, Simonian A. Biosens. Bioelectron. 2007;22(12):3001–3007. doi: 10.1016/j.bios.2006.12.029. [DOI] [PubMed] [Google Scholar]
- Rascher D, Geerlof A, Kremmer E, Krämer P, Schmid M, Hartmann A, Rieger M. Biosens. Bioelectron. 2014;59:251–258. doi: 10.1016/j.bios.2014.03.052. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Mozaz S, López de Alda MJ, Barceló D. Handbook of Environmental Chemistry 5 J. 2009:33–46. [Google Scholar]
- Saaski EW. Proc. SPIE. 2009;7553:716704–716710. [Google Scholar]
- Sagmeister M, Lamprecht B, Kraker E, Köstler S, Ribitsch V, Galler N, Ditlbacher H, Krenn J, Ungerböck B, Abel T, Mayr T. Appl. Phys. B. 2013;111(4):647–650. [Google Scholar]
- Sakamuri RM, Capek P, Dickerson TJ, Barry CE, III, Mukundan H, Swanson BI. J. Microbiol. l Methods. 2014;103:112–117. doi: 10.1016/j.mimet.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapsford KE, Taitt CR, Fertig S, Moore MH, Lassman ME, Maragos C, Shriver-Lake LC. Biosens. Bioelectron. 2006;21(12):2298–2305. doi: 10.1016/j.bios.2005.10.021. [DOI] [PubMed] [Google Scholar]
- Sapsford KE, Taitt CR, Loo N, Ligler FS. Appl. Environ. Microbiol. 2005;71(9):5591–5592. doi: 10.1128/AEM.71.9.5590-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Menges B, Knoll W. J. Appl. Phys. 2009;105(1):014701. [Google Scholar]
- Saturno G, Pesenti M, Cavazzoli C, Rossi A, Giusti AM, Bierke B, Pawlak M, Venturi M. Toxicologic Pathology. 2007;35(7):972–983. doi: 10.1080/01926230701748271. [DOI] [PubMed] [Google Scholar]
- Schaudies RP. State of the art for autonomous detection systems using immunoassays and protein signatures. In: Hook-Barnard I, Posey Norris SM, Alper J, editors. Technologies to Enable Autonomous Detection for BioWatch: Ensuring Timely and Accurate Information for Public Health Officials : Workshop Summary. Washington, DC: National Academies Press; 2013. pp. 173–196. [PubMed] [Google Scholar]
- Schuller JA, Barnard ES, Cai WS, Jun YC, White JS, Brongersma ML. Nature Materials. 2010;9(3):193–204. doi: 10.1038/nmat2630. [DOI] [PubMed] [Google Scholar]
- Schumacher S, Lüdecke C, Ehrentreich-Förster E, Bier F. Platform technologies for molecular diagnostics near the patient’s bedside. In: Seitz H, Schumacher S, editors. Molecular Diagnostics. Berlin Heidelberg: Springer; 2013. pp. 75–87. [DOI] [PubMed] [Google Scholar]
- Schumacher S, Nestler J, Otto T, Wegener M, Ehrentreich-Forster E, Michel D, Wunderlich K, Palzer S, Sohn K, Weber A, Burgard M, Grzesiak A, Teichert A, Brandenburg A, Koger B, Albers J, Nebling E, Bier FF. Lab Chip. 2012;12(3):464–473. doi: 10.1039/c1lc20693a. [DOI] [PubMed] [Google Scholar]
- Shi H-C, Song B-D, Long F, Zhou X-H, He M, Lv Q, Yang H-Y. Environ. Sci. Tech. 2013;47(9):4434–4441. doi: 10.1021/es305196f. [DOI] [PubMed] [Google Scholar]
- Shriver-Lake LC, Donner BL, Ligler FS. Environ. Sci. Tech. 1997;31(3):837–841. [Google Scholar]
- Shriver-Lake LC, Erickson J, Sapsford KE, Ngundi MM, Shaffer KM, Kulagina NV, Hu J, Gray S, Golden J, Ligler FS, Taitt CR. Anal. Letts. 2007a;40:3219–3231. [Google Scholar]
- Shriver-Lake LC, Turner S, Taitt CR. Anal. Chim. Acta. 2007b;584:66–71. doi: 10.1016/j.aca.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Smolyaninova V, Smolyaninov I, Kildishev A, Shalaev V. Applied Physics B. 2012;106(3):577–581. [Google Scholar]
- Sobel J, Khan AS, Swerdlow DL. Lancet. 2002;359(9309):874–880. doi: 10.1016/S0140-6736(02)07947-3. [DOI] [PubMed] [Google Scholar]
- Taitt C, Anderson G, Ligler F. Biosens. Bioelectron. 2005;20(12):2470–2487. doi: 10.1016/j.bios.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Taitt CR, Shriver-Lake LC, Ngundi MM, Ligler FS. Sensors. 2008;8:8361–8377. doi: 10.3390/s8128361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Oh YS, Li H, Song J, Chen PS, Lin SF. Heart Rhythm. 2007;4(9):1208–1213. doi: 10.1016/j.hrthm.2007.05.031. [DOI] [PubMed] [Google Scholar]
- Tegnebratt T, Ruge E, Bader S, Ishii N, Aida S, Yoshimura Y, Ooi C-H, Lu L, Mitsios N, Meresse V, Mulder J, Pawlak M, Venturi M, Tessier J, Stone-Elander EJNMMI Research. 2014;4(34) doi: 10.1186/s13550-014-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RB, Villarruel CA. Waveguide-binding sensor for use with assays. 5061857S. US patent. 1991
- Ton X-A, Acha V, Bonomi P, Tse Sum Bui B, Haupt K. Biosens. Bioelectron. 2015;64:359–366. doi: 10.1016/j.bios.2014.09.017. [DOI] [PubMed] [Google Scholar]
- Trévisan M, Schawaller M, Quapil G, Souteyrand E, Mérieux Y, Cloarec J-P. Biosens. Bioelectron. 2010;26(4):1631–1637. doi: 10.1016/j.bios.2010.08.038. [DOI] [PubMed] [Google Scholar]
- Tschmelak J, Kappel N, Gauglitz G. Anal. Bioanal. Chem. 2005;382(8):1895–1903. doi: 10.1007/s00216-005-3261-x. [DOI] [PubMed] [Google Scholar]
- Tschmelak J, Kumpf M, Käppel N, Proll G, Gauglitz G. Talanta. 2006;69(2):343–350. doi: 10.1016/j.talanta.2005.09.048. [DOI] [PubMed] [Google Scholar]
- Tschmelak J, Proll G, Gauglitz G. Biosens. Bioelectron. 2004;20(4):743–752. doi: 10.1016/j.bios.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Updike SJ, Hicks GP. Nature. 1967;214:968–988. doi: 10.1038/214986a0. [DOI] [PubMed] [Google Scholar]
- Van Oostrum J, Calonder C, Rechsteiner D, Ehrat M, Mestan J, Fabbro D, Voshol H. Proteomics Clin. Appl. 2009;3:412–422. doi: 10.1002/prca.200800070. [DOI] [PubMed] [Google Scholar]
- Viveros L, Paliwal S, McCrae D, Wild J, Simonian A. Sens. Act. B: Chemical. 2006;115(1):150–157. [Google Scholar]
- Voshol H, Ehrat M, Traenkle J, Bertrand E, van Oostrum J. FEBS Journal. 2009;276(23):6871–6879. doi: 10.1111/j.1742-4658.2009.07395.x. [DOI] [PubMed] [Google Scholar]
- Wang CW, Manne U, Reddy VB, Oelschlager DK, Katkoori VR, Grizzle WE, Kapoor R. J. Biomed. Optics. 2010;16(1):019801. doi: 10.1117/1.3523368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xiang Y, Zhou X, Liu L-h, Shi H. Biosens. Bioelectron. 2015;66:11–18. doi: 10.1016/j.bios.2014.10.079. [DOI] [PubMed] [Google Scholar]
- Wei H, Guo Z, Zhu Z, Tan Y, Du Z, Yang R. Sens. Act. B: Chemical. 2007a;127(2):525–530. [Google Scholar]
- Wei H, Zhao Y, Bi Y, Liu H, Guo Z, Song Y, Zhai J, Huang H, Yang R. Direct detection of Yersinia pestis from the infected animal specimens by a fiber optic biosensor. Sens. Act. B: Chemical. 2007b;123(1):204–210. [Google Scholar]
- Weissenstein U, Schneider MJ, Pawlak M, Cicenas J, Eppenberger-Castori S, Oroszlan P, Ehret S, Geurts-Moespot A, Sweep FCGJ, Eppenberger U. Proteomics. 2006;6(5):1427–1436. doi: 10.1002/pmic.200500078. [DOI] [PubMed] [Google Scholar]
- World Health Organization. World Health Organization; 2008. Terrorist threats to food: Guidance for establishing and strengthening prevention and response systems. [Google Scholar]
- Xiao-Hong Z, Bao-Dong S, Han-Chang S, Lan-Hua L, Hong-li G, Miao H. Sens. Act. B. 2014;198:150–156. [Google Scholar]
- Xiao-hong Z, Lan-hua L, Wei-qi X, Bao-dong S, Jian-wu S, Miao H, Han-chang S. Scientific Reports. 2014;4 doi: 10.1038/srep04572. article 4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Persson B, Löfås S, Knoll W. J. Am. Chem. Soc. 2004;126(29):8902–8903. doi: 10.1021/ja048583q. [DOI] [PubMed] [Google Scholar]
- Yue J, Zhang L, Yang Z. Internat. J. Environ. Anal. Chem. 2009;89(8–12):821–833. [Google Scholar]
- Zhu P, Shelton DR, Li S, Adams DL, Karns JS, Amstutz P, Tang C-M. Biosens. Bioelectron. 2011;30(1):337–341. doi: 10.1016/j.bios.2011.09.029. [DOI] [PubMed] [Google Scholar]