Summary
Epigenetic targets are exciting new avenues for cancer drug discovery. Zhang and colleagues have designed the open-source EZH2 inhibitor JQEZ5 and shown anti-tumor efficacy in vitro and in vivo in preclinical studies in murine and human lung adenocarcinomas models expressing high levels of EZH2 (1).
Cancer cell growth depends upon selective gene expression. While mutations in “driver” oncogenes and tumor suppressor genes can do this, other important mechanisms involve changes to the cancer epigenome. If these changes can be identified and thwarted, such approaches would represent important new tools for the early detection, prevention, and rational treatment of cancers, such as lung cancer, the biggest cancer killer of men and women in the USA. Zhang et al. addresses one important such mechanism (and target) involving EZH2 of the polycomb repressive complex 2 (PRC2) (1). As part of their studies they developed genetically engineered mouse models (GEMM) of lung cancer driven by overexpression of EZH2 in target tissues and examined human lung cancer lines and databases to study lung adenocarcinomas that greatly overexpress EZH2. EZH2 was then targeted genetically and chemically with the new open-source EZH2 inhibitor probe JQEZ5 they developed, to demonstrate anti-cancer effects. While the compound (or its derivatives) will need additional refinement for optimal clinical applications, it serves as an excellent probe for research and reinforces EZH2 as an important target in lung adenocarcinoma.
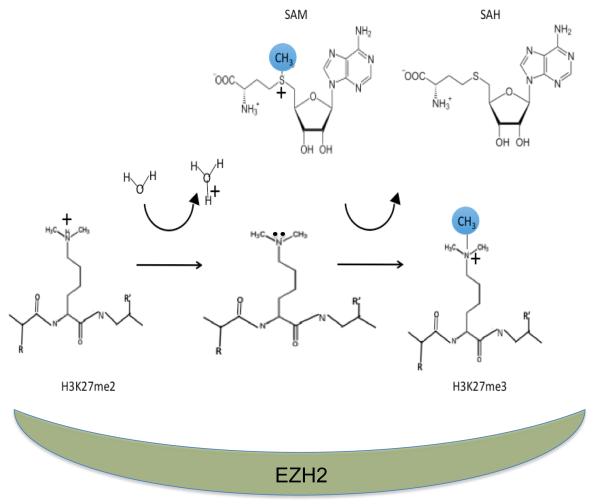
Chromatin-associated complexes can alter the chromosomal DNA or its associated histones leading to altered transcription. PRC2 is an assembly of polycomb group—PcG proteins including embryonic ectoderm development protein—EED, suppressor of zeste protein—SUZ12, retinoblastoma associated protein—RbAp46/48 and the enhancer of zeste homolog polypeptide—EZH1 or EZH2 (2). EZH2 but not EZH1 protein is expressed in proliferating cells. The 746 amino acid EZH2 has domains providing scaffolding for PRC2 proteins and a C-terminal Su(var)3-9 enhancer of zeste trithorax—SET enzymatic domain (3-5). Chemical biology is at the heart of the Zhang et al. work requiring understanding of the enzymology involved. The EZH2 SET domain has S-adenosyl-methionine (SAM) and H3 histone N-terminal tail lysine binding sites connected by a narrow channel for methyl transfer. After binding of positively charged S-adenosyl-methionine and positively charged histone tail lysine to SET, water-mediated deprotonation of the H3K27 ε-amino group occurs at the active site (6). The H3K27 ε-amino group then performs a nucleophilic attack on the methyl donor SAM. The methyl group from SAM is transferred to the ε-amino group of the substrate lysine by an SN2 substitution reaction sequentially producing S-adenosyl-homocysteine (SAH) and H3K27me3 (Figure 1A). Notably, PRC2 contains an unusual split catalytic domain consisting of the SET activation loop (SAL) from the N-terminal portion of Ezh2 and the SET domain at its C-terminus (4).
Figure 1A.
Enzymatic activity of PRC2. The residues in the active site including Y726 lower the pKa of the substrate lysine side chain and provide a solvent channel so water can deprotonate the ε-amino group. The amino group can then perform a nucleophilic attack on nearby S-adenosyl-methionine yielding methylated H3K27 and S-adenosyl-homocysteine. The EZH2 and its partner EED and SUZ12 polypeptides catalyze these reactions.
H3K27me3-marked chromatin recruits PRC1 complex with H2AK119 monoubiquitination activity, DNA methyltransferases that methylate CpG islands and histone deacetylase-containing complexes. The result is steric hindrance for transcription machinery binding, chromatin compaction and gene silencing which leads to changes in gene expression in cancer. While non-enzymatic transcriptional activator functions of EZH2 have been observed in some breast and prostate cancers, most research has analyzed the impact of altered H3K27me3. Global H3K27me3 increase leads to blocks in expression of differentiation genes and tumor suppressor genes and malignant transformation. Zhang demonstrated shifts in super enhancer signatures from H3K27ac to H3K27me3 by ChIP-seq with associated decreases in gene expression in these regions by RNA-seq (1).
Prior definitive evidence for the role of EZH2 in carcinogenesis leading up to the current study, emerged with the discovery of activating mutations in the SET domain of EZH2 in B-cell lymphomas, overexpression of EZH2 in lung adenocarcinomas, and mutational inactivation of UTX histone demethylase and of SWI/SNF complex in over 20% of cancers (7, 8). UTX mutations occur in the JmjC catalytic domain yielding increased H3K27me3. SWI/SNF is an ATP-dependent antagonist of PRC2 gene repression such that mutational loss of one of its components—ARID1A, PBRM1, SMARCB1, or SMARCA4 also leads to H3K27me3 markers. These classes of mutations mimic EZH2 over-activity. As shown by Zhang, preclinical models—mouse transgenic and human lung cancer xenograft models treated with EZH2 shRNAs, inhibit tumor growth, and thus confirm the importance of EZH2 in the establishment and preservation of EZH2(+)(but pAKT(−) and pERK(−)) lung adenocarcinomas.
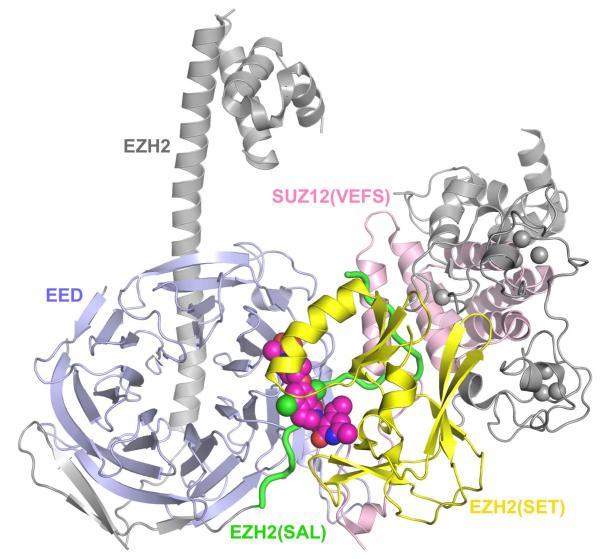
Because of such EZH2 pathway mutations, previously high throughput screens had been used to identify S-adenosylmethionine-competitive inhibitors of EZH2 (9). Analogs were optimized for oral bioavailability and improved pharmacokinetics and three—tazemetostat, GSK2816126, and CPI-1205 have entered clinical trials (NCT01897571, NCT02395601, NCT02601937, NCT02601950 and NCT02082977) (Figure 1B). The first two have a pyridine-amide core, and the last has a tetramethylpiperidinylbenzamide framework. Three other reports describe pyridine-amide based EZH2 inhibitor chemical probes for preclinical experiments—inhibitor 1, EI1 and JQEZ5 (1, 3, 10). The structure of the latter differs from G2816126 by four methyl groups (Figure 1B). Both co-crystallization x-ray structures of inhibitor with a human EED, human SUZ12(VEFS) domain and engineered American chameleon EZH2 subunits and in silico docking with EZH2 homology models built on the x-ray crystal structure of GLP were used. A step-by-step description of “hit” optimization was reported for tazemetostat (11). The pyridine-amide type of inhibitors bind to the unique split catalytic domain of PRC2 on the interface between the SAL and SET, which accounts for the remarkable selectivity displayed by these inhibitors (Figure 1C) (3).
Figure 1B.
EZH2 inhibitors. Each of the preclinical and clinical molecules shown has a pyridine amide core except CPI-1205. The compounds in clinical studies are tazemetostat, GSK281626, and CPI-1205.
Figure 1C.
Structure of PRC2 bound to an inhibitor (Inhibitor 1). The structure model was made based on PDB 5IJ7 (3) and the image was rendered by PyMOL (www.pymol.org). EED is shown in light blue and SUZ12(VEFS) in light pink. EZH2 is represented by grey except for the SAL and SET regions, which are highlighted in green and yellow, respectively. The bound EZH2 inhibitor is shown as a space-filling model.
Clinical results with each of the three EZH2 inhibitors (Tazemetostat, CPI-1205, and GSK2816126) are preliminary but encouraging. Tazemetostat was associated with cytopenias, hypertension, anorexia and transaminasemia with a recommended phase 2 dose of 800 mg po twice daily. 13/47 evaluable non-Hodgkin’s lymphoma (NHL) patients achieved objective responses including four complete remissions and nine partial remissions (12). Complete and partial responses have also been achieved in malignant rhabdoid tumor patients. CPI-1205 has been administered in a dose escalation schedule po twice daily, with low-grade diarrhea as a side effect, and disease stabilization in B cell NHL patients. Finally, GSK2816126 has been given on a dose escalation twice weekly IV schedule to NHL, multiple myeloma and solid tumor patients and has been well tolerated to date (13).
Patient selection based on likely tumor EZH2 dependency will be critical for the first phase of lung cancer clinical trials, and analyses for NSCLC tumors using CLIA certified molecular tests which are becoming routine will facilitate this. Similarly, lymphomas with EZH2 Y641 mutants, UTX histone demethylase mutations, and tumors with SWI-SNF core subunit inactivation should be more likely respond – but time will tell. Resistance mechanisms need to be studied in preclinical models, and should be anticipated such as secondary mutations in EZH2 or development of non-PRC2 MAPK pathway or AKT pathway activation (8, 14). Solutions to overcome such resistance may include combinations of EZH2 inhibitors or combinations with cytotoxic chemotherapy or other epigenetics or tyrosine kinase targeted agents. The Zhang et al study indicates EZH2 inhibitors will likely be at center stage of epigenetic cancer therapy development, and point the way to new opportunities and provide important reagents for further preclinical studies.
Acknowledgements
XL was supported by the Welch Foundation research grant I-1790, CPRIT research grant R1119, Rita Allen Foundation research grant, and NIH grant GM114576; JDM by NCI SPORE P50CA70907, CTD2N CA176284, and CPRIT RP110708.
Footnotes
Disclosure: The authors state they have no conflicts of interest to disclose.
References
- 1.Zhang H. Oncogenic deregulation of EZH2 as an opportunity for targeted therapy in lung cancer. Cancer Discovery. 2016 doi: 10.1158/2159-8290.CD-16-0164. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gall Troselj K, Novak Kujundzic R, Ugarkovic D. Polycomb repressive complex's evolutionary conserved function: the role of EZH2 status and cellular background. Clin Epigenetics. 2016;8:55. doi: 10.1186/s13148-016-0226-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooun A, Gajiwala KS, Deng YL, Liu W, Bolanos B, Bingham P, et al. Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance. Nat Commun. 2016;7:11384. doi: 10.1038/ncomms11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiao L, Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science. 2015;350:aac4383. doi: 10.1126/science.aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Justin N, Zhang Y, Tarricone C, Martin SR, Chen S, Underwood E, et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun. 2016;7:11316. doi: 10.1038/ncomms11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kipp DR, Quinn CM, Fortin PD. Enzyme-dependent lysine deprotonation in EZH2 catalysis. Biochemistry. 2013;52:6866–78. doi: 10.1021/bi400805w. [DOI] [PubMed] [Google Scholar]
- 7.Brien GL, Valerio DG, Armstrong SA. Exploiting the Epigenome to Control Cancer-Promoting Gene-Expression Programs. Cancer Cell. 2016;29:464–76. doi: 10.1016/j.ccell.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22:128–34. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaniskan HU, Konze KD, Jin J. Selective inhibitors of protein methyltransferases. J Med Chem. 2015;58:1596–629. doi: 10.1021/jm501234a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci U S A. 2012;109:21360–5. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuntz KW, Campbell JE, Keilhack H, Pollock RM, Knutson SK, Porter-Scott M, et al. The Importance of Being Me: Magic Methyls, Methyltransferase Inhibitors, and the Discovery of Tazemetostat. J Med Chem. 2016;59:1556–64. doi: 10.1021/acs.jmedchem.5b01501. [DOI] [PubMed] [Google Scholar]
- 12.Morera L, Lubbert M, Jung M. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin Epigenetics. 2016;8:57. doi: 10.1186/s13148-016-0223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yap T. A phase 1, open-label study of GSK2816126, an enhancer of zeste homolog 2 (EZH2) inhibitor, in patients with relapsed/refractory diffuse large B-cell lymphoma (DLBCL), transformed follicular lymphoma (tFL), other non-Hodgkin’s lymphomas (NHL), multiple myeloma (MM) and solid tumor. J Clin Oncol. 2016;34 Abstract TPS2595. [Google Scholar]
- 14.Gibaja V, Shen F, Harari J, Korn J, Ruddy D, Saenz-Vash V, et al. Development of secondary mutations in wild-type and mutant EZH2 alleles cooperates to confer resistance to EZH2 inhibitors. Oncogene. 2016;35:558–66. doi: 10.1038/onc.2015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]