Abstract
We propose an algorithm to distinguish 3D+t images of healthy from diseased subjects by solving logistic regression based on cardinality constrained, group sparsity. This method reduces the risk of overfitting by providing an elegant solution to identifying anatomical regions most impacted by disease. It also ensures that consistent identification across the time series by grouping each image feature across time and counting the number of non-zero groupings. While popular in medical imaging, group cardinality constrained problems are generally solved by relaxing counting with summing over the groupings. We instead solve the original problem by generalizing a penalty decomposition algorithm, which alternates between minimizing a logistic regression function with a regularizer based on the Frobenius norm and enforcing sparsity. Applied to 86 cine MRIs of healthy cases and subjects with Tetralogy of Fallot (TOF), our method correctly identifies regions impacted by TOF and obtains a statistically significant higher classification accuracy than logistic regression without and relaxed grouped sparsity constraint.
1 Introduction
Identifying image phenotypes by automatically classifying 3D+t MRIs is popular in medical image analysis, such as for cardiac motion [1, 2] and brain development [3, 4]. Automatic classification is difficult due to the complex information captured by MRIs coupled with small sample sizes of most MRI studies. Besides avoiding overfitting, the image phenotypes extracted by the classifiers need to be meaningful to clinicians. To address both issues, group sparsity constrained logistic regression models [5, 6] first reduce the dense image information to a small number of features by grouping image features via the l2-norm and counting the number of non-zero groupings (l0 “norm”). The identified groupings then often can be directly linked to specific anatomy or function. Motivated by [5, 7], these methods generally find a solution by relaxing the l0 “norm” to the sum over the l2-norm values. However, this solution relates to the original sparse problem only in specific conditions, e.g. compressed sensing [8], which generally do not hold for medical image applications. We now propose an algorithm for solving the logistic regression problem with the group cardinality constraint and show on a data set of cine MRIs that it produces statistically significant better results than the corresponding approach based on the relaxed norm.
Specifically, we model consistency constraints of disease patterns across the series of images via group sparsity, i.e. we group the weights associated with each image feature across time and then apply the l0 “norm” to those groupings. We find a solution within this model by generalizing Penalty Decomposition (PD) [9] from solving sparse logistic regression problem with group size one to more than one. In detail, we decouple the logistic regression function from enforcing the group sparsity constraint by introducing a penalty term based on the Forbenius norm. We minimize the smooth and convex logistic regression with the penalty term via gradient descent and derive a closed form solution for the modified group sparsity problem. We then find a local minimum of the original problem by iteratively solving the two subproblems via block coordinate descent.
We apply our method to cine MRI of 40 patients with reconstructive surgery of Tetralogy of Fallot (TOF) and 46 healthy volunteers, whose diagnosis we view as ground truth. As the residual effects of TOF mostly impact the shape of the RV [10], the regions impacted by TOF should not change across time, i.e. an ideal test bed for our logistic regression approach. We encode each cine MRI by first non-rigidly registering the image series to a 3D+t template, which divides the left and right ventricle into subregions. For each of those regions, we then record the average of the Jacobian determinant of the deformation map.
We measure the accuracy of our model via five-fold cross-validation. During training, we automatically set all important parameters of our approach by first creating a separate classifier for each setting of the parameter space. We then reduce the risk of overfitting by combining those classifiers into a single ensemble of classifiers [11], i.e. we compute a weighted average across the outputs of the individual classifiers. As we will show later, the ensemble of classifiers correctly favors subregions of the ventricles most likely impacted by TOF. Furthermore, the accuracy of the ensemble of classifiers is statistically significant higher than the outcomes obtained by relaxing and omitting group cardinality constraint, i.e. the classical logistic regression model that keeps all regions for disease detection.
2 Solving Sparse Group Logistic Regression
Our aim is to train a classifier to correctly assign subjects to cohorts based on their corresponding image data. To do so, we first describe the logistic regression model with group cardinality constraint. We then find a solution within that model by generalizing the PD approach.
Model
We assume that our training set consists of N subjects that are represented by their corresponding diagnosis {b1, … , bN} and the encoding of 3D+t medical images {Z1, … ,ZN } with T time points. The value of bs ∈ {−1, +1} depends on the subject ‘s’ being healthy (+1) or diseased (−1). The corresponding feature matrix of that subject is composed of vectors encoding the tth frame with M features.
Next, we train the classifier on that data by solving the logistic regression problem confined by group sparsity. To define the problem, we introduce the variable As := bs · Zs for s = 1, … ,N, the weight matrix defining the importance of each feature in correctly classifying 3D+t images, the trace of a matrix Tr(·), the logistic function θ(y) := log(1 + exp(−y)), and the average logistic loss function with respect to the label weight
| (1) |
Ignoring the time point associated with each feature, we now assume that the disease is characterized by features, i.e. ‘r’ rows of the feature matrix Zs subject to (s.t.) r ≤ M. Then the logistic regression problem with group sparsity constraint is defined as
| (2) |
where groups the weight vectors over time as is the ith row of matrix W. Note, equals the number of nonzero components of , i.e. the model accounts for the relationship of the same features across all the time points. Intuitively, if the model chooses a feature in one frame, the corresponding features in other frames should also be chosen since the importance of a feature should be similar across time. Replacing with (or in the case of T = 1) results in the more common sparse logistic regression problem, which, in contrast, chooses individual features of W ignoring any temporal dependency, which is not desired for our application.
Algorithm
We find a local minimum to Eq. (2) by decoupling the minimization of lavg(·, ·) from the sparsity constraint
Specifically, we approximate the sparse weights via the unconstrained variable so that Eq. (2) changes to
| (3) |
Denoting with a penalty parameter and the matrix Frobenius norm with , we solve Eq. (3) by generalizing PD proposed in [9] from enforcing the cardinality constraint for group size T = 1 to T ≥ 1, i.e. we obtain a local solution of the following nonconvex problem
| (4) |
Ascending at each iteration, we use Block Coordinate Descent (BCD) to determine the solution by alternating between minimizing Eq. (4) with fixed W and by fixing v and Y. When W is set to W′, finding the minimum with respect to v and Y turns into a smooth and convex problem
| (5) |
which can be solved via a gradient descent. In turn, minimizing the objective function just with respect to W, i.e.
| (6) |
can now be solved in closed form. We derive the closed form solution by assuming (without loss of generality) that the rows of Y′ are nonzero and listed in descending order according to their l2-norm, i.e. let (Y′)i be the ith row of Y′ for i = 1, … , M then . It can then be easily shown (see 1) that W′ is defined by the first ‘r’ rows of Y′, i.e.
| (7) |
In theory, the global solution derived above is not unique for Eq. (6), which we have not experienced in practice. One can also prove (similar to [9]) that the method converges with respect to to a local minimum of Eq. (2).
3 Characterizing TOF Based on Cine MRIs
We apply our approach to cine MRIs of 40 TOF cases and 46 healthy controls. We choose this dataset due to the ground-truth diagnosis, i.e. each subject received reconstructive surgery for TOF during infancy or not. Furthermore, refining the quantitative analysis of these scans could lead to improved timing for follow-up surgeries in TOF patients. Finally, the assumption of our group sparsity model, i.e. the features extracted from each time point of the image series are sample descriptions of the same phenomena, fits well to this dataset. As we describe next, the features of this experiment are deformation-based shape encodings of the heart. As the residual effects of TOF reconstructive surgery mostly impact the shape of the right ventricle [10], our encoding should capture differences between the two groups in the same heart regions across time. We not only show that this is the case but our approach defined by Eq. (2) achieves significantly higher accuracy than logistic regression with relaxed sparsity constraints, i.e. given the sparse regularizing parameter λ solving
| (8) |
and without sparsity constraints, i.e., λ = 0. Note we omit comparison to implementations replacing the l2-norm in Eq. (8) with the l1-norm since the resulting regularizor ignores consistency of the features across time. In other words, the regions identified by those approaches most likely have no relevance with respect to TOF, whose residual effects should be consistent across time.
Extract Image Feature
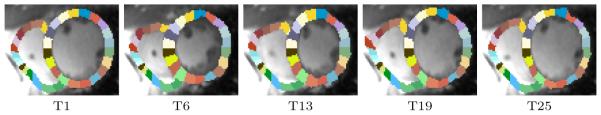
All 86 cine MR scans are defined by 30 time points. Each time point of a cine MRI is represented by a 3D image and a semiautomatically segmented label map of the right ventricular blood pool (RV) and the myocardium of the left ventricle (LV). Based on those segmentations, we omitted non cardiac structures from the scans and corrected each scan for image inhomogeneity and slice motion [12]. Next, we randomly choose the first time point of a healthy subject as a template and rigidly registered each case to the template. We encode the shape of the RV and LV by non-rigidly registering (via Demon’s approach [13]) each time point of the remaining 85 scans to the template. We then compute the Jacobian determinant of the deformation maps [14]. We confine the determinant maps to the LV segmentations and for the RV reduce the maps to a 7mm band along its boundary, which is similar to the width of the LV myocardium. We also parcellate the RV and LV into smaller regions by first computing its corresponding mass center and then sectoring it into region of interests by the subtended angles as shown in Fig. (1). We finalize our shape descriptor by mapping the refined Jacobians into the template space and computing their average value with respect to individual sections. The size of the sections are defined with respect to degrees.
Fig. 1.
Example of partitioning the template at different time points (T1 to T25) of the heart cycle. Each section of the RV is subtended by 9° and by 18° for the LV.
Measure Accuracy
We measure the accuracy of our approach via five-fold cross validation. We define the parameter space of the smoothness parameter p = {0, 0.5, … , 5.0} of the Demon’s algorithm, the subtended angle d = {45°, 36°, 30°, 24°, 20°, 18°, 15°, 12°, 10°, 8°, 6°, 5°, 4° } of each section of the LV and RV, and the sparsity constraint r ∈ {5, 10, … , 50}, i.e. the number of average regional values chosen by our approach. Note, that we choose such a broad parameter space to allow for a fair comparison with other logistic regression models. For each parameter setting, we specify a regressor by computing the optimal weights with respect to training data. We do so by initializing the penalty parameter ρ at 0.1 and increase ρ by a factor of at each PD iteration until convergence, i.e., . We also compute its training accuracy with respect to correctly assigning cine MRIs to each patient group. We account for the imbalance in cohort size by computing the normalized accuracy (nAcc), i.e. we separately compute the accuracy for each cohort and then average their values. On our dataset, parameter exploration on the training set resulted in multiple settings with 100% training accuracy. We therefore cross-validate a classifier based on training a single ensemble of classifiers [11]. We do so by treating the training nAcc score of a regressor as its weight in the decision of the ensemble of classifiers. In other words, the final label is then determined by the weighted average across the set of regressors, all of whom have different parameter settings. We also refer to this ensemble of classifier as l0-Grp. Fig. (2) shows the weight of each region according to the l0-Grp across time. We notice that the weights are fairly consistent across time due to the group sparsity term. In addition, the maps correctly point out (in red and yellow) the importance of the RV (left side) and more specifically the right ventricular outflow tract (the area where RV meets the LV) in identifying TOF.
Fig. 2.
Regions weighted by importance according to the proposed l0-Grp approach. The sparsity constraints correctly identified the RV (left side) and specifically the right ventricular outflow tract (the regions where RV and LV meet) as important markers for identifying the residual TOF effects.
Alternative Models
For comparison, we also generated an ensemble of logistic regression classifiers omitting the sparsity constraint (called No-Grp) and one by relaxing the group cardinality constraint (called Rlx-Grp) as defined by Eq. (8) [15]. These ensembles of classifiers used the same parameter space for the smoothness parameter p and the subtended angle d of each section. For Rlx-Grp, the sparsity parameter λ of Eq. (8) was automatically set so that the number of chosen group features were similar to the group cardinality, i.e. {5, 10, … , 50}. We applied each ensemble implementation to the dataset of just the LV, RV, and both ventricles.
Classification Results
The accuracy scores in % of all three ensembles are listed in Table 1 with respect to the encoding of the LV, RV, and both ventricles (LV&RV). Scores in bold represent results, which are significantly better (p-value < 0.05) than those of the other two methods. We compute the p-value based on the DeLongs Test [16] of the methods’ ROC curves.
Table 1.
Accuracy scores (nAcc) in % of No-Grp, Rlx-Grp, and our proposed model (l0-Grp).
| LV | RV | LV&RV | |
|---|---|---|---|
|
| |||
| No-Grp | 74.5 | 85.1 | 86.2 |
| Rlx-Grp | 74.8 | 84.7 | 83.8 |
| l0-Grp | 74.1 | 91.8 | 94.0 |
As expected, all methods achieve higher accuracy scores based on RV encoding vs. LV encoding. Including both ventricles in the analysis, leads to higher accuracy scores for No-Grp and the proposed l0-Grp. The bold accuracy scores of l0-Grp are significantly higher (p-value < 0.05) than the other two implementations, which was the case for the two experiments including the RV.
All three methods have similar accuracy scores for the LV (around 74.5%). As expected, the accuracy scores of all methods improve (by at least 9.9%) when the classification is based on the RV. The scores further improve for No-Grp and l0-Grp by including both ventricles in the analysis. The increase in the score of l0-Grp is explained by looking at the weight map of LV&RV in Fig. (3) (a), which, compared to the map of RV only in Fig. (3) (b), also highlights part of the LV. The impact of TOF on the LV was also recently echoed by [10].
Fig. 3.
Regions of the first time point weighted by importance according to different implementations: l0-Grp based on RV&LV (a), just based on the RV (b), based only on the first time point (c), and l0-Grp based on RV&LV (d). Note, implementations putting more emphasis on the RV (a+b) are those with the higher accuracy scores.
Not only does the proposed l0-Grp achieve the overall best score with 94% but the scores involving the RV are significantly better than those of the other two methods (p-values ≤ 0.036). While these results further validate the proposed group cardinality constraint model for logistic regression, the relatively poor accuracy scores by Rlx-Grp is somewhat surprising (84.7%). Comparing the regional weight maps of the two approaches (see Fig. (3) (a) + (d)), the map of Rlx-Grp mostly ignores the RV failing to identify regions impacted by TOF. It not only explains the relatively low score but further highlights the superiority of the group cardinality over the l2-norm relaxation with respect to this experiment.
To study the importance of the group-sparsity over just sparsity, we confined l0-Grp to the first time point, i.e. reducing the sparsity term to the cardinality constraint without group constraint. The accuracy of l0-Grp dropped for all three experiments by a minimum of 4.5%. The weight map in Fig. (3) (c) reveals the reason for the drop in accuracy as it puts less emphasis on the RV. Furthermore, its scores are now similar to those of No-Grp (less than 2.5% difference). We therefore conclude that the l0 group sparsity takes proper advantage of the repeat samples of the same disease patterns provided by the image time series.
4 Conclusion
We proposed an algorithm for solving logistic regression based on the group cardinality constraint. Unlike existing approaches, our algorithm did not relax the l0-norm regularizer but instead used penalty decomposition to solve the original problem. Applied to 86 cine MRIs of healthy cases and subjects with TOF, our method not only correctly identified regions impacted by TOF but also obtains statistically significant higher classification accuracy than logistic regression without and with relaxed group sparsity constraints.
Acknowledgement
We would like to thank DongHye Ye for his help on generating the cardiac 515 dataset. This project was supported in part by the NIH grant R01HL127661. It was also supported by the Creative and Novel Ideas in HIV Research Program through a supplement to NIH P30 AI027763. This funding was made possible by collaborative efforts of the Office of AIDS Research, the National Institutes of Allergies and Infectious Diseases, and the International AIDS Society.
Footnotes
References
- 1.Yu Y, Zhang S, Li K, Metaxas D, Axel L. Deformable models with sparsity constraints for cardiac motion analysis. Med. Image Anal. 2014;18(6):927–937. doi: 10.1016/j.media.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Assen HC, Danilouchkine MG, Frangi AF, Ordás S, Westenberg JJ, Reiber JH, Lelieveldt BP. SPASM: segmentation of sparse and arbitrarily oriented cardiac MRI data using a 3D-ASM. 2005:33–43. doi: 10.1016/j.media.2005.12.001. In: FIMH. LNCS 3504. [DOI] [PubMed] [Google Scholar]
- 3.Serag A, Gousias I, Makropoulos A, Aljabar P, Hajnal J, Boardman J, Counsell S, Rueckert D. Unsupervised learning of shape complexity: Application to brain development. 2012:88–99. In: STIA. LNCS 7570. [Google Scholar]
- 4.Bernal-Rusiel JL, Reuter M, Greve DN, Fischl B, Sabuncu MR. Spatiotemporal linear mixed effects modeling for the mass-univariate analysis of longitudinal neuroimage data. NeuroImage. 2013;81:358–370. doi: 10.1016/j.neuroimage.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meier L, Van De Geer S, Bühlmann P. The group lasso for logistic regression. J. Roy. Soc. Ser. B. 2008;70(1):53–71. [Google Scholar]
- 6.Wu F, Yuan Y, Zhuang Y. Heterogeneous feature selection by group lasso with logistic regression. 2010:983–986. In: ACM-MM. [Google Scholar]
- 7.Friedman J, Hastie T, Tibshirani R. A note on the group lasso and a sparse group lasso. 2010 arXiv preprint arXiv:1001.0736. [Google Scholar]
- 8.Cand`es EJ, Romberg J, Tao T. Robust uncertainty principles: Exact signal reconstruction from highly incomplete frequency information. IEEE Trans. Inf. Theory. 2006;52(2):489–509. [Google Scholar]
- 9.Lu Z, Zhang Y. Sparse approximation via penalty decomposition methods. SIAM J. Optim. 2013;23(4):2448–2478. [Google Scholar]
- 10.Atrey PK, Hossain MA, El Saddik A, Kankanhalli MS. Multimodal fusion for multimedia analysis: a survey. Multimedia Syst. 2010;16(6):345–379. [Google Scholar]
- 11.Rokach L. Ensemble-based classifiers. Artif. Intell. Rev. 2010;33(1-2):1–39. [Google Scholar]
- 12.Ye D, Desjardins B, Hamm J, Litt H, Pohl K. Regional manifold learning for disease classification. IEEE T. Med. Imaging. 2014;33(6):1236–1247. doi: 10.1109/TMI.2014.2305751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vercauteren T, Pennec X, Perchant A, Ayache N. Diffeomorphic demons: Efficient non-parametric image registration. NeuroImage. 2009;45(1):S61–S72. doi: 10.1016/j.neuroimage.2008.10.040. [DOI] [PubMed] [Google Scholar]
- 14.Shen D, Davatzikos C. Very high-resolution morphometry using mass-preserving deformations and hammer elastic registration. NeuroImage. 2003;18(1):28–41. doi: 10.1006/nimg.2002.1301. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Ji S, Ye J. SLEP: Sparse Learning with Efficient Projections. Arizona State University. 2009 [Google Scholar]
- 16.DeLong E, DeLong D, Clarke-Pearson D. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]