Abstract
Gene fusions have been described in approximately one-third of soft tissue tumors (STT); of the 142 different fusions that have been reported, more than half are recurrent in the same histologic subtype. These gene fusions constitute pivotal driver mutations, and detailed studies of their cellular effects have provided important knowledge about pathogenetic mechanisms in STT. Furthermore, most fusions are strongly associated with a particular histotype, serving as ideal molecular diagnostic markers. In recent years, it has also become apparent that some chimeric proteins, directly or indirectly, constitute excellent treatment targets, making the detection of gene fusions in STT ever more important. Indeed, pharmacological treatment of STT displaying fusions that activate protein kinases, such as ALK and ROS1, or growth factors, such as PDGFB, is already in clinical use. However, the vast majority (52/78) of recurrent gene fusions create structurally altered and/or deregulated transcription factors, and a small but growing subset develops through rearranged chromatin regulators. The present review provides an overview of the spectrum of currently recognized gene fusions in STT, and, on the basis of the protein class involved, the mechanisms by which they exert their oncogenic effect are discussed.
GENE FUSIONS ARE IMPORTANT DRIVER MUTATIONS IN SOFT TISSUE TUMORS
Gene fusions, i.e., the juxtapositioning of two genes leading to the translation of a deregulated and/or chimeric protein, have been described in all major types of neoplasia, including benign as well as malignant tumors of hematologic, epithelial, and mesenchymal origin (Mertens et al., 2015; Mitelman et al., 2015). While some of the fusions appear to be passenger mutations caused by the increased genetic instability that is a hallmark of many malignant neoplasms, others constitute strong driver alterations (Mertens et al., 2015; Yoshihara et al., 2015). Similar to other types of mutation, the classification of a gene fusion as driver or passenger mutation can be achieved through careful evaluation in experimental models, but strong indirect support for a significant pathogenetic role is also suggested when a certain gene fusion is recurrent, by revealing that it is associated with relatively few other mutations, or by establishing that it is restricted to one or a few morphologic subtypes.
The characterization of neoplasia-associated gene fusions has increased our general understanding of fundamental pathogenetic mechanisms as well as improved patient management through refined tumor classification and stratification, thus being highly rewarding for scientists and clinicians dealing with the clinically heterogeneous group of neoplasms known as soft tissue tumors (STT). STT currently encompass more than 100 separate morphologic entities (Fletcher et al., 2013), many of which are exceedingly rare and thus require review by expert soft tissue pathologists for a correct classification. In addition, an increasing number of STT patients is considered for neoadjuvant treatment, further increasing the need for a precise diagnosis (Linch et al., 2014; Reichardt, 2014). Still, even highly experienced pathologists with access to an extensive battery of immunohisto-chemical (IHC) markers sometimes face differential diagnostic challenges; in such cases, genetic analyses investigating the presence of particular gene fusions may prove pivotal (Antonescu and Dal Cin, 2014). In the early 1980s, when characteristic chromosomal aberrations were first detected in STT, it emerged that certain subtypes display one or more recurrent, often pathognomonic, structural chromosome aberrations, notably translocations. With time, these rearrangements could be characterized at the molecular level, revealing that they often result in a gene fusion (Mitelman et al., 2007). Once the molecular genetic correlates of the chromosomal aberrations were established, they became identifiable also by directed analyses such as fluorescence in situ hybridization (FISH) or RT-PCR. Moreover, formalin-fixed paraffin-embedded tissue could now be used for their detection, thus increasing the number of cases that could be tested and circumventing the need for fresh tumor material. In recent years, the identification of gene fusions has been further improved by the introduction of next-generation sequencing (NGS) tools, in the clinic as well as in the research setting (Mertens and Tayebwa, 2014; Mertens et al., 2015).
During the last two decades—starting with the EWSR1-FLI1 fusion in Ewing sarcoma in 1992 (Delattre et al., 1992)—142 different gene fusions have been identified in STT, revealing pathogenetic mechanisms that involve a variety of different genes and pathways. These fusions have become highly useful diagnostic tools, either as such or by providing a rationale for novel IHC markers (Hornick, 2014). Furthermore, the molecular characterization has revealed new pathways in tumorigenesis and disclosed unexpected molecular links among different subtypes of STT or with carcinomas, lymphomas, and leukemias. While there are recent reviews on general (Mertens et al., 2015), technological (Mertens and Tayebwa, 2014), or clinical (Antonescu and Dal Cin, 2014) aspects of gene fusions in STT and other neoplasms, a comprehensive review of their biological features is missing. Hence, the current review aims at presenting the distribution of gene fusions among different morphologic subtypes, examine the types of gene involved and how they interact with each other, and provide an overview of patho-genetic mechanisms in these STT.
DISTRIBUTION OF GENE FUSIONS IN STT
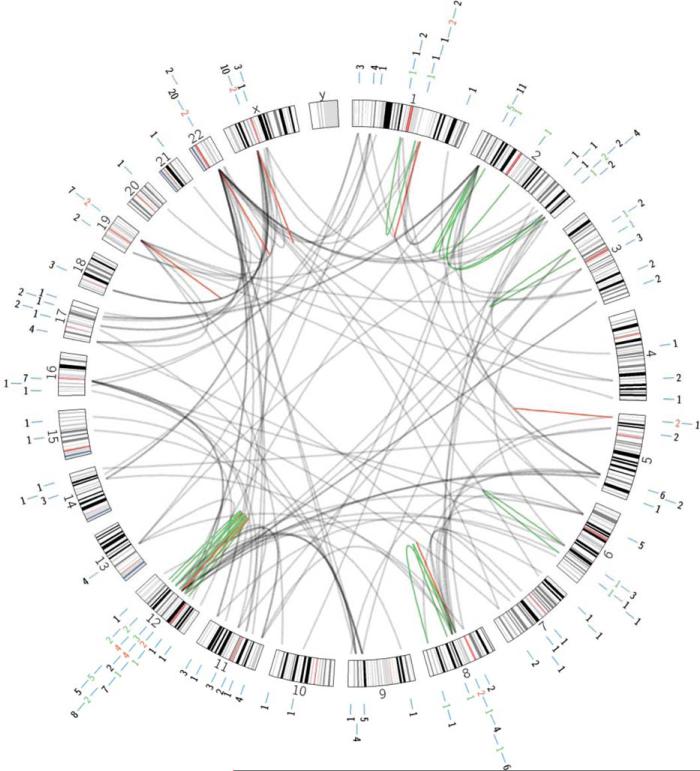
The Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer (Mitelman et al., 2015) was queried on September 24, 2015 for gene fusions reported in tumors that are included in the latest WHO classification of soft tissue and bone tumors and that originated in soft tissues (Fletcher et al., 2013). In addition, three recently delineated STT—pericytoma with t(7;12), angiofibroma of soft tissue, and biphenotypic sinonasal sarcoma—were included (Dahlén et al., 2004; Mariño-Enriquez and Fletcher, 2012; Wang et al., 2014). Using these criteria, gene fusions were found in 40 different tumor types, representing all major subgroups except smooth muscle tumors, gastrointestinal stromal tumors, and peripheral nerve sheath tumors (Table 1). All chromosomes except the Y chromosome are involved, but with a distinctly non-random distribution of affected gene loci (Fig. 1). For instance, six separate genes (ATF1, DDIT2, GLI1, HMGA2, NAB2, and STAT6) that map to chromosome bands 12q13-q15 are recurrently involved in different types of STT; in addition, there are numerous nonrecurrent gene fusions affecting other genes in chromosome arm 12q, typically resulting from the intrachromosomal rearrangements that are associated with the amplification events that are characteristic for certain subtypes of STT (Table 1). Otherwise, there is a relative lack of gene fusions occurring through intrachromosomal rearrangements in STT, probably reflecting the fact that these tumors remain poorly investigated by NGS techniques (Mertens and Tayebwa, 2014). Indeed, several recurrent gene fusions that were recently detected by NGS, such as the NAB2-STAT6 fusion in solitary fibrous tumor, strongly suggest that fusions of neighboring genes are pathogenetically significant also in STT (Chmielecki et al., 2013; Mohajeri et al., 2013; Robinson et al., 2013).
TABLE 1.
Gene Fusions in Soft Tissue Tumorsa
| Tumor | Recurrent fusionb | Single case |
|---|---|---|
| Adipocyte tumors | ||
| Lipoma | EBF1-RPSAP52 | HMGA2-EBF1 |
| HMGA2-ACKR3 | HMGA2-LHFP | |
| HMGA2-LPP | LPP-C12orf9 | |
| HMGA2-NFIB | ||
| HMGA2-PPAP2B | ||
| Lipoblastoma | COL1A2-PLAG1 | RAB2A-PLAG1 |
| COL3A1-PLAG1 | RAD51B-PLAG1 | |
| HAS2-PLAG1 | ||
| Chondroid lipoma | C11orf95-MKL2 | |
| Myxoid/round cell liposarcoma | FUS-DDIT3 | |
| EWSR1-DDIT3 | ||
| Atypical lipomatous tumor | METTL25-AMDHD1c | |
| METTL25-FGD6c | ||
| LRTOMT-METTL25c | ||
| NELL1-NAV3c | ||
| NELL1-SYT1c | ||
| PAWR-AMDHD1c | ||
| SYT1-METTL25c | ||
| SYT1-FGD6c | ||
| TBC1D15-METTL25c | ||
| UHMK1-DDR2c | ||
| ZDHHC14-PAWRc | ||
| Dedifferentiated liposarcoma | TRIO-TERT | CNOT2-ASTN2 |
| CTDSP2-FAM19A2 | ||
| NR6A1-TRHDE | ||
| NUP107-LGR5 | ||
| NUP107-PAPPA | ||
| RCOR1-WDR70 | ||
| Fibroblastic/Myofibroblastic tumors | ||
| Nodular fasciitis | MYH9-USP6 | |
| Soft tissue angiofibroma | AHRR-NCOA2 | GTF2I-NCOA2 |
| Dermatofibrosarcoma protuberansd | COL1A1-PDGFB | |
| Solitary fibrous tumor | NAB2-STAT6 | |
| Infantile fibrosarcoma | ETV6-NTRK3 | |
| Low-grade fibromyxoid sarcoma | EWSR1-CREB3L1 | |
| FUS-CREB3L1 | ||
| FUS-CREB3L2 | ||
| Sclerosing epithelioid fibrosarcoma | EWSR1-CREB3L1 | |
| EWSR1-CREB3L2 | ||
| FUS-CREB3L2 | ||
| Inflammatory myofibroblastic tumor | CARS-ALK | ATIC-ALK |
| CLTC-ALK | LMNA-ALK | |
| EML4-ALK | NAB2-PDGFRB | |
| FN1-ALK | PRKAR1A-ALK | |
| PPFIBP1-ALK | TFG-ALK | |
| RANBP2-ALK | YWHAE-ROS1 | |
| SEC31A-ALK | ||
| TFG-ROS1 | ||
| TPM3-ALK | ||
| TPM4-ALK | ||
| So-called fibrohistiocytic tumors | ||
| Tenosynovial giant cell tumor | COL6A3-CSF1 | CSF1-S100A10 |
| Benign fibrous histiocytoma | PDPN-PRKCB | KIRREL-PRKCA |
| LAMTOR1-PRKCD | ||
| RDH5-PRKCD | ||
| SQSTM1-ALK | ||
| VCL-ALK | ||
| Pericytic (perivascular) tumors | ||
| Glomus tumor | MIR143-NOTCH2 | MIR143-NOTCH1 |
| MIR143-NOTCH3 | ||
| NOTCH2-CEP128 | ||
| Pericytoma with t(7;l2) | ACTB-GLI1 | |
| Skeletal muscle tumors | ||
| Alveolar rhabdomyosarcoma | PAX3-FOXO1 | FOXO1-FGFR1 |
| PAX3-NCOA1 | PAX3-FOXO4 | |
| PAX3-NCOA2 | ||
| PAX7-FOXO1 | ||
| Congenital spindle cell rhabdomyosarcoma | TEAD1-NCOA2 | SRF-NCOA2 |
| VGLL2-CITED2 | ||
| VGLL2-NCOA2 | ||
| Vascular tumors | ||
| Epithelioid hemangioma | ZFP36-FOSB | WWTR1-FOSB |
| Pseudomyogenic hemangioendothelioma | SERPINE1-FOSB | |
| Epithelioid hemangioendothelioma | WWTR1-CAMTA1 | |
| YAP1-TFE3 | ||
| Tumors of uncertain differentiation | ||
| Angiomatoid fibrous histiocytoma | EWSR1-CREB1 | |
| FUS-ATF1 | ||
| EWSR1-ATF1 | ||
| Biphenotypic sinonasal sarcoma | PAX3-MAML3 | PAX3-FOXO1 |
| PAX3-NCOA1 | ||
| Ossifying fibromyxoid tumor | EP400-PHF1 | ZC3H7B-BCOR |
| MEAF6-PHF1 | ||
| Myoepithelioma/mixed tumor | EWSR1-PBX1 | EWSR1-ATF1 |
| EWSR1-POU5F1 | EWSR1-KLF17 | |
| EWSR1-ZNF444 | EWSR1-PBX3 | |
| FUS-KLF17 | LIFR-PLAG1 | |
| Phosphaturic mesenchymal tumor | FN1-FGFR1 | |
| Synovial sarcoma | SS18-SSX1 | SS18L1-SSX1 |
| SS18-SSX2 | ||
| SS18-SSX4 | ||
| Alveolar soft part sarcoma | ASPSCR1-TFE3 | |
| Clear cell sarcoma | EWSR1-CREB1 | |
| EWSR1-ATF1 | ||
| Extraskeletal myxoid chondrosarcoma | EWSR1-NR4A3 | FUS-NR4A3 |
| TAF15-NR4A3 | TCF12-NR4A3 | |
| TFG-NR4A3 | ||
| Desmoplastic small round cell tumor | EWSR1-WT1 | EWSR1-ERG |
| PEComa | RAD51B-OPHN1 | DVL2-TFE3 |
| RAD51B-RRAGB | HTR4-ST3GAL1 | |
| SFPQ-TFE3 | RASSF1-PDZRN3 | |
| Ewing sarcoma and undifferentiated/unclassified sarcomas | ||
| Ewing sarcoma | EWSR1-ERG | EWSR1-PATZ1 |
| EWSR1-ETV1 | EWSR1-SMARCA5 | |
| EWSR1-ETV4 | ||
| EWSR1-FEV | ||
| EWSR1-FLI1 | ||
| Undifferentiated/unclassified sarcomas | BCOR-CCNB3 | CITED2-PRDM10 |
| CIC-DUX4 | EWSR1-POU5F1 | |
| CIC-DUX4L10 | EWSR1-SP3 | |
| CIC-FOXO4 | KMT2B-GPS2 | |
| MED12-PRDM10 | ||
| Spindle cell sarcoma | HMGA2-DYRK2c | |
| PTGES3-PTPRBc | ||
| TMBIM4-MSRB3c | ||
| USP15-CNTN1c | ||
| Chondro-osseous tumors | ||
| Soft tissue chondroma | HMGA2-LPP | |
| Mesenchymal chondrosarcoma | HEY1-NCOA2 | IRF2BP2-CDX1 |
Gene fusions were retrieved from Mitelman F, Johansson B, Mertens F (Eds.). Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer, http://cgap.nci.nih.gov/Chromosomes/Mitelman. Queried on September 24, 2015. Gene fusions occurring in the same morphologic entity but originating in bone are not included.
Recurrent gene fusions have been reported in at least two cases of the same morphology. Gene fusions in bold are present in at least 10% of the tumors.
These fusion genes were all detected in the same case of atypical lipomatous tumor and spindle cell sarcoma, respectively, both displaying extensive gene amplification.
Includes giant cell fibroblastoma.
Figure 1.
Circos plot showing the chromosomal distribution of genes involved in recurrent and non-recurrent gene fusiosn in soft tissue tumors. Black lines 5 fusions involving genes on different chromosomes; green lines 5 fusions arising through intra-chromosomal rearrangements; red lines 5 fusion of genes located in the same chromosome band. The numbers outside the circos plot refer to the number of different gene fusions per chromosome band. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
A total of 142 different gene fusions were found, 78 of which have been reported in at least two cases of the same morphological STT subtype and are here defined as recurrent; these gene fusions and references to the studies in which they were first described are given in Table 2. Recurrent gene fusions seem to be particularly prevalent among tumors of uncertain differentiation or histo-genesis, involving close to half of the tumor types (Table 1). However, it should be emphasized that many of the gene fusions were only very recently described, and their association(s) with different morphologic subtypes remains poorly explored. Furthermore, several of the gene fusions so far described only in single cases will undoubtedly become recurrent in the near future. Attempts were also made to estimate, on the basis of all available literature data, the frequency of each recurrent gene fusion (Table 2). Gene fusions estimated to occur in at least 10% of a specific STT are indicated in bold in Table 1.
TABLE 2.
Recurrent Gene Fusions in Soft Tissue Tumorsa
| Gene fusion | Tumor typeb | Other tumorsc | First described in STT |
|---|---|---|---|
| ACTB-GLI1 | Pericytoma with t(7;12) (100%) | Bone | Dahleen et al., 2004 |
| AHRR-NCOA2 | Soft tissue angiofibroma (>50%) | Jin et al., 2012 | |
| ASPSCR1-TFE3 | Alveolar soft part sarcoma (100%) | KC | Ladanyi et al., 2001 |
| BCOR-CCNB3 | URCS (>20%) | Bone | Pierron et al., 2012 |
| C11ORF95-MKL2 | Chondroid lipoma (>90%) | Huang et al., 2010 | |
| CARS-ALK | Inflammatory myofibroblastic tumor | Cools et al., 2002 | |
| CIC-DUX4 | URCS (>25%) | Kawamura-Saito et al., 2006 | |
| CIC-DUX4L10 | URCS (>25%) | Italiano et al., 2012 | |
| CIC-FOXO4 | URCS | Bone | Brohl et al., 2014; Sugita et al., 2014 |
| CLTC-ALK | Inflammatory myofibroblastic tumor (15%) | ML | Bridge et al., 2001 |
| COL1A1-PDGFB | Dermatofibrosarcoma protuberansb (>95%) | Simon et al., 1997 | |
| COL1A2-PLAG1 | Lipoblastoma | Hibbard et al., 2000 | |
| COL3A1-PLAG1 | Lipoblastoma (>10%) | Yoshida et al., 2014 | |
| COL6A3-CSF1 | Tenosynovial giant cell tumor (30%) | West et al., 2006 | |
| EBF1-RPSAP52 | Lipoma | Nilsson et al., 2006 | |
| EML4-ALK | Inflammatory myofibroblastic tumor (10%) | AL, BC,CC, LC, ML, TC | Lovly et al., 2014 |
| EP400-PHF1 | Ossifying fibromyxoid tumor (45%) | Gebre-Medhin et al., 2012 | |
| ETV6-NTRK3 | Infantile fibrosarcoma (>90%) | AC, AL, BC, ES, ML, MN, SGC | Knezevich et al., 1998; Rubin et al., 1998 |
| EWSR1-ATF1 | Clear cell sarcoma (>90%) | SGC, URCS, | Zucman et al., 1993a |
| Angiomatoid fibrous histiocytoma | Bone, MET | Hansén Hallor et al., 2005 | |
| EWSR1-CREB1 | Clear cell sarcoma (>50%) | PMS | Antonescu et al., 2006 |
| Angiomatoid fibrous histiocytoma (>80%) | Antonescu et al., 2007; Rossi et al., 2007 | ||
| EWSR1-CREB3L1 | Low-grade fibromyxoid sarcoma | URCSB | Lau et al., 2013 |
| Sclerosing epithelioid fibrosarcoma (>60%) | Arbajian et al., 2014; Stockman et al., 2014 | ||
| EWSR1-CREB3L2 | Sclerosing epithelioid fibrosarcoma (>10%) | Prieto-Granada et al., 2015; Puls et al., in press | |
| EWSR1-DD1T3 | Myxoid liposarcoma | Panagopoulos et al., 1996 | |
| EWSR1-ERG | Ewing sarcoma | URCSB, DSCRT | Zucman et al., 1993b |
| EWSR1-ETV1 | Ewing sarcoma | Bone | Jeon et al., 1995 |
| EWSR1-ETV4 | Ewing sarcoma | Kaneko et al., 1996; Urano et al., 1996 | |
| EWSR1-FEV | Ewing sarcoma | Bone | Peter et al., 1997 |
| EWSR1-FLI1 | Ewing sarcoma (90%) | Bone | Delattre et al., 1992 |
| EWSR1-NR4A3 | ESMCS (70%) | Labelle et al., 1995 | |
| EWSR1-PBX1 | Myoepithelial tumor | Bone | Brandal et al., 2008 |
| EWSR1-POU5F1 | Myoepithelial tumor | HS, SGC, URCS, URCSB | Antonescu et al., 2010 |
| EWSR1-WT1 | DSCRT (>95%) | Ladanyi and Gerald, 1994 | |
| EWSR1-ZNF444 | Myoepithelial tumor | Brandal et al., 2009 | |
| FN1-ALK | Inflammatory myofibroblastic tumor | Ouchi et al., 2015 | |
| FN1-FGFR1 | PMT (60%) | Lee et al., 2015 | |
| FUS-ATF1 | Angiomatoid fibrous histiocytoma | Waters et al., 2000 | |
| FUS-CREB3L1 | Low-grade fibromyxoid sarcoma | Mertens et al., 2005 | |
| FUS-CREB3L2 | Low-grade fibromyxoid sarcoma (90%) | Storlazzi et al., 2003 | |
| Sclerosing epithelioid fibrosarcoma | Guillou et al., 2007 | ||
| FUS-DDIT3 | Myxoid liposarcoma (95%) | ||
| Crozat et al., 1993; Rabbitts et al., 1993 | |||
| FUS-KLF17 | Myoepithelial tumor | Bone | Huang et al., 2015 |
| HAS2-PLAG1 | Lipoblastoma (>10%) | Hibbard et al., 2000 | |
| HEY1-NCOA2 | Mesenchymal chondrosarcoma (65%) | Nyquist et al., 2012; Wang et al., 2012 | |
| HMGA2-ACKR3 | Lipoma | Broberg et al., 2002 | |
| HMGA2-LPP | Lipoma (15%) | CHL | Schoenmakers et al., 1995 |
| HMGA2-NFIB | Lipoma | SGA | Nilsson et al., 2005 |
| HMGA2-PPAP2B | Lipoma | Bianchini et al., 2013 | |
| MEAF6-PHF1 | Ossifying fibromyxoid tumor | ESS | Antonescu et al., 2014b |
| MED12-PRDM10 | UPS | Hofvander et al., 2015 | |
| MIR143-NOTCH2 | Glomus tumor (35%) | Mosquera et al., 2013 | |
| MYH9-USP6 | Nodular fasciitis (75%) | Erickson-Johnson et al., 2011 | |
| NAB2-STAT6 | Solitary fibrous tumor (>95%) | Chmielecki et al., 2013; Mohajeri et al., 2013; Robinson et al., 2013 | |
| PAX3-FOXO1 | Alveolar RMS (65%) | Galili et al., 1993; Shapiro et al., 1993 | |
| PAX3-MAML3 | Biphenotypic sinonasal sarcoma (80%) | Wang et al., 2014 | |
| PAX3-NCOA1 | Alveolar RMS | Wachtel et al., 2004 | |
| Biphenotypic sinonasal sarcoma | Huang et al., in press | ||
| PAX3-NCOA2 | Alveolar RMS | Sumegi et al., 2010 | |
| PAX7-FOXO1 | Alveolar RMS (20%) | Davis et al., 1994 | |
| PDPN-PRKCB | Benign fibrous histiocytoma | Plaszczyca et al., 2014 | |
| PPFIBP1-ALK | Inflammatory myofibroblastic tumor | Takeuchi et al., 2011 | |
| RAD51B-OPHNI | PEComa | Agaram et al., 2015 | |
| RAD51B-RRAGB | PEComa | Agaram et al., 2015 | |
| RANBP2-ALK | Inflammatory myofibroblastic tumor | AL, MDS, ML | Ma et al., 2003 |
| SEC31A-ALK | Inflammatory myofibroblastic tumor | ML | Panagopoulos et al., 2006 |
| SERPINE1-FOSB | PMy hemangioendothelioma (100%) | Bone | Walther et al., 2014 |
| SFPQ-TFE3 | PEComa | KC | Tanaka et al., 2009 |
| SS18-SSX1 | Synovial sarcoma (60%) | Clark et al., 1994; Crew et al., 1995; de Leeuw et al., 1995 | |
| SS18-SSX2 | Synovial sarcoma (35%) | Clark et al., 1994; Crew et al., 1995; de Leeuw et al., 1995 | |
| SS18-SSX4 | Synovial sarcoma | Skytting et al., 1999 | |
| TAF15-NR4A3 | ESMCS (25%) | Attwool et al., 1999; Panagopoulos et al., 1999; Sjögren et al., 1999 | |
| TEAD1-NCOA2 | Congenital spindle cell RMS (20%) | Mosquera et al., 2013 | |
| TFG-ROS1 | Inflammatory myofibroblastic tumor | Lovly et al., 2014 | |
| TPM3-ALK | Inflammatory myofibroblastic tumor (15%) | ML | Lawrence et al., 2000 |
| TPM4-ALK | Inflammatory myofibroblastic tumor | ML | Lawrence et al., 2000 |
| TRIO-TERT | Dedifferentiated liposarcoma | Stransky et al., 2014 | |
| VGLL2-CITED2 | Congenital spindle cell RMS (30%) | Alaggio et al., in press | |
| VGLL2-NCOA2 | Congenital spindle cell RMS (20%) | Alaggio et al., in press | |
| WWTR1-CAMTA1 | Epithelioid hemangioendothelioma (90%) | Errani et al., 2011; Tanas et al., 2011 | |
| YAP1-TFE3 | Epithelioid hemangioendothelioma | Antonescu et al., 2013 | |
| ZFP36-FOSB | Epithelioid hemangioma (15%) | Bone | Antonescu et al., 2014a |
Only gene fusions that have been detected in multiple cases of a tumor type included in the WHO classification of soft tissue and bone tumors (Fletcher et al., 2013) are included; also angiofibroma of soft tissue, pericytoma with t(7;12), and biphenotypic sinonasal sarcoma were included. When a gene fusion is present in at least 10% of a specific tumor type, the approximate frequency (as suggested by cytogenetic, molecular, and/or FISH studies) is given in parentheses. Fusions between genes transcribed in the same direction, i.e., both are transcribed from the centromere towards the telomere or vice versa, are in green; fusions of genes transcribed in opposite directions are in red.
DSCRT, desmoplastic small round cell tumor; ESMCS, extraskeletal myxoid chondrosarcoma; PEComa, perivascular epithelioid-cell tumor; PMT, phosphaturic mesenchymal tumor; PMy, pseudomyogenic; UPS, undifferentiated pleomorphic sarcoma; URCS, undifferentiated round cell sarcoma.
Tumor(s) with same morphology and same gene fusion also described in: AC, astrocytoma; AL, acute leukemia; BC, breast carcinoma; Bone, primary skeletal lesion; CC, colon carcinoma; CHL, chondroid hamartoma of the lung; DSCRT, desmoplastic small round cell tumor; ES, Ewing sarcoma; ESS, endometrial stromal cell sarcoma of the uterus; HS, hidradenoma of the skin; KC, kidney carcinoma; LC, lung carcinoma; MDS, myelodysplastic syndrome; MET, myoepithelial tumor; ML, malignant lymphoma; MN, mesoblastic nephroma; PMS, pulmonary myxoid sarcoma; RMS, rhabdomyosarcoma; SGA, salivary gland adenoma; SGC, salivary gland carcinoma; TC, thyroid carcinoma; URCS, undifferentiated round cell sarcoma; URCSB, URCS of bone.
Why have gene fusions been reported in only one-third of all STT subtypes? In general, the absence or presence of gene fusions is not related to basic clinicopathologic features such as grade of malignancy or degree of differentiation: gene fusions may be found in completely benign, highly differentiated lesions, such as conventional lipoma, as well as in undifferentiated, extremely aggressive tumors, like Ewing sarcoma and undifferentiated small round cell sarcomas (Table 1). Instead, one obvious explanation for the lack of gene fusions in some STT entities may be sought in the trivial fact that they have not been investigated in sufficient detail. For some tumors, like inclusion body fibromatosis or benign Triton tumor, there are simply no published genetic data at all, while for other more common, highly malignant lesions, like angiosarcoma or leiomyosarcoma, the search for potential gene fusions might have been thwarted by the typically very complex genomic rearrangements detected at chromosome banding or array-based studies (Guillou and Aurias, 2010). Although it was for a long time assumed that gene fusions were restricted to neoplasms with balanced chromosomal rearrangements, NGS-based studies of numerous malignancies with complex genomic changes have unequivocally shown that pathogenetically important gene fusions may be found also in tumor types that generally have heavily rearranged genomes (Mertens et al., 2015; Yoshihara et al., 2015). Thus, it would be highly surprising if they were not to be detected also in, e.g., angiosarcoma or leiomyosarcoma once these tumor types are subjected to large-scale transcriptome or whole-genome sequencing; support for this notion may be derived from the recent description of gene fusions in a small subset of undifferentiated pleomorphic sarcoma (Hofvander et al., 2015). Whether such gene fusions will turn out to be rare, like in lung cancer (McCoach and Doebele, 2014), or frequent, like in prostate cancer (Rubin et al., 2011), remains to be elucidated. A third explanation for the lack of gene fusions in some entities is that they may develop through other types of mutation. Indeed, for some STT types for which comprehensive data have emerged it seems as if gene fusions are rare or absent. For instance, in-depth genomic analysis of embryonal rhabdomyosarcomas has disclosed that they, in contrast to alveolar rhabdomyosarcomas, develop through characteristic combinations of point mutations and chromosomal and allelic imbalances (Shern et al., 2014), while some tumors, like desmoid-type fibromatosis or gastrointestinal stromal tumor, display near-universal mutations of a limited set of genes (Miettinen and Lasota, 2013; Crago et al., 2015).
In this context, it should be emphasized that not all gene fusions or fusion transcripts are pathogenetically relevant (Mertens et al., 2015). This could be illustrated by the three tumor types—atypical lipomatous tumor, dedifferentiated liposarcoma, and unclassified spindle cell sarcoma—in which mainly nonrecurrent gene fusions have been reported (Table 1). In all three tumor types, the gene fusions that have been reported derive from NGS studies of one or a few cases. The findings in these tumors are in line with the observation that gene fusions often are formed as a by-product of the extensive reshuffling of genetic material that is associated with gene amplification (Yoshihara et al., 2015).
ONE VERSUS MULTIPLE GENE FUSIONS IN THE SAME STT
Some of the STT subtypes with known gene fusions currently have only one highly prevalent variant, like COL1A1-PDGFB in dermatofibrosarcoma protuberans and giant cell fibroblastoma, ETV6-NTRK3 in infantile fibrosarcoma, NAB2-STAT6 in solitary fibrous tumor, or EWSR1-WT1 in desmoplastic small round cell tumor, indicating that these particular gene combinations are crucial for tumorigenesis (Tables 1 and 2). The extreme example here would be pericytoma with t(7;12), which by definition will always have an ACTB-GLI1 fusion (Dahlén et al., 2004). At present, however, this remains the only STT defined by its genetic features, in contrast to the current view on hematopoietic neoplasms, where genetic markers are mandatory for several diagnoses (Swerdlow et al., 2008). The lesson learnt from leukemias, which have been analyzed in far greater detail than STT, is that eventually all morphological sub-types will display some heterogeneity, either in the form of alternative gene fusions or by other types of mutation affecting the same critical pathway (Mitelman et al., 2007). In line with this, recent studies combining IHC for STAT6 expression and genetic tools for the detection of the NAB2-STAT6 fusion strongly indicate that there must, in rare cases, exist alternate ways of activating STAT6 expression in solitary fibrous tumor (Vogels et al., 2014; Tai et al., 2015).
Many STT are already known to display several related, recurrent as well as nonrecurrent, gene fusions, a variation that often seems to have little or no discernible impact on the morphology or biology of the tumor. For instance, there are no proved clinico-pathologic differences among lipomas with different 3′-partners to the HMGA2 gene or between myxoid liposarcomas carrying the FUS-DDIT3 or the EWSR1-DDIT3 fusion (Dal Cin et al., 1997; Bartuma et al., 2007). In other STT, however, such variation does seem to be of significance. The best example is probably alveolar rhabdomyosarcoma, in which the PAX3-FOXO1 and PAX7-FOXO1 fusions account for some 65 and 20%, respectively, of all cases (Shern et al., 2014). While children with PAX-fusion positive aleolar rhabdomyosarcoma have significantly worse prognosis than those with fusion-negative tumors, patients with PAX3-FOXO1 positive tumors have a particularly poor outcome (Missiaglia et al., 2012); in addition, the patterns of secondary changes differ between PAX3-FOXO1 and PAX7-FOXO1 positive cases (Shern et al., 2014). Additional examples include synovial sarcomas with SS18-SSX1 vs. SS18-SSX2, where the former fusion is relatively more common in biphasic tumors (Ladanyi et al., 2002), benign fibrous histiocytoma, where ALK fusions seem to be restricted to the epithelioid sub-type whereas PRKC fusions have been detected in several subtypes (Doyle et al., 2015; Walther et al., 2015), and Ewing sarcoma/undifferentiated round cell sarcomas, where the fusion partners to EWSR1 (FLI1 and ERG vs less common Ets family genes) seem to be associated with patient age and tumor location (Wang et al., 2007). However, the data are still scarce, and it will require large collaborative efforts to clarify the clinicopathologic relevance of the gene fusion heterogeneity in STT.
In STT with multiple recurrent gene fusions, one is typically (much) more common than the other(s). It has been suggested that this bias is due to the transcriptional direction of the respective genes, i.e., gene fusions that could arise through only two DNA double strand breaks (DSB; trans-location, inversion, interstitial deletion) would be more common than those (insertion, complex rearrangements) that require more breaks. Indeed, of the recurrent gene fusions, 51 involved two genes that are transcribed in the same direction, i.e., both from the telomere towards the centromere or vice versa, while 27 involved genes that are transcribed in opposite directions (Table 2). This could then explain why, e.g., FUS-DDIT3 is so much more common than EWSR1-DDIT3 in myxoid liposarcoma and an SS18-SSX3 has never been found in synovial sarcoma. However, most likely, this explanation is too simplistic because some STT have more recurrent fusions arising through complex rearrangements than from rearrangements requiring only two DNA DSB. For instance, of the 10 recurrent gene fusions in inflammatory myofibroblastic tumors, seven involve genes transcribed in opposite directions, as is the case also for all known recurrent gene fusions in SFT, sclerosing epithelioid fibrosarcoma, and ossifying fibromyxoid tumor. In some instances, as for SFT, this could be explained by the close location of the two genes, but in inflammatory myofibroblastic tumors, sclerosing epithelioid fibrosarcoma, and ossifying fibromyxoid tumor there is no obvious explanation for the abundance of gene fusions arising through complex genomic rearrangements. It could here be mentioned that the fusions arising through more complex rearrangements often are more difficult to detect by FISH, because the fusion event is typically seen on only one of the two derivate chromosomes.
OVERLAP AMONG STT AND WITH OTHER TUMORS
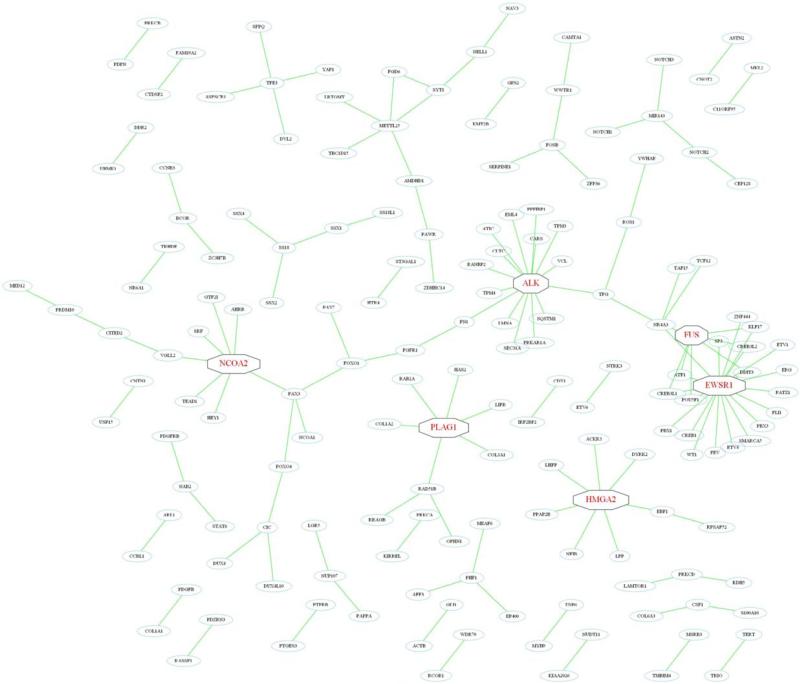
Regardless of its causes and consequences, the main drawback in the routine molecular work-up is related to the increasing number of gene fusions that have to be assessed in order to support or refute a diagnosis. To some extent this problem is reduced by investigating only one of the two gene partners at a time, instead of the chimeric fusion product. Thus, interphase FISH with a break-apart probe for EWSR1, DDIT3 or PLAG1 readily identifies all recurrent gene fusions in Ewing sarcoma, myxoid liposarcoma, or lipoblastoma, respectively. However, as some genes are more promiscuous, being recurrently involved in many different STT, this approach opens up for other diagnostic problems (Table 1). The most prominent example here is EWSR1, which is involved in gene fusions in 10 different STT, followed by FUS in six entities; several other genes also serve as hubs in extensive networks of gene fusions (Fig. 2). While this overlap among STT is of little clinical concern in some cases, e.g., an angiomatoid fibrous histiocytoma is not morphologically mistaken for a myxoid liposarcoma; it becomes a problem when the differential diagnoses are more similar, like desmoplastic small round cell tumor and Ewing sarcoma. This problem should be reduced once NGS-based approaches replace the currently used technologies, allowing for detailed and comprehensive detection of gene fusions (Mertens and Tayebwa, 2014).
Figure 2.
Network of gene fusions in soft tissue tumors. Some genes—ALK, EWSR1, FUS, HMGA2, NCOA2, and PLAG1 (indicated in red)—are promiscuous in the sense that they recombine with more than five different partners, leading to the formation of interconnected networks. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
From a biological point of view, an even more intriguing situation is when the identical gene fusion is shared by many STT, like EWSR1-CREB1 in angiomatoid fibrous histiocytoma and clear cell sarcoma [as well as pulmonary myxoid sarcoma (Thway et al., 2011)] or FUS-CREB3L2 in low-grade fibromyxoid sarcoma and sclerosing epithelioid fibrosarcoma (Table 2). Because there is good reason to believe that these gene fusions are strong driver mutations, the morphologic and clinical differences between the tumor types must be explained either by different sets of additional mutations and/or different cellular origins. While the effect of the mutational background of gene fusion-positive STT remains poorly investigated and thus difficult to evaluate (e.g., Robinson et al., 2013; Joseph et al., 2014; Shern et al., 2014), some experimental data suggest that the cell of origin is critical for tumor cell morphology. For instance, Straessler et al. (2013) demonstrated that conditional expression of EWSR1-ATF1 in a mouse model resulted in slightly different tumor phenotypes depending on the cell type, and its differentiation stage, in which the fusion was expressed. Possibly, this could also explain why some fusion partners occur in several tumors, with different malignant potential, of the same line of differentiation. For instance, FOSB is involved in benign epithelioid hemangiomas as well as borderline malignant pseudomyogenic hemangioendotheliomas (Table 1).
As expected, many STT display the same type of fusion(s) as morphologically identical lesions arising in bone (Table 2). Thus, no less than 14 of the recurrent gene fusions that have been described in STT have also been described in primary intraskeletal tumors. More unexpectedly, and again emphasizing the importance of the cell type in which the mutation occurs, some gene fusions in STT are shared with neoplasms of other lineages, such as leukemias, lymphomas, and carcinomas. For example, some ALK fusions in inflammatory myofibroblastic tumor are shared with malignant lymphomas and lung cancer, and TFE3 fusions in alveolar soft part sarcoma, PEComa, and epithelioid hemangioendothelioma are found also in kidney cancer (Table 2). The most versatile gene fusion in this context is ETV6-NTRK3, the molecular hallmark of infantile fibrosarcoma. The same gene fusion has been identified also in acute myeloid leukemia, mesoblastic nephroma, secretory carcinoma of the breast and salivary glands, adenocarcinoma of the colon and thyroid, pediatric glioma, and Ewing sarcoma (Mitelman et al., 2015).
GENE CLASSES
Transcription Factors
While almost all known gene fusions in STT ultimately affect the transcription of multiple other genes, the mechanisms by which they do so vary. Most human genes and their encoded proteins have more than one function, depending on how, when and where they are expressed, making it difficult to classify gene fusions into distinct, separate subgroups. For instance, the EWSR1 protein is usually classified as RNA-binding, but is in addition known to interact also with other proteins as well as with DNA (Tan and Manley, 2009). Furthermore, the part contributed to the chimeric protein may lack the domain(s) used for categorizing the wild type protein, e.g., NCOA2 is formally classified as a transcription factor (TF), but the DNA-binding part of the protein is never included in the pathogenetically important fusion proteins (see below). Still, an attempt to subdivide broadly the recurrent gene fusions in STT according to type of protein affected shows that most (52/78) fusions involve a proper TF or another type of protein, such as transcriptional co-activators or corepressors, directly involved in DNA transcription (Table 3). The Human Protein Atlas (http://www.proteinatlas.org/) currently lists more than 1,500 human genes that encode TFs, which can be further subdivided into superclasses, classes, families, and subfamilies on the basis of their DNA-binding domains (Wingender et al., 2012; http://www.edgar-wingender.de/huTF_classification.html). The 41 different TF-encoding genes that are recurrently involved in gene fusions in STT belong to 7 of the 10 superclasses and to 13 of the 40 classes, with some classes and families being more commonly affected than others: six genes each encode basic leucine zipper factors (bZIP; ATF1, CREB1, CREB3L1, CREB3L2, DDIT3, and FOSB), C2H2 zinc finger factors (C2H2; GLI1, KLF17, PLAG1, PRDM10, WT1, and ZNF444), or tryptophan cluster factors (TCf; ERG, ETV1, ETV4, ETV6, FEV, and FLI1), and five code for basic helix-loop-helix factors (bHLH; AHRR, HEY1, NCOA1, NCOA2, and TFE3); notably, all six TCf belong to the family of Ets-related factors, all C2H2 except KLF17 are included in the “more than three adjacent zinc finger factors” family, and the first four bZIP belong to the family of CREB-related factors (Table 3). Thus, the repertoire of TFs involved in fusions in STT is clearly nonrandom, and the involvement of a specific TF is typically seen in only one tumor type (for exceptions, see above); for instance, only Ewing sarcoma shows recurrent fusions involving an Ets-related factor as the carboxy-terminal partner.
TABLE 3.
Type of Gene Involved in Recurrent Gene Fusions in Soft Tissue Tumorsa
Why, then, are some TFs specifically associated with only some morphologies, and why are some transcription factors never involved? One explanation would be that only a limited set of TFs, and their target genes, are of relevance to the cells in which the tumors originate, i.e., only some developmental lineages are permissive for neoplastic transformation. Indeed, the few STT-associated chimeric TFs that have been analyzed in experimental systems, such as EWSR1-FLI1 or EWSR1-ATF1, show that only certain cell types can be transformed, that different genetic programs are affected in different cell types, and that the phenotypic effects vary dependent on in which cell it is expressed (Patel et al., 2012; Straessler et al., 2013; Riggi et al., 2014). Some further support for this notion could perhaps also be derived from STT showing differentiation towards a “normal” lineage, like fat or striated muscle, as in lipoma, myxoid liposarcoma, and alveolar rhabdomyosarcoma. Studies on normal, physiological development have disclosed critical roles for HMGA2 and DDIT3 in adipogenesis (Batchvarova et al., 1995; Thies et al., 2014) and for PAX3 and PAX7 in rhabdomyogenesis (Buckingham and Relaix, 2007). Furthermore, highly recurrent gene fusions in congenital spindle cell rhabdomyosarcoma involve critical transcriptional activators of muscle-specific genes, such as VGLL2, TEAD1, and SRF (Alaggio et al., in press). However, many STT do not display any discernible or distinct line of differentiation. In addition, the cell of origin is unknown in most cases, making it difficult to speculate on why a specific TF is associated with a specific STT subtype. Still, it seems clear that the effect of the chimeric TF is not limited to activation or repression of genes normally regulated by the wild-type TF, and that the role of the amino-terminal partner is more complex than simply potentiating the transcription of TF target genes (Riggi et al., 2014).
TFs typically occur as carboxy-terminal partners in the fusions, where they retain their DNA-binding domain(s) in the chimeric protein. In some fusions, like those involving the Ets family of TFs, the concomitant loss of regulatory amino-terminal sequences seems to be necessary for a tumorigenic effect (Hollenhorst et al., 2011; Cooper et al., 2014), whereas in others, like DDIT3 in myxoid liposarcoma or PLAG1 in lipoblastoma, the complete coding sequence is included in the fusion. The role of the amino-terminal partner is then usually to provide a more active promoter and/or transactivating domain(s) but, as discussed above, additional effects may be achieved. In some subtypes the gene fusion involves two TFs (Tables 1 and 2). Notably, NCOA2 is the 3′-partner in several recurrent fusions with other TF-encoding genes: AHRR in angiofibroma of soft tissue, HEY1 in mesenchymal chondrosarcoma, and PAX3 in alveolar rhabdomyosarcoma, as well as non-recurrent ones (GTF2I in angiofibroma of soft tissue and SRF in spindle cell rhabdomyosarcoma). NCOA2 encodes a member of the p160 steroid receptor coactivator (SRC) family and contains three structural domains: an N-terminal bHLH/PAS domain that can bind to DNA as well as heterodimerize with a variety of proteins, a central region with three LXXLL motifs that interact with nuclear hormone receptors, and a C-terminal portion with two transcriptional activation domains (Xu et al., 2009). One of them binds other coactivators, such as CREBBP and EP300, which leads to chromatin remodelling, while the other is responsible for interaction with histone methyltransferases, coactivator-associated arginine methyltransferase 1, and PRMT1 (Xu et al., 2009). In the STT-associated fusions involving NCOA2, the chimeric protein always retains the DNA-binding parts of the 5′ TF and adds the two transcriptional activation domains from NCOA2. Thus, in some subtypes the important impact on differentiation is derived from the spec-ificity of the amino-terminal TF. Similar to NCOA2, CITED2 has recently been reported as a recurrent 3′ partner in congenital spindle cell rhabdomyosarcoma, preserving its CREBBP/EP300 interaction domain in the fusion protein (Alaggio et al., in press).
There is also an example of a TF serving as the amino-terminal partner to a protein kinase (PK): the ETV6-NTRK3 fusion in infantile fibrosarcoma. Here, the TF ETV6 contributes with a SAM domain, corresponding to the PNT domain in the Est family of TF, allowing for polymerization and constitutive activation of the tyrosine kinase domain of NTRK3 (Cetinbas et al., 2013). Yet another scenario is illustrated by conventional lipoma; some 75% of these tumors have a rearrangement of the HMGA2 gene, resulting in either an in-frame fusion transcript or in truncation of the gene, typically after the first three exons that encode the AT-hook domains (Lee and Dutta, 2007). It was shown that functional inactivation of the 3′ untranslated region of HMGA2, which contains multiple binding sites for the let-7 family of microRNAs, results in upregulation of HMGA2 by reducing HMGA2 mRNA degradation in the cytoplasm. Functional studies of different HMGA2 constructs have shown that the cellular consequences are the same (Fedele et al., 1998), raising questions about why some gene fusions are recurrent. In summary, TFs are involved in a variety of ways in STT.
The numerous gene fusions encoding chimeric TFs make them appealing treatment targets in STT. However, this task has been notoriously difficult, and therapeutical efforts have thus shifted toward down-stream targets of the chimeric proteins. An additional challenge in developing new drugs for STT treatment is that these tumors are rare. Thus, whenever gene fusions are shared with other, more common neoplasms, the chances of developing new, efficient drugs increase. Indeed, the high incidence of fusions involving Ets family proteins also in prostate cancer has spurred interest, and several promising studies are on-going (Cooper et al., 2014; Feng et al., 2014).
Protein Kinases
The second largest group of proteins involved in gene fusions in STT comprises PKs, all of which, except PRKCB (a serine/threonine kinase), are receptor tyrosine kinases. In all cases, the PK-encoding gene is the 3′-partner and the result of the fusion at the protein level is a constitutive activation of the kinase domain of a truncated PK (Table 3). The tumor types showing such gene fusions—notably inflammatory myofibroblastic tumor and benign fibrous histiocytoma—typically display a large variety of different 5′-partners, reflecting that the main role of the 5′-partner is to ensure a high level of transcription by providing a more active promoter, but also loss of regulatory elements from the wild-type PK may be important (Table 1). These aspects could explain why no single gene fusion makes up more than some 15% of the cases of inflammatory myofibroblastic tumor or benign fibrous histiocytoma (Table 2). The only exception at the moment—phosphaturic mesenchymal tumor, in which 60% of reported cases display a FN1-FGFR1 fusion—can perhaps be explained by the limited number of cases (n = 15) that has been reported. In addition to providing an active promoter, the amino-terminal partner may be important also through contributing oligomerization domains or by ensuring a particular subcellular localization of the chimeric protein. An indication that the involvement of a particular PK is less tissue-specific than for TFs could be gleaned from the fact that several of the STT-associated gene fusions are seen in many other types of neoplasia, ETV6-NTRK3 and EML4-ALK constituting the most prominent examples (Table 2). Furthermore, the PK that is most often involved in STT-associated chimeras—ALK—is activated by numerous other fusions, as well as other types of mutation, in a large number of neoplasms (Mariño-Enriquez and Dal Cin, 2013; Mitelman et al., 2015).
Another feature shared by inflammatory myofibroblastic tumor and benign fibrous histiocytoma is that the type of PK involved in the fusions varies, i.e., either ALK, ROS1, or PDGFRB in the former and either ALK or protein kinase C in the latter. As mentioned above, this variation correlates with morphological subtype in benign fibrous histiocytoma; whether clinicopathological differences caused by type of PK fusion exist also among inflammatory myofibroblastic tumor remains to be seen. For obvious reasons, this tumor type is the STT subtype with PK-encoding gene fusions that has attracted most clinical attention. While benign fibrous histiocytoma and phospathaturic mesenchymal tumor are considered benign and metastases only rarely develop in infantile fibrosarcoma, inflammatory myofibroblastic tumor is more aggressive (Fletcher et al., 2013). Thus, the chimeric PKs here, as in other malignancies with receptor tyrosine kinases activated by gene fusions/mutations, offer an excellent therapeutic target (Shaw et al., 2013; Lovly et al., 2014 Stransky et al., 2014).
Chromatin Regulators
Proteins that are involved in chromatin modification and remodeling have emerged as important players in tumorigenesis (Chen and Dent, 2014). Gene fusions resulting in distorted chromatin regulation have been recognized in leukemias for two decades, with fusions involving the KMT2A (previously known as MLL) gene as a well-known example (Krivtsov and Armstrong, 2007). As expected, the cellular effects of such fusion proteins are more global than for fusions involving TFs with their specific sets of target genes. Thus, STT with gene fusions affecting chromatin regulation are either undifferentiated, as exemplified by undifferentiated round cell sarcomas with the BCOR-CCNB3 fusion, or display disparate lines of differentiation, such as synovial sarcoma with SS18-SSX fusions or ossifying fibromyxoid tumor with PHF1 fusions.
Most likely, the significance of chromatin remodeling extends far beyond what is suggested by the crude classification in Table 3; several of the fusions listed among TFs probably to some extent exert their pathogenetic impact by also affecting the chromatin configuration. For instance, the part of the NAB2 protein that replaces the carboxy-terminal part of STAT6 in the NAB2-STAT6 fusion in solitary fibrous tumor is known to interact with the NuRD complex. Furthermore, in all the fusions combining an amino-terminal TF with the carboxy-terminal part of NCOA1 or NCOA2, the histone methyltransferase-interacting domains of the NCOA proteins will be included, and also the EWSR1-FLI1 chimera can alter chromatin (Patel et al., 2012). There is thus reason to hope that drugs, such as DNA methylation and his-tone deacetylase inhibitors, that were developed for other neoplasms might be useful for epigenetic treatment of many malignant STT, too (Højfeldt et al., 2013).
Altered Growth Factor Signaling and Other Mechanisms
At first glance, a few fusions in STT seem to affect neither TFs, PKs, nor chromatin modifiers (Table 3). Indirectly, however, their effect is similar to what has been described above. Two gene fusions involve growth factors—the COL1A1-PDGFB fusion in dermatofibrosarcoma protuberans/giant cell fibroblastoma and the COL6A3-CSF1 fusion in tenosyno-vial giant cell tumor—that result in constitutive activation of their respective tyrosine kinase receptors, PDGFRB and CSF1R. Hence, in line with targeted therapies for fusions involving PKs, the use of a tyrosine kinase inhibitor, such as Imatinib, is of clinical value in unresectable or metastatic cases of dermatofibrosarcoma protuberans (Mentzel et al., 2013). Also activated CSF1R, which belongs to the same family of tyrosine kinases as PDGFRB, should respond to treatment with tyrosine kinase inhibitors, but the benefits of pharmacological treatment of tenosynovial giant cell tumors are less obvious (Cassier et al., 2012). USP6, which is activated through promoter-swapping with MYH9 in some 75% of cases of nodular fasciitis (Table 2), encodes an ubiquitin-specific protease. Similar USP6 fusions have been described also in primary aneurysmal bone cyst and in giant cell reparative granuloma of the hands and feet, but with other or unknown 5′-partners (Oliveira et al., 2004; Agaram et al., 2014). In all cases, it seems as if the contribution of the 5′-partner is restricted to providing enhanced transcription of USP6. Preliminary data suggest that overex-pressed USP6 results in activation of the NF-κB TF complex, but the exact mechanisms remain to be elucidated. If also malignant STT, as has been shown for other types of malignancy (Mitelman et al., 2015), turn out to display fusions activating ubiquitin-specific proteases, promising treatment options have emerged (Pal et al., 2014).
An exception to the rule that recurrent fusion proteins in STT affect the transcription of other genes was recently provided by Stransky et al. (2014), who identified a TRIO-TERT chimera in two dedifferentiated liposarcomas. STT typically maintain telomere length through mutations in the promoter region of the TERT gene (Killela et al., 2013) or by alternative lengthening of telomeres (Taylor et al., 2011). The fusion described by Stransky et al. (2014), which involved the nonenzymatic part of the PK TRIO, resulted in highly increased expression of the TERT gene, and thus represents a novel mechanism to ensure the telomere integrity of STT cells. Another intriguing mechanism is exemplified by glomus tumors, in which MIR143-NOTCH fusions have been detected in both benign and malignant lesions (Mosquera et al., 2013). Most likely, the strong promoter of MIR143 drives the expression of the NOTCH intracellular domain, which translocates to the nucleus and induces the transcriptional program associated with NOTCH activation. Thus, such tumors might potentially be sensitive to targeting with γ-secretase inhibitors.
CONCLUSIONS
As the NGS technologies continue to improve technically and to become cheaper and more user-friendly, they will gradually replace old-fashioned methods for detecting gene fusions not only in research projects but also in the clinical setting. Hence, the number of known gene fusions in STT can within the next few years be expected to show the same dramatic increase as has been witnessed for carcinomas (Mertens et al., 2015). Because probably only a few of them will be path-ogenetically relevant, the sorting out of driver mutations from passenger mutations will be an arduous task, hopefully facilitated by the development of more sophisticated algorithms for data analysis (Yoshihara et al., 2015). Much also remains to be done when it comes to elucidating the pathogenetic mechanisms involved, the interaction between gene fusions and other mutations, and the cellular contexts in which they arise. An important step in this endeavour is the creation of transgenic mouse models, which so far exist for only four gene fusions (EWSR1-ATF1, FUS-DDIT3, PAX3-FOXO1, and SS18-SSX2) and truncated HMGA2; all of these models develop the corresponding human tumors (Arlotta et al., 2000; Pérez-Losada et al., 2000; Keller et al., 2004; Haldar et al., 2007; Straessler et al., 2013). Most importantly, however, the new data need to be evaluated with regard to clinical significance, i.e., their diagnostic impact and their usefulness for treatment stratification and targeted therapy will have to be established. That gene fusion status is already of seminal importance for the management of cancer patients, such as chronic myeloid leukemia with BCR-ABL1 or lung cancer with ALK fusions, is well known but, as recently shown for inflammatory myofibroblastic tumor (Lovly et al., 2014), many new actionable targets are likely to be identified also in malignant STT for which currently no cure is available unless presenting as a localized disease.
ACKNOWLEDGMENT
The authors are indebted to Prof. Göran Stenman for critical reading of the manuscript.
Supported by: The Swedish Cancer Society, the Gunnar Nilsson Cancer Foundation, the IngaBritt and Arne Lundberg Foundation, the Swedish Childhood Cancer Foundation, the Medical Faculty of Lund University, and Governmental Funding of Clinical Research within the National Health Service.
REFERENCES
- Agaram NP, LeLoarer FV, Zhang L, Hwang S, Athanasian EA, Hameed M, Antonescu CR. USP6 gene rearrangements occur preferentialy in giant cell reparative granulomas of the hands and feet but not in gnathic location. Hum Pathol. 2014;45:1147–1152. doi: 10.1016/j.humpath.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agaram NP, Sung Y-S, Zhang L, Chen C-L, Chen H-W, Singer S, Dickson MA, Berger MF, Antonescu CR. Dichotomy of genetic abnormalities in PEComas with therapeutic implications. Am J Surg Pathol. 2015;39:813–825. doi: 10.1097/PAS.0000000000000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaggio R, Zhang L, Sung Y-S, Huang S-C, Chen C-L, Bisogno G, Zin A, Agaram NP, LaQuaglia MP, Wexler LH, Antonescu CR. A molecular study of pediatric spindle and sclerosing rhabdomyosarcoma: Identification of novel and recurrent VGLL2-related fusions in infantile cases. Am J Surg Pathol. doi: 10.1097/PAS.0000000000000538. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu CR, Dal Cin P. Promiscuous genes involved in recurrent chromosomal translocations in soft tissue tumours. Pathol. 2014;46:105–112. doi: 10.1097/PAT.0000000000000049. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Nafa K, Segal NH, Dal Cin P, Ladanyi M. EWS-CREB1: A recurrent variant fusion in clear cell sarcoma—Association with gastrointestinal location and absence of melanocytic differentiation. Clin Cancer Res. 2006;12:5356–5362. doi: 10.1158/1078-0432.CCR-05-2811. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Dal Cin P, Nafa K, Teot LA, Surti U, Fletcher CD, Ladanyi M. EWSR1-CREB1 is the predominant gene fusion in angiomatoid fibrous histiocytoma. Genes Chromosomes Cancer. 2007;46:1051–1060. doi: 10.1002/gcc.20491. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Zhang L, Chang N-E, Pawel BR, Travis W, Katabi N, Edelman M, Rosenberg AE, Nielsen GP, Dal Cin P, Fletcher CDM. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–1124. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu CR, Le Loarer F, Mosquera J-M, Sboner A, Zhang L, Chen C-L, Chen H-W, Pathan N, Krausz T, Dickson BC, Weinreb I, Rubin MA, Hameed M, Fletcher CDM. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosomes Cancer. 2013;52:775–784. doi: 10.1002/gcc.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu CR, Chen HW, Zhang L, Sung Y-S, Panicek D, Agaram NP, Dickson BC, Krausz T, Fletcher CD. ZFP36-FOSB fusion defines a subset of epithelioid hemangioma with atypical features. Genes Chromosomes Cancer. 2014a;53:951–959. doi: 10.1002/gcc.22206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu CR, Sung Y-S, Chen C-L, Zhang L, Chen H-W, Singer S, Agaram NP, Sboner A, Fletcher CD. Novel ZC3H7B-BCOR, MEAF6-PHF1, and EPC1-PHF1 fusions in ossifying fibromyxoid tumors-molecular characterization shows genetic overlap with endometrial stromal sarcoma. Genes Chromosomes Cancer. 2014b;53:183–193. doi: 10.1002/gcc.22132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbajian E, Puls F, Magnusson L, Thway K, Fisher C, Sumathi VP, Tayebwa J, Nord KH, Kindblom L-G, Mertens F. Recurrent EWSR1-CREB3L1 gene fusions in sclerosing epithelioid fibrosarcoma. Am J Surg Pathol. 2014;38:801–808. doi: 10.1097/PAS.0000000000000158. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Tai AK-F, Manfioletti G, Clifford C, Jay G, Ono SJ. Transgenic mice expressing a truncated form of the high mobility group I-C protein develop adiposity and an abnormally high prevalence of lipomas. J Biol Chem. 2000;275:14394–14400. doi: 10.1074/jbc.m000564200. [DOI] [PubMed] [Google Scholar]
- Attwooll C, Tariq M, Harris M, Coyne JD, Telford N, Varley JM. Identification of a novel fusion gene involving hTAFII68 and CHN from a t(9;17)(q22;q11.2) translocation in an extraskeletal myxoid chondrosarcoma. Oncogene. 1999;18:7599–7601. doi: 10.1038/sj.onc.1203156. [DOI] [PubMed] [Google Scholar]
- Bartuma H, Hallor KH, Panagopoulos I, Collin A, Rydholm A, Gustafson P, Bauer HCF, Brosjö O, Domanski HA, Mandahl N, Mertens F. Assessment of the clinical and molecular impact of different cytogenetic subgroups in a series of 272 lipomas with abnormal karyotype. Genes Chromosomes Cancer. 2007;46:594–606. doi: 10.1002/gcc.20445. [DOI] [PubMed] [Google Scholar]
- Batchvarova N, Wang XZ, Ron D. Inhibition of adipogenesis by the stress-induced protein CHOP (Gadd153). EMBO J. 1995;14:4654–4661. doi: 10.1002/j.1460-2075.1995.tb00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchini L, Birtwisle L, Saada E, Bazin A, Long E, Roussel J-F, Michiels J-F, Forest F, Dani C, Myklebost O, Birtwisle-Peyrottes I, Pedeutour F. Identification of PPAP2B as a novel recurrent translocation partner gene of HMGA2 in lipomas. Genes Chromosomes Cancer. 2013;52:580–590. doi: 10.1002/gcc.22055. [DOI] [PubMed] [Google Scholar]
- Brandal P, Panagopoulos I, Bjerkehagen B, Gorunova L, Skjeldal S, Micci F, Heim S. Detection of a t(1;22)(q23;q12) trans-location leading to an EWSR1-PBX1 fusion gene in a myoepithelioma. Genes Chromosomes Cancer. 2008;47:558–564. doi: 10.1002/gcc.20559. [DOI] [PubMed] [Google Scholar]
- Brandal P, Panagopoulos I, Bjerkehagen B, Heim S. t(19; 22)(q13;q12) translocation leading to the novel fusion gene EWSR1-ZNF444 in soft tissue myoepithelial carcinoma. Genes Chromosomes Cancer. 2009;48:1051–1056. doi: 10.1002/gcc.20706. [DOI] [PubMed] [Google Scholar]
- Bridge JA, Kanamori M, Ma Z, Pickering D, Hill DA, Lydiatt W, Lui MY, Colleoni GWB, Antonescu CR, Ladanyi M, Morris SW. Fusion of the ALK gene to the clathrin heavy chain gene, CLTC, in inflammatory myofibroblastic tumor. Am J Pathol. 2001;159:411–415. doi: 10.1016/S0002-9440(10)61711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broberg K, Zhang M, Strömbeck B, Isaksson M, Nilsson M, Mertens F, Mandahl N, Panagopoulos I. Fusion of RDC1 with HMGA2 in lipomas as the result of chromosome aberrations involving 2q35-37 and 12q13-15. Int J Oncol. 2002;21:321–326. [PubMed] [Google Scholar]
- Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, Patidar R, Hurd L, Chen L, Shern JF, Liao H, Wen X, Gerard J, Kim J-S, Lopez Guerrero JA, Machado I, Wai DH, Picci P, Triche T, Horvai AE, Miettinen M, Wei JS, Catchpool D, Llombart-Bosch A, Waldman T, Khan J. The genomic landscape of the Ewing sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 2014;10:e1004475. doi: 10.1371/journal.pgen.1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol. 2007;23:645–673. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- Cassier PA, Gelderblom H, Stacchiotti S, Thomas D, Maki RG, Kroep JR, van der Graaf WT, Italiano A, Seddon B, Dômont J, Bompas E, Wagner AJ, Blay JY. Efficacy of imatinib mesylate for the treatment of locally advanced and/or metastatic tenosynovial giant cell tumor/pigmented villonodular synovitis. Cancer. 2012;118:1649–1655. doi: 10.1002/cncr.26409. [DOI] [PubMed] [Google Scholar]
- Cetinbas N, Huang-Hobbs H, Tognon C, Leprivier G, An J, McKinney S, Bowden M, Chow C, Gleave M, McIntosh LP, Sorensen PH. Mutation of the salt bridge-forming residues in the ETV6-SAM domain interface blocks ETV6-NTRK3-indiced cellular transformation. J Biol Chem. 2013;288:27940–27950. doi: 10.1074/jbc.M113.475301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Dent SYR. Chromatin modifiers and remodellers: Regulators of cellular differentiation. Nat Rev Genet. 2014;15:93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmielecki J, Crago AM, Rosenberg M, O'Connor R, Walker SR, Ambrogio L, Auclair D, McKenna A, Heinrich MC, Frank DA, Meyerson M. Whole-exome sequencing identifies a recurrent NAB2-STAT6 fusion in solitary fibrous tumors. Nat Genet. 2013;45:131–132. doi: 10.1038/ng.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J, Rocques PJ, Crew AJ, Gill S, Shipley J, Chan AM-L, Gusterson BA, Cooper CS. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet. 1994;7:502–508. doi: 10.1038/ng0894-502. [DOI] [PubMed] [Google Scholar]
- Cools J, Wlodarska I, Somers R, Mentens N, Pedeutour F, Maes B, De Wolf-Peeters C, Pauwels P, Hagemeijer A, Marynen P. Identification of novel fusion partners of ALK, the anaplastic lymphoma kinase, in anaplastic large-cell lymphoma and inflammatory myofibroblastic tumor. Genes Chromosomes Cancer. 2002;34:354–362. doi: 10.1002/gcc.10033. [DOI] [PubMed] [Google Scholar]
- Cooper CDO, Newman JA, Gileadi O. Recent advances in the structural molecular biology of Ets transcription factors: Interactions, interfaces and inhibition. Biochem Soc Trans. 2014;42:130–138. doi: 10.1042/BST20130227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crago AM, Chmielecki J, Rosenberg M, O'Connor R, Byrne C, Wilder FG, Thorn K, Agius P, Kuk D, Socci ND, Qin LX, Meyerson M, Hameed M, Singer S. Near universal detection of alterations in CTNNB1 and Wnt pathway regulators in desmoid-type fibromatosis by whole-exome sequencing and genomic analysis. Genes Chromosomes Cancer. 2015;54:606–615. doi: 10.1002/gcc.22272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crew AJ, Clark J, Fisher C, Gill S, Grimer R, Chand A, Shipley J, Gusterson BA, Cooper CS. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. EMBO J. 1995;14:2333–2340. doi: 10.1002/j.1460-2075.1995.tb07228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat A, Åman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- Dahlén A, Fletcher CDM, Mertens F, Fletcher JA, Perez-Atayde AR, Hicks MJ, Debiec-Rychter M, Sciot R, Wejde J, Wedin R, Mandahl N, Panagopoulos I. Activation of the GLI oncogene through fusion with the beta-actin gene (ACTB) in a group of distinctive pericytic neoplasms: Pericytoma with t(7; 12). Am J Pathol. 2004;164:1645–1653. doi: 10.1016/s0002-9440(10)63723-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Cin P, Sciot R, Panagopoulos I, Åman P, Samson I, Mandahl N, Mitelman F, Van den Berghe H, Fletcher CDM. Additional evidence of a variant translocation t(12;22) with EWS/CHOP fusion in myxoid liposarcoma: Clinicopathological features. J Pathol. 1997;182:437–441. doi: 10.1002/(SICI)1096-9896(199708)182:4<437::AID-PATH882>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Davis RJ, D'Cruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) trans-location in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869–2872. [PubMed] [Google Scholar]
- Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, Aurias A, Thomas G. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- de Leeuw B, Balemans M, Olde Weghuis D, Geurts van Kessel A. Identification of two alternative fusion genes, SYT-SSX1 and SYT-SSX2, in t(X;18)(p11.2;q11.2)-positive synovial sarcomas. Hum Mol Genet. 1995;4:1097–1099. doi: 10.1093/hmg/4.6.1097. [DOI] [PubMed] [Google Scholar]
- Doyle LA, Mariño-Enriquez A, Fletcher CDM, Hornick JL. ALK rearrangement and overexpression in epithelioid fibrous histiocytoma. Mod Pathol. 2015;28:904–912. doi: 10.1038/modpathol.2015.49. [DOI] [PubMed] [Google Scholar]
- Erickson-Johnson MR, Chou MM, Evers BR, Roth CW, Seys AR, Jin L, Ye Y, Lau AW, Wang X, Oliveira AM. Nodular fasciitis: A novel model of transient neoplasia induced by MYH9-USP6 gene fusion. Lab Invest. 2011;91:1427–1433. doi: 10.1038/labinvest.2011.118. [DOI] [PubMed] [Google Scholar]
- Errani C, Zhang L, Sung YS, Hajdu M, Singer S, Maki RG, Healey JH, Antonescu CR. A novel WWTR1-CAMTA1 gene fusion is a consistent abnormality in epithelioid hemangioendothelioma of different anatomic sites. Genes Chromosomes Cancer. 2011;50:644–653. doi: 10.1002/gcc.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedele M, Berlingieri MT, Scala S, Chiarotti L, Viglietto G, Rippel V, Bullerdiek J, Santoro M, Fusco A. Truncated and chimeric HMGI-C genes induce neoplastic transformation of NIH3T3 murine fibroblasts. Oncogene. 1998;17:413–418. doi: 10.1038/sj.onc.1201952. [DOI] [PubMed] [Google Scholar]
- Feng FY, Brenner JC, Hussain M, Chinnaiyan AM. Molecular pathways: Targeting ETS gene fusions in cancer. Clin Cancer Res. 2014;20:4442–4448. doi: 10.1158/1078-0432.CCR-13-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F. WHO Classification of Tumours of Soft Tissue and Bone. IARC; Lyon: 2013. p. 468. [Google Scholar]
- Galili N, Davis RJ, Fredericks WJ, Mukhopadhyay S, Rauscher FJ, III, Emanuel BS, Rovera G, Barr FG. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;5:230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- Gebre-Medhin S, Nord KH, Möller E, Mandahl N, Magnusson L, Nilsson J, Jo VY, Vult von Steyern F, Brosjö O, Larsson O, Domanski HA, Sciot R, Debiec-Rychter M, Fletcher CDM, Mertens F. Recurrent rearrangement of the PHF1 gene in ossifying fibromyxoid tumors. Am J Pathol. 2012;181:1069–1077. doi: 10.1016/j.ajpath.2012.05.030. [DOI] [PubMed] [Google Scholar]
- Guillou L, Aurias A. Soft tissue sarcomas with complex genomic profiles. Virch Arch. 2010;456:201–217. doi: 10.1007/s00428-009-0853-4. [DOI] [PubMed] [Google Scholar]
- Guillou L, Benhattar J, Gengler C, Gallagher G, Ranchere-Vince D, Collin F, Terrier P, Terrier-Lacombe M-J, Leroux A, Marques B, de Saint Aubain Somerhausen N, Keslair F, Pedeutour F, Coindre J-M. Translocation-positive low-grade fibromyxoid sarcoma: Clinicopathologic and molecular analysis of a series expanding the morphologic spectrum and suggesting potential relationship to sclerosing epithelioid fibrosarcoma: A study from the French Sarcoma Group. Am J Surg Pathol. 2007;31:1387–1402. doi: 10.1097/PAS.0b013e3180321959. [DOI] [PubMed] [Google Scholar]
- Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. A conditional mouse model of synovial sarcoma: Insights into a myogenic origin. Cancer Cell. 2007;11:375–388. doi: 10.1016/j.ccr.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Hansén Hallor K, Mertens F, Jin Y, Meis-Kindblom JM, Kindblom L-G, Behrendtz M, Kalén A, Mandahl N, Panagopoulos I. Fusion of the EWSR1 and ATF1 genes without expression of the MITF-M transcript in angiomatoid fibrous histiocytoma. Genes Chromosomes Cancer. 2005;44:97–102. doi: 10.1002/gcc.20201. [DOI] [PubMed] [Google Scholar]
- Hibbard MK, Kozakewich HP, Dal Cin P, Sciot R, Tan X, Xiao S, Fletcher JA. PLAG1 fusion oncogenes in lipoblastoma. Cancer Res. 2000;60:4869–4872. [PubMed] [Google Scholar]
- Hofvander J, Tayebwa J, Nilsson J, Magnusson L, Brosjö O, Larsson O, Vult von Steyern F, Mandahl N, Fletcher CDM, Mertens F. Recurrent PRDM10 gene fusions in undifferentiated pleomorphic sarcoma. Clin Cancer Res. 2015;21:864–869. doi: 10.1158/1078-0432.CCR-14-2399. [DOI] [PubMed] [Google Scholar]
- Højfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nat Rev Drug Discov. 2013;12:917–930. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem. 2011;80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornick JL. Novel uses of immunohistochemistry in the diagnosis and classification of soft tissue tumors. Mod Pathol. 2014;27:S47–S63. doi: 10.1038/modpathol.2013.177. [DOI] [PubMed] [Google Scholar]
- Huang D, Sumegi J, Dal Cin P, Reith JD, Yasuda T, Nelson M, Muirhead D, Bridge JA. C11orf95-MKL2 is the resulting fusion oncogene of t(11;16)(q13;p13) in chondroid lipoma. Genes Chromosomes Cancer. 2010;49:810–818. doi: 10.1002/gcc.20788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S-C, Chen H-W, Zhang L, Sung Y-S, Agaram NP, Davis M, Edelman M, Fletcher CDM, Antonescu CR. Novel FUS-KLF17 and EWSR1-KLF17 fusions in myoepithelial tumors. Genes Chromosomes Cancer. 2015;54:267–275. doi: 10.1002/gcc.22240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S-C, Ghossei RA, Bishop JA, Zhang L, Chen TC, Huang HY, Antonescu CR. Novel PAX3-NCOA1 fusions in biphenotypic sinonasal sarcoma with focal rhabdomyoblastic differentiation. Am J Surg Pathol. doi: 10.1097/PAS.0000000000000492. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiano A, Sung YS, Zhang L, Singer S, Maki RG, Coindre J-M, Antonescu CR. High prevalence of CIC fusion with double-homeobox (DUX4) transcription factors in EWSR1-negative undifferentiated small blue round cell sarcomas. Genes Chromosomes Cancer. 2012;51:207–218. doi: 10.1002/gcc.20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon I-S, Davis JN, Braun BS, Sublett JE, Roussel MF, Denny CT, Shapiro DN. A variant Ewing's sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene. 1995;10:1229–1234. [PubMed] [Google Scholar]
- Jin Y, Möller E, Nord KH, Mandahl N, Vult von Steyern F, Domanski HA, Mariño-Enriquez A, Magnusson L, Nilsson J, Sciot R, Fletcher CDM, Debiec-Rychter M, Mertens F. Fusion of the AHRR and NCOA2 genes through a recurrent translocation t(5;8)(p15;q13) in soft tissue angiofibroma results in upregulation of aryl hydrocarbon receptor target genes. Genes Chromosomes Cancer. 2012;51:510–520. doi: 10.1002/gcc.21939. [DOI] [PubMed] [Google Scholar]
- Joseph CG, Hwang H, Jiao Y, Wood LD, Kinde I, Wu J, Mandahl N, Luo J, Hruban RH, Diaz LAj, He T-C, Vogelstein B, Kinzler KW, Mertens F, Papadopoulos N. Exomic analysis of myxoid liposarcomas, synovial sarcomas and osteosarcomas. Genes Chromosomes Cancer. 2014;53:15–24. doi: 10.1002/gcc.22114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Yoshida K, Handa M, Toyoda Y, Nishihira H, Tanaka Y, Sasaki Y, Ishida S, Higashino F, Fujinaga K. Fusion of an ETS-family gene, EIAF, to EWS by t(17;22)(q12;q12) chromosome translocation in an undifferentiated sarcoma of infancy. Genes Chromosomes Cancer. 1996;15:115–121. doi: 10.1002/(SICI)1098-2264(199602)15:2<115::AID-GCC6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Kawamura-Saito M, Yamazaki Y, Kaneko K, Kawaguchi N, Kanda H, Mukai H, Gotoh T, Motoi T, Fukayama M, Aburatani H, Takizawa T, Nakamura T. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet. 2006;15:2125–2137. doi: 10.1093/hmg/ddl136. [DOI] [PubMed] [Google Scholar]
- Keller C, Hansen MS, Coffin CM, Capecchi MR. Pax3:Fkhr interferes with embryonic Pax3 and Pax7 function: Implications for alveolar rhabdomyosarcoma cell of origin. Genes Dev. 2004;18:2608–2613. doi: 10.1101/gad.1243904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, Grollman AP, He T-C, He Y, Hruban RH, Jallo GI, Mandahl N, Meeker AK, Mertens F, Netto GJ, Rasheed BA, Riggins GJ, Rosenquist TA, Schiffman M, Shih I-M, Theodorescu D, Torbenson MS, Velculescu VE, Wang T-L, Wentzensen N, Wood LD, Zhang M, McLendon RE, Bigner DD, Kinzler KW, Vogelstein B, Papadopoulos N, Yan H. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci USA. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevich SR, McFadden DE, Tao W, Lim JF, Sorensen PHB. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet. 1998;18:184–187. doi: 10.1038/ng0298-184. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modofactions and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Labelle Y, Zucman J, Stenman G, Kindblom L-G, Knight J, Turc-Carel C, Dockhorn-Dworniczak B, Mandahl N, Desmaze C, Peter M, Aurias A, Delattre O, Thomas G. Oncogenic conversion of a novel orphan nuclear receptor by chromosome translocation. Hum Mol Genet. 1995;4:2219–2226. doi: 10.1093/hmg/4.12.2219. [DOI] [PubMed] [Google Scholar]
- Ladanyi M, Gerald W. Fusion of the EWS and WT1 genes in the desmoplastic small round cell tumor. Cancer Res. 1994;54:2837–2840. [PubMed] [Google Scholar]
- Ladanyi M, Lui MY, Antonescu CR, Krause-Boehm A, Meindl A, Argani P, Healey JH, Ueda T, Yoshikawa H, Meloni-Ehrig A, Sorensen PHB, Mertens F, Mandahl N, van den Berghe H, Sciot R, Dal Cin P, Bridge J. The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene. 2001;20:48–57. doi: 10.1038/sj.onc.1204074. [DOI] [PubMed] [Google Scholar]
- Ladanyi M, Antonescu CR, Leung DH, Woodruff JM, Kawai A, Healey JH, Brennan MF, Bridge JA, Neff JR, Barr FG, Goldsmith JD, Brooks JSJ, Goldblum JR, Ali SZ, Shipley J, Cooper CS, Fisher C, Skytting B, Larsson O. Impact of SYT-SSX fusion type on the clinical behavior of synovial sarcoma: A multi-institutional retrospective study of 243 patients. Cancer Res. 2002;62:135–140. [PubMed] [Google Scholar]
- Lau PPL, Lui PCW, Lau GTC, Yau DTW, Cheung ETY, Chan JKC. EWSR1-CREB3L1 gene fusion: A novel alternative molecular aberration of low-grade fibromyxoid sarcoma. Am J Surg Pathol. 2013;37:734–738. doi: 10.1097/PAS.0b013e31827560f8. [DOI] [PubMed] [Google Scholar]
- Lawrence B, Perez-Atayde A, Hibbard MK, Rubin BP, Dal Cin P, Pinkus JL, Pinkus GS, Xiao S, Yi ES, Fletcher CDM, Fletcher JA. TPM3-ALK and TPM4-ALK oncogenes in inflammatory myofibroblastic tumors. Am J Pathol. 2000;157:377–384. doi: 10.1016/S0002-9440(10)64550-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-C, Jeng Y-M, Su S-Y, Wu C-T, Tsai K-S, Lee C-H, Lin C-Y, Carter JM, Huang J-W, Chen S-H, Shih S-R, Marino-Enriquez A, Chen C-C, Folpe AL, Chang Y-L, Liang C-W. Identification of a novel FN1-FGFR1 genetic fusion as a frequent event in phosphaturic mesenchymal tumour. J Pathol. 2015;235:539–545. doi: 10.1002/path.4465. [DOI] [PubMed] [Google Scholar]
- Linch M, Miah AB, Thway K, Judson IR, Benson C. Systemic treatment of soft-tissue sarcoma—Gold standard and novel therapies. Nat Rev Clin Oncol. 2014;11:187–202. doi: 10.1038/nrclinonc.2014.26. [DOI] [PubMed] [Google Scholar]
- Lovly CM, Gupta A, Lipson D, Otto G, Brennan T, Chung CT, Borinstein SC, Ross JS, Stephens PJ, Miller VA, Coffin CM. Inflammatory myofibroblastic tumors harbor multiple potentially actionable kinase fusions. Cancer Discov. 2014;4:889–895. doi: 10.1158/2159-8290.CD-14-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Hill DA, Collins MH, Morris SW, Sumegi J, Zhou M, Zuppan C, Bridge JA. Fusion of ALK to the Ran-binding protein 2 (RANBP2) gene in inflammatory myofibroblastic tumor. Genes Chromosomes Cancer. 2003;37:98–105. doi: 10.1002/gcc.10177. [DOI] [PubMed] [Google Scholar]
- Mariño-Enriquez A, Fletcher CDM. Angiofibroma of soft tissue: Clinicopathologic characterization of a distinctive benign fibrovascular neoplasm in a series of 37 cases. Am J Surg Pathol. 2012;26:500–508. doi: 10.1097/PAS.0b013e31823defbe. [DOI] [PubMed] [Google Scholar]
- Mariño-Enriquez A, Dal Cin P. ALK as a paradigm of oncogenic promiscuity: Different mechanisms of activation and different fusion partners drive tumors of different lineages. Cancer Genet. 2013;206:357–373. doi: 10.1016/j.cancergen.2013.07.001. [DOI] [PubMed] [Google Scholar]
- McCoach CE, Doebele RC. The minority report: Targeting the rare oncogenes in NSCLC. Curr Treat Options Oncol. 2014;15:644–657. doi: 10.1007/s11864-014-0310-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentzel T, Pedeutour F, Lazar A, Coindre J-M. Dermatofibrosarcoma protuberans. In: Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F, editors. WHO Classification of Tumours of Soft Tissue and Bone. IARC; Lyon: 2013. p. 468. [Google Scholar]
- Mertens F, Tayebwa J. Evolving techniques for gene fusion detection in soft tissue tumors. Histopathol. 2014;64:151–162. doi: 10.1111/his.12272. [DOI] [PubMed] [Google Scholar]
- Mertens F, Fletcher CDM, Antonescu CR, Coindre JM, Colecchia M, Domanski HA, Downs-Kelly E, Fisher C, Goldblum JR, Guillou L, Reid R, Rosai J, Sciot R, Mandahl N, Panagopoulos I. Clinicopathologic and molecular genetic characterization of low-grade fibromyxoid sarcoma, and cloning of a novel FUS/CREB3L1 fusion gene. Lab Invest. 2005;85:408–415. doi: 10.1038/labinvest.3700230. [DOI] [PubMed] [Google Scholar]
- Mertens F, Johansson B, Fioretos T, Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15:371–381. doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Lasota J. Gastrointestinal stromal tumors. Gastroeneterol Clin North Am. 2013;42:399–415. doi: 10.1016/j.gtc.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missiaglia E, Williamson D, Chisholm J, Wirapati P, Pierron G, Petel F, Concordet JP, Thway K, Oberlin O, Pritchard-Jones K, Delattre O, Delorenzi M, Shipley J. PAX3/FOXO1 fusion gene status is the key prognostic molecular marker in rhabdomyosarcoma and significantly improves current risk stratification. J Clin Oncol. 2012;30:1670–1677. doi: 10.1200/JCO.2011.38.5591. [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2015 Available at: http://cgap.nci.nih.gov/Chromosomes/Mitelman.
- Mohajeri A, Tayebwa J, Collin A, Nilsson J, Magnusson L, Vult von Steyern F, Brosjö O, Domanski HA, Larsson O, Sciot R, Debiec-Rychter M, Hornick JL, Mandahl N, Nord KH, Mertens F. Comprehensive genetic analysis identifies a pathognomonic NAB2/STAT6 fusion gene, nonrandom secondary genomic imbalances, and a characteristic gene expression profile in solitary fibrous tumor. Genes Chromosomes Cancer. 2013;52:873–886. doi: 10.1002/gcc.22083. [DOI] [PubMed] [Google Scholar]
- Mosquera J-M, Sboner A, Zhang L, Chen C-L, Sung Y-S, Chen H-W, Agaram NP, Briskin D, Basha BM, Singer S, Rubin MA, Tuschl T, Antonescu CR. Novel MIR143-NOTCH fusions in benign and malignant glomus tumors. Genes Chromosomes Cancer. 2013;52:1075–1087. doi: 10.1002/gcc.22102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M, Panagopoulos I, Mertens F, Mandahl N. Fusion of the HMGA2 and NFIB genes in lipoma. Virchows Arch. 2005;447:855–858. doi: 10.1007/s00428-005-0037-9. [DOI] [PubMed] [Google Scholar]
- Nilsson M, Mertens F, Höglund M, Mandahl N, Panagopoulos I. Truncation and fusion of HMGA2 in lipomas with rearrangements of 5q32->q33 and 12q14->q15. Cytogenet Genome Res. 2006;112:60–66. doi: 10.1159/000087514. [DOI] [PubMed] [Google Scholar]
- Nyquist KB, Panagopoulos I, Thorsen J, Haugom L, Gorunova L, Bjerkehagen B, Fosså A, Guriby M, Nome T, Lothe RA, Skotheim RI, Heim S, Micci F. Whole-transcriptome sequencing identifies novel IRF2BP2-CDX1 fusion gene brought about by translocation t(1;5)(q42;q32) in mesenchymal chondrosarcoma. PLoS One. 2012;7:e49705. doi: 10.1371/journal.pone.0049705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AM, Hsi BL, Weremowicz S, Rosenberg AE, Dal Cin P, Joseph N, Bridge JA, Perez-Atayde AR, Fletcher JA. USP6 (Tre2) fusion oncogenes in aneurysmal bone cyst. Cancer Res. 2004;64:1920–1923. doi: 10.1158/0008-5472.can-03-2827. [DOI] [PubMed] [Google Scholar]
- Ouchi K, Miyachi M, Tsuma Y, Tsuchiya K, Iehara T, Konishi E, Yanagisawa A, Hosoi H. FN1: A novel fusion partner of ALK in an inflammatory myofibroblastic tumor. Pediatr Blood Cancer. 2015;62:909–911. doi: 10.1002/pbc.25424. [DOI] [PubMed] [Google Scholar]
- Pal A, Young MA, Donato MJ. Emerging potential of therapeutic targeting of ubiquitin-specific proteases in the treatment of cancer. Cancer Res. 2014;74:4955–4966. doi: 10.1158/0008-5472.CAN-14-1211. [DOI] [PubMed] [Google Scholar]
- Panagopoulos I, Höglund M, Mertens F, Mandahl N, Mitelman F, Åman P. Fusion of the EWS and CHOP genes in myxoid liposarcoma. Oncogene. 1996;12:489–494. [PubMed] [Google Scholar]
- Panagopoulos I, Mencinger M, Dietrich CU, Bjerkehagen B, Saeter G, Mertens F, Mandahl N, Heim S. Fusion of the RBP56 and CHN genes in extraskeletal myxoid chondrosarcomas with translocation t(9;17)(q22;q11). Oncogene. 1999;18:7594–7598. doi: 10.1038/sj.onc.1203155. [DOI] [PubMed] [Google Scholar]
- Panagopoulos I, Nilsson T, Domanski HA, Isaksson M, Lindblom P, Mertens F, Mandahl N. Fusion of the SEC31L1 and ALK genes in an inflammatory myofibroblastic tumor. Int J Cancer. 2006;118:1181–1186. doi: 10.1002/ijc.21490. [DOI] [PubMed] [Google Scholar]
- Patel M, Simon JM, Iglesia MD, Wu SB, McFadden AW, Lieb JD, Davis IJ. Tumor-specific retargeting of an oncogenic transcription factor chimera results in dysregulation of chromatin and transcription. Genome Res. 2012;22:259–270. doi: 10.1101/gr.125666.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Losada J, Sánchez-Martin M, Rodríguez-García MA, Pérez-Mancera PA, Pintado B, Flores T, Battaner E, Sánchez-García I. Liposarcoma initiated by FUS/TLS-CHOP: The FUS/TLS domain plays a critical role in the pathogenesis of liposarcoma. Oncogene. 2000;19:6015–6022. doi: 10.1038/sj.onc.1204018. [DOI] [PubMed] [Google Scholar]
- Peter M, Couturier J, Pacquement H, Michon J, Thomas G, Magdelenat H, Delattre O. A new member of the ETS family fused to EWS in Ewing tumors. Oncogene. 1997;14:1159–1164. doi: 10.1038/sj.onc.1200933. [DOI] [PubMed] [Google Scholar]
- Pierron G, Tirode F, Lucchesi C, Reynaud S, Ballet S, Cohen-Gogo S, Perrin V, Coindre J-M, Delattre O. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat Genet. 2012;44:461–466. doi: 10.1038/ng.1107. [DOI] [PubMed] [Google Scholar]
- Plaszczyca A, Nilsson J, Magnusson L, Brosjö O, Larsson O, Vult von Steyern F, Domanski HA, Lilljebjörn H, Fioretos T, Tayebwa J, Mandahl N, Nord KH, Mertens F. Fusions involving protein kinase C and membrane-associated proteins in benign fibrous histiocytoma. Int J Biochem Cell Biol. 2014;53:475–481. doi: 10.1016/j.biocel.2014.03.027. [DOI] [PubMed] [Google Scholar]
- Prieto-Granada C, Zhang L, Chen H-W, Sung Y-S, Agaram NP, Jungbluth AA, Antonescu CR. A genetic dichotomy between pure sclerosing epithelioid fibrosarcoma (SEF) and hybrid SEF/low-grade fibromyxoid sarcoma: A pathologic and molecular study of 18 cases. Genes Chromosomes Cancer. 2015;54:28–38. doi: 10.1002/gcc.22215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls F, Magnusson L, Niblett A, Douis H, Peake D, Taniere P, Kindblom LG, Mertens F. Non-fibrosing sclerosing epithelioid fibrosarcoma: An unusual variant. Histopathol. doi: 10.1111/his.12791. (in press) [DOI] [PubMed] [Google Scholar]
- Rabbitts TH, Forster A, Larson R, Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignantnt liposarcoma. Nat Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- Reichardt P. Soft tissue sarcomas, a look into the future: Different treatments for different subtypes. Future Oncol. 2014;10:19–27. doi: 10.2217/fon.14.116. [DOI] [PubMed] [Google Scholar]
- Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suvá ML, Rossetti ME, Boonseng WE, Oksuz O, Cook EB, Formey A, Patel A, Gymrek M, Thapar V, Deshpande V, Ting DT, Hornicek FJ, Nielsen GP, Stamenkovic I, Aryee MJ, Bernstein BE, Rivera MN. EWS-FLI1 utilizes divergent chromatin remodelling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell. 2014;28:668–681. doi: 10.1016/j.ccell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DR, Wu Y-M, Kalyana-Sundaram S, Cao X, Lonigro RJ, Sung Y-S, Chen C-L, Zhang L, Wang R, Su F, Iyer MK, Roychowdhury S, Siddiqui J, Pienta KJ, Kunju LP, Talpaz M, Mosquera JM, Singer S, Schuetze SM, Antonescu CR, Chinnaiyan AM. Identification of recurrent NAB2-STAT6 gene fusions in solitary fibrous tumor by integrative sequencing. Nat Genet. 2013;45:180–185. doi: 10.1038/ng.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S, Szuhai K, Ijszenga M, Tanke HJ, Zanatta L, Sciot R, Fletcher CDM, Dei Tos AP, Hogendoorn PCW. EWSR1-CREB1 and EWSR1-ATF1 fusion genes in angiomatoid fibrous histiocytoma. Clin Cancer Res. 2007;13:7322–7328. doi: 10.1158/1078-0432.CCR-07-1744. [DOI] [PubMed] [Google Scholar]
- Rubin BP, Chen C-J, Morgan TW, Xiao S, Grier HE, Kozakewich HP, Perez-Atayde AR, Fletcher JA. Congenital mesoblastic nephroma t(12;15) is associated with ETV6-NTRK3 gene fusion. Cytogenetic and molecular relationship to congenital (infantile) fibrosarcoma. Am J Pathol. 1998;153:1451–1458. doi: 10.1016/S0002-9440(10)65732-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clin Oncol. 2011;29:3659–3668. doi: 10.1200/JCO.2011.35.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenmakers EFPM, Wanschura S, Mols R, Bullerdiek J, Van den Berghe H, Van de Ven WJM. Recurrent rearrangements in the high mobility group protein gene, HMGI-C, in benign mesenchymal tumours. Nat Genet. 1995;10:436–444. doi: 10.1038/ng0895-436. [DOI] [PubMed] [Google Scholar]
- Shapiro DN, Sublett JE, Li B, Downing JR, Naeve CW. Fusion of PAX3 to a member of the forkhead family of transcription factors in human alveolar rhabdomyosarcoma. Cancer Res. 1993;53:5108–5112. [PubMed] [Google Scholar]
- Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer. 2013;13:772–787. doi: 10.1038/nrc3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, Ambrogio L, Auclair D, Wang J, Song YK, Tolman C, Hurd L, Liao H, Zhang S, Bogen D, Brohl AS, Sindiri S, Catchpoole D, Badgett T, Getz G, Mora J, Anderson JR, Skapek SX, Barr FG, Meyerson M, Hawkins DS, Khan J. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014;4:216–231. doi: 10.1158/2159-8290.CD-13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M-P, Pedeutour F, Sirvent N, Grosgeorge J, Minoletti F, Coindre J-M, Terrier-Lacombe M-J, Mandahl N, Craver RD, Blin N, Sozzi G, Turc-Carel C, O'Brien KP, Kedra D, Fransson I, Guilbaud C, Dumanski JP. Deregulation of the platelet-derived growth factor B-chain gene via fusion with collagen gene COL1A1 in dermatofibrosarcoma protuberans and giant-cell fibroblastoma. Nat Genet. 1997;15:95–98. doi: 10.1038/ng0197-95. [DOI] [PubMed] [Google Scholar]
- Sjögren H, Meis-Kindblom J, Kindblom L-G, Åman P, Stenman G. Fusion of the EWS-related gene TAF2N to TEC in extraskeletal myxoid chondrosarcoma. Cancer Res. 1999;59:5064–5067. [PubMed] [Google Scholar]
- Skytting B, Nilsson G, Brodin B, Xie Y, Lundeberg J, Uhlén M, Larsson O. A novel fusion gene, SYT-SSX4, in synovial sarcoma. J Natl Cancer Inst. 1999;91:974–975. doi: 10.1093/jnci/91.11.974. [DOI] [PubMed] [Google Scholar]
- Stockman DL, Ali SM, He J, Ross JS, Meis JM. Sclerosing epithelioid fibrosarcoma presenting as intraabdominal sarcomatosis with a novel EWSR1-CREB3L1 gene fusion. Hum Pathol. 2014;45:2173–2178. doi: 10.1016/j.humpath.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Storlazzi CT, Mertens F, Nascimento A, Isaksson M, Wejde J, Brosjö O, Mandahl N, Panagopoulos I. Fusion of the FUS and BBF2H7 genes in low grade fibromyxoid sarcoma. Hum Mol Genet. 2003;12:2349–2358. doi: 10.1093/hmg/ddg237. [DOI] [PubMed] [Google Scholar]
- Straessler KM, Jones KB, Hu H, Jin H, van de Rijn M, Capecchi MR. Modeling clear cell sarcomagenesis in the mouse: Cell of origin differentiation state impacts tumor characteristics. Cancer Cell. 2013;23:215–227. doi: 10.1016/j.ccr.2012.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita S, Arai Y, Tonooka A, Hama N, Totoki Y, Fujii T, Aoyama T, Asanuma H, Tsukahara T, Kaya M, Shibata T, Hasegawa T. A novel CIC-FOXO4 gene fusion in undifferentiated small round cell sarcoma: A genetically distinct variant of Ewing-like sarcoma. Am J Surg Pathol. 2014;38:1571–1576. doi: 10.1097/PAS.0000000000000286. [DOI] [PubMed] [Google Scholar]
- Sumegi J, Streblow R, Frayer RW, Dal Cin P, Rosenberg A, Meloni-Ehrig A, Bridge JA. Recurrent t(2;2) and t(2;8) translocations in rhabdomyosarcoma without the canonical PAX-FOXO1 fuse PAX3 to members of the nuclear receptor transcriptional coactivator family. Genes Chromosomes Cancer. 2010;49:224–236. doi: 10.1002/gcc.20731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC; Lyon: 2008. p. 439. [Google Scholar]
- Tai H-C, Chuang I-C, Chen T-C, Li C-F, Huang S-C, Kao Y-C, Lin P-C, Tsai J-W, Lan J, Yu S-C, Yen S-L, Jung S-M, Liao K-C, Fang F-M, Huang H-Y. NAB2-STAT6 fusion types account for clinicopathological variations in solitary fibrous tumors. Mod Pathol. 2015;28:1324–1335. doi: 10.1038/modpathol.2015.90. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Soda M, Togashi Y, Sugawara E, Hatano S, Asaka R, Okumura S, Nakagawa K, Mano H, Ishikawa Y. Pulmonary inflammatory myofibroblastic tumor expressing a novel fusion, PPFIBP1-ALK: Reappraisal of anti-ALK immunohisto-chemistry as a tool for novel ALK fusion identification. Clin Cancer Res. 2011;17:3341–3348. doi: 10.1158/1078-0432.CCR-11-0063. [DOI] [PubMed] [Google Scholar]
- Tan AY, Manley JL. The TET family of proteins: Functions and roles in disease. J Mol Cell Biol. 2009;1:82–92. doi: 10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Kato K, Gomi K, Matsumoto M, Kudo H, Shinkai M, Ohama Y, Kigasawa H, Tanaka Y. Perivascular epithelioid cell tumor with SFPQ/PSF-TFE3 gene fusion in a patient with advanced neuroblastoma. Am J Surg Pathol. 2009;33:1416–1420. doi: 10.1097/PAS.0b013e3181a9cd6c. [DOI] [PubMed] [Google Scholar]
- Tanas MR, Sboner A, Oliveira AM, Erickson-Johnson MR, Hespelt J, Hanwright PJ, Flanagan J, Luo Y, Fenwick K, Natrajan R, Mitsopoulos C, Zvelebil M, Hoch BL, Weiss SW, Debiec-Rychter M, Sciot R, West RB, Lazar AJ, Ashworth A, Reis-Filho JS, Lord CJ, Gerstein MB, Rubin MA, Rubin BP. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci Transl Med. 2011;3:98ra82. doi: 10.1126/scitranslmed.3002409. [DOI] [PubMed] [Google Scholar]
- Taylor BS, Barretina J, Maki RG, Antonescu CR, Singer S, Ladanyi M. Advances in sarcoma genomics and new therapeutic targets. Nat Rev Cancer. 2011;11:541–557. doi: 10.1038/nrc3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thies HW, Nolte I, Wenk H, Mertens F, Bullerdiek J, Markowski DN. Permanent activation of HMGA2 in lipomas mimics its temporal physiological activation linked to the gain of adipose tissue. Obesity. 2014;22:141–150. doi: 10.1002/oby.20137. [DOI] [PubMed] [Google Scholar]
- Thway K, Nicholson AG, Lawson K, Gonzalez D, Rice A, Balzer B, Swansbury J, Min T, Thompson L, Adu-Poku K, Campbell A, Fisher C. Primary pulmonary myxoid sarcoma with EWSR1-CREB1 fusion: A new tumor entity. Am J Surg Pathol. 2011;35:1722–1732. doi: 10.1097/PAS.0b013e318227e4d2. [DOI] [PubMed] [Google Scholar]
- Urano F, Umezawa A, Hong W, Kikuchi H, Hata J. A novel chimera gene between EWS and E1A-F, encoding the adenovirus E1A enhancer-binding protein, in extraosseous Ewing's sarcoma. Biochem Biophys Res Commun. 1996;219:608–612. doi: 10.1006/bbrc.1996.0281. [DOI] [PubMed] [Google Scholar]
- Vogels RJC, Vlenterie M, Versleijen-Jonkers YMH, Ruijter E, Bekers EM, Verdijk MAJ, Link MM, Bonenkamp JJ, van der Graaf WTA, Slootweg PJ, Suurmeijer AJH, Groenen PJTA, Flucke U. Solitary fibrous tumor—Clinicopathologic, immunohistochemical and molecular analysis of 28 cases. Diagn Pathol. 2014;9:224. doi: 10.1186/s13000-014-0224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtel M, Dettling M, Koscielniak E, Stegmaier S, Treuner J, Simon-Klingenstein K, Buhlmann P, Niggli FK, Schäfer BW. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res. 2004;64:5539–5545. doi: 10.1158/0008-5472.CAN-04-0844. [DOI] [PubMed] [Google Scholar]
- Walther C, Tayebwa J, Lilljebjörn H, Magnusson L, Nilsson J, Vult von Steyern F, Öra I, Domanski HA, Fioretos T, Nord KH, Fletcher CDM, Mertens F. A novel SERPINE1-FOSB fusion gene results in transcriptional up-regulation of FOSB in pseudomyogenic haemangioendothelioma. J Pathol. 2014;232:534–540. doi: 10.1002/path.4322. [DOI] [PubMed] [Google Scholar]
- Walther C, Hofvander J, Nilsson J, Magnusson L, Domanski HA, Gisselsson D, Tayebwa J, Doyle LA, Fletcher CDM, Mertens F. Gene fusion detection in formalin-fixed paraffin-embedded benign fibrous histiocytomas using fluorescence in situ hybridization and RNA sequencing. Lab Invest. 2015;95:1071–1076. doi: 10.1038/labinvest.2015.83. [DOI] [PubMed] [Google Scholar]
- Wang L, Bhargava R, Zheng T, Wexler L, Collins MH, Roulston D, Ladanyi M. Undifferentiated small round cell sarcomas with rare EWS gene fusions: Identification of a novel EWS-SP3 fusion and of additional cases with the EWS-ETV1 and EWS-FEV fusions. J Mol Diagn. 2007;9:498–509. doi: 10.2353/jmoldx.2007.070053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Motoi T, Khanin R, Olshen A, Mertens F, Bridge J, Dal Cin P, Antonescu CR, Singer S, Hameed M, Bovee JVMG, Hogendoorn PCW, Socci N, Ladanyi M. Identification of a novel, recurrent HEY1-NCOA2 fusion in mesenchymal chondrosarcoma based on a genome-wide screen of exon-level expression data. Genes Chromosomes Cancer. 2012;51:127–139. doi: 10.1002/gcc.20937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bledsoe KL, Graham RP, Asmann YW, Viswanatha DS, Lewis JE, Lewis JT, Chou MM, Yaszemski MJ, Jen J, Westendorf JJ, Oliveira AM. Recurrent PAX3-MAML3 fusion in biphenotypic sinonasal sarcoma. Nat Genet. 2014;46:666–668. doi: 10.1038/ng.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters BL, Panagopoulos I, Allen EF. Genetic characterization of angiomatoid fibrous histiocytoma identifies fusion of the FUS and ATF-1 genes induced by a chromosomal translocation involving bands 12q13 and 16p11. Cancer Genet Cytogenet. 2000;121:109–116. doi: 10.1016/s0165-4608(00)00237-5. [DOI] [PubMed] [Google Scholar]
- West RB, Rubin BP, Miller MA, Subramanian S, Kaygusuz G, Montgomery K, Zhu S, Marinelli RJ, De Luca A, Downs-Kelly E, Goldblum JR, Corless CL, Brown PO, Gilks CB, Nielsen TO, Huntsman D, van de Rijn M. A landscape effect in tenosynovial giant-cell tumor from activation of CSF1 expression by a translocation in a minority of tumor cells. Proc Natl Acad Sci USA. 2006;103:690–695. doi: 10.1073/pnas.0507321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Schoeps T, Dönitz J. TFClass: An expandable hierarchical classification of human transcription factors. Nucl Acids Res. 2012;41:D165–D170. doi: 10.1093/nar/gks1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wu R-C, O'Malley BW. Normal and cancer-related functions of the p160 steroid receptor coactivator (SRC) family. Nat Rev Cancer. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Miyachi M, Ouchi K, Kuwahara Y, Tsuchiya K, Iehara T, Konishi E, Yanagisawa A, Hosoi H. Identification of COL3A1 and RAB2A as novel translocation partner genes of PLAG1 in lipoblastoma. Genes Chromosomes Cancer. 2014;53:606–611. doi: 10.1002/gcc.22170. [DOI] [PubMed] [Google Scholar]
- Yoshihara K, Wang Q, Torres-Garcia W, Zheng S, Vegesna R, Kim H, Verhaak RG. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34:4845–4854. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucman J, Delattre O, Desmaze C, Epstein AL, Stenman G, Speleman F, Fletcher CDM, Aurias A, Thomas G. EWS and ATF-1 gene fusion induced by t(12;22) translocation in malignant melanoma of soft parts. Nat Genet. 1993a;4:341–345. doi: 10.1038/ng0893-341. [DOI] [PubMed] [Google Scholar]
- Zucman J, Melot T, Desmaze C, Ghysdael J, Plougastel B, Peter M, Zucker JM, Triche TJ, Sheer D, Turc-Carel C, Ambros P, Combaret V, Lenoir G, Aurias A, Thomas G, Delattre O. Combinatorial generation of variable fusion proteins in the Ewing family of tumours. EMBO J. 1993b;12:4481–4487. doi: 10.1002/j.1460-2075.1993.tb06137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]