Abstract
RecBCD enzyme is a complex, three-subunit protein machine essential for the major pathway of DNA double-strand break repair and homologous recombination in Escherichia coli. Upon encountering a Chi recombination-hotspot during DNA unwinding, RecBCD nicks DNA to produce a single-stranded DNA end onto which it loads RecA protein. Conformational changes that regulate RecBCD’s helicase and nuclease activities are induced upon its interaction with Chi, defined historically as 5′ GCTGGTGG 3′. Chi is thought to be recognized as single-stranded DNA passing through a tunnel in RecC. To define the Chi recognition-domain in RecC and thus the mechanism of the RecBCD-Chi interaction, we altered by random mutagenesis eight RecC amino acids lining the tunnel. We screened for loss of Chi activity with Chi at one site in bacteriophage λ. The 25 recC mutants analyzed thoroughly had undetectable or strongly reduced Chi-hotspot activity with previously reported Chi sites. Remarkably, most of these mutants had readily detectable, and some nearly wild-type, activity with Chi at newly generated Chi sites. Like wild-type RecBCD, these mutants had Chi activity that responded dramatically (up to fivefold, equivalent to Chi’s hotspot activity) to nucleotide changes flanking 5′ GCTGGTGG 3′. Thus, these and previously published RecC mutants thought to be Chi-recognition mutants are actually Chi context-dependence mutants. Our results fundamentally alter the view that Chi is a simple 8-bp sequence recognized by the RecC tunnel. We propose that Chi hotspots have dual nucleotide sequence interactions, with both the RecC tunnel and the RecB nuclease domain.
Keywords: chromosomal sites, recombination hotspots, Chi, RecBCD enzyme, DNA context-dependence
HOMOLOGOUS recombination, like other aspects of chromosome metabolism, is controlled by special DNA sites. The sites controlling homologous recombination that are best understood at the molecular level are the Chi hotspots of Escherichia coli (Smith 2012). Chi sites control RecBCD enzyme, a complex enzyme with both nuclease and helicase activities essential for the major pathway of DNA double-strand break repair and genetic recombination. These sites, called Chi for crossover hotspot instigator (Lam et al. 1974), locally stimulate recombination by altering the multiple activities of RecBCD. Here, we address the question of how RecBCD recognizes Chi, the first step in its stimulation of recombination. Our results force a reanalysis of both the Chi nucleotide sequence and how RecBCD recognizes Chi.
Chi was discovered as a set of mutations that increase the plaque size of phage λ red gam mutants, which rely on the E. coli RecBCD pathway for growth (Lam et al. 1974; McMilin et al. 1974; Henderson and Weil 1975). In the absence of its own recombination functions (Red) and of the RecBCD inhibitor (Gam), λ replication forms only monomeric circles. Phage DNA packaging, however, requires dimeric or higher order concatemers, which can be formed by RecBCD-promoted recombination between monomers. Wild-type λ lacks Chi sites; thus, the RecBCD pathway operates on λ red gam mutants only at low level, only a few packaged phage are produced, and burst and plaque sizes are small. Mutations creating a Chi hotspot arise at multiple, widely scattered locations in the genome (Henderson and Weil 1975; Stahl et al. 1975); increase the frequency of RecBCD-promoted concatemer formation; and result in larger burst and plaque sizes. Analysis of six Chi sites in λ and one in the E. coli lacZ gene revealed a sequence, 5′ GCTGGTGG 3′, common to all (Smith et al. 1984). Mutations creating or inactivating Chi are all in this octamer. The flanking sequences have no discernible similarity, and it was concluded that Chi is 5′ GCTGGTGG 3′, its complement, or the duplex (Smith et al. 1981a). Indeed, insertion of synthetic DNA with 5′ GCTGGTGG 3′ results in Chi activity both with purified enzyme and in cells (Dixon and Kowalczykowski 1991; Dabert et al. 1992; Dabert and Smith 1997). Analysis of heteroduplex DNA showed that only the strand with 5′ GCTGGTGG 3′ is needed to activate purified RecBCD (Bianco and Kowalczykowski 1997).
Since Chi acts on the host RecBCD pathway but not on the λ Red pathway or on two host pathways activated by suppressors of recBC null mutations (Gillen and Clark 1974; Stahl and Stahl 1977), it seemed likely that Chi interacts with RecBCD enzyme, the component unique to the RecBCD pathway. This hypothesis was bolstered by the isolation of intragenic pseudorevertants of a recC null mutant that regained at least partial recombination proficiency but lacked detectable Chi activity (Schultz et al. 1983).
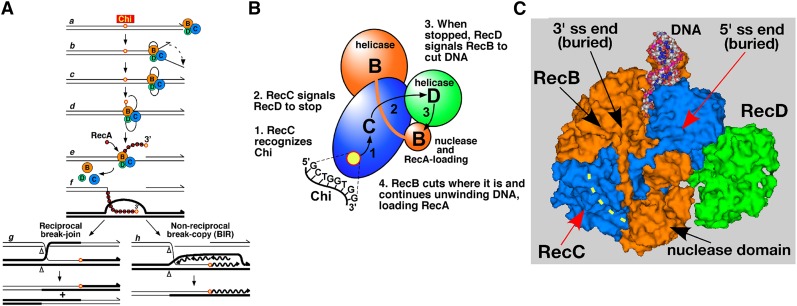
Direct evidence that RecBCD recognizes Chi came from the demonstration that wild-type RecBCD enzyme, but not that from the pseudorevertants, nicks DNA at Chi during DNA unwinding (Ponticelli et al. 1985; Taylor et al. 1985). Collectively, these observations supported a model of recombination (Figure 1A) (Smith et al. 1981b) in which RecBCD initiates DNA unwinding at a free double-stranded (ds) DNA end, rapidly unwinds the DNA with the production of single-stranded (ss) DNA loops, nicks one strand at a properly oriented Chi site, and continues unwinding. The newly generated 3′ ss end, with Chi near its end, is coated by RecA strand-exchange protein and invades an intact homologous duplex to generate a D-loop. The D-loop is converted into a Holliday junction, which may be resolved into reciprocal recombinants; alternatively, the D-loop may prime DNA replication and generate nonreciprocal recombinants (Smith 1991, 2012). Later reports showed that RecBCD actively loads RecA onto the newly generated 3′ end in a Chi-dependent manner (Anderson and Kowalczykowski 1997).
Figure 1.
Models for recombination and regulation of RecBCD enzyme by Chi hotspots, and RecBCD structural features involved in its context-dependent response to Chi. See text for further explanations. (A) Model for RecBCD-promoted recombination (Amundsen et al. 2007). Break-induced replication (BIR). (B) Intersubunit signal transduction model for regulation of RecBCD enzyme activities in response to Chi (Amundsen et al. 2007). (C) The RecBCD-ds DNA complex shown in a surface representation (PDB 1W36; Singleton et al. 2004). The RecB, C, and D subunits are colored as in panels A and B. The bound DNA has four terminal unwound bp; the 3′ terminus extends into the RecB helicase domain and the 5′ terminus extends into RecC headed toward the RecD helicase domain. Dotted yellow line represents the RecC tunnel in which Chi is putatively recognized. See Figure 2 for additional views.
The multiple activities of RecBCD are altered by Chi, as noted above. The phenotypes of a class of special missense mutations altering the RecB helicase domain led to the “signal transduction” model of Chi’s control of RecBCD (Figure 1B) (Amundsen et al. 2007). In this model, when Chi is in a tunnel in RecC, this subunit signals the RecD helicase to stop unwinding, which in turn signals the RecB nuclease domain to nick the DNA at Chi and to begin loading RecA onto the newly generated 3′-ended ss DNA as unwinding by RecB continues. Conformational changes in RecBCD upon binding of DNA and again upon encountering Chi have supported this model (Taylor et al. 2014), which is consistent with current genetic and biochemical data.
Central to this model is recognition of Chi by RecC. In the crystal structure of RecBCD bound to ds DNA (Figure 1C and Figure 2), the 5′ end of the DNA heads toward the RecD helicase and the 3′ end is in the RecB helicase domain headed toward a tunnel in RecC (Singleton et al. 2004; Saikrishnan et al. 2008). Amino acids lining this tunnel are altered in the pseudorevertants noted above. A set of RecC mutants, each with 1 of 11 amino acids lining the RecC tunnel changed to alanine, was reported to have reduced or abolished Chi activity (Handa et al. 2012), further implicating the RecC tunnel in Chi recognition. That report used Chi at one locus in λ. We report here a different, but overlapping, set of RecC tunnel mutants and that these, as well as some of the previously reported RecC tunnel mutants, in fact have Chi activity; but that this activity depends strongly (up to fivefold) on the nucleotide-sequence context of Chi. Our results force a reanalysis of the essential sequence of Chi and how this sequence interacts with RecBCD enzyme.
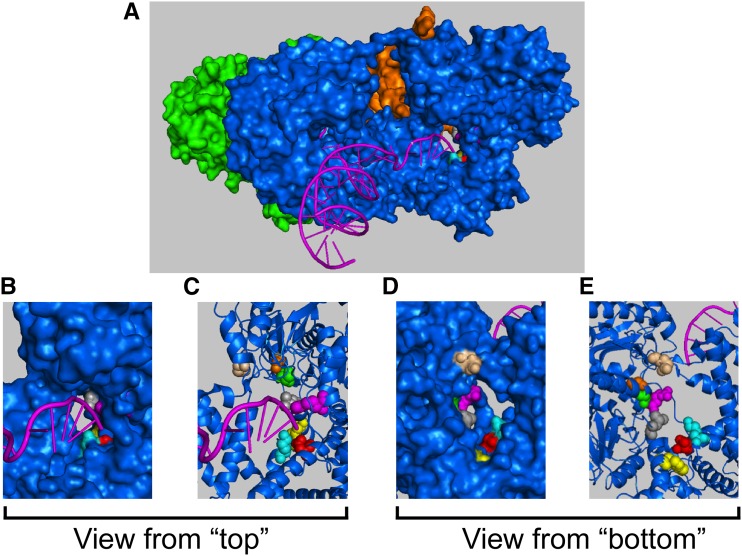
Figure 2.
The RecC tunnel in which Chi is putatively recognized. (A) A portion of the RecBCD-ds DNA complex in surface view (PDB 1K70; Saikrishnan et al., 2008). RecC (blue), the RecB nuclease domain (orange), and RecD (green) are shown. One ss DNA end (magenta) enters a tunnel (right) in RecC leading to the region implicated in Chi recognition. Amino acid residues altered by mutation, seen best in B–E, are colored as follows: S39 (green), G41 (orange), L64 (gray), W70 (yellow), K88 (cyan), D136 (red), Q652 (wheat), R708 (magenta). The other ss DNA end (left) enters a second tunnel in RecC leading to RecD, a helicase. Additional panels show (B and D) enlarged surface and (C and E) ribbon representations of the tunnel. Views B and C are oriented as in A; views D and E are from the opposite side.
Materials and Methods
Bacterial strains, phage, and plasmids
E. coli strains, plasmids, and phage are listed in Supplemental Material, Table S1 with their genotypes and sources. Many plasmids are derivatives of plasmid pSA607, which contains recB+, recD+, and recC2773 (recC with six histidine codons added to the C terminus) and encodes a fully functional RecBCD enzyme (Taylor et al. 2014). Methods to prepare λ phage libraries with 25-bp inserts and various χ+L alleles inserted into the λ gam gene by recombineering (Thomason et al. 2009) are in File S1.
Growth media
Tryptone broth (TB) and agar (TBA), LB broth (LB) and agar (LBA), and suspension medium (SM) have been described (Schultz et al. 1983; Cheng and Smith 1989). TB top agar contained 0.75% Bacto-Agar (Becton Dickinson) and TB bottom agar contained 1.0% Bacto-Agar. Tryptone agar plates (BBL YE) for the detection of clear and turbid plaques contained 0.2% yeast extract (Becton Dickinson). Media were supplemented with ampicillin (100 µg/ml) or streptomycin (25 µg/ml) as needed.
Mutagenesis and screen for mutants with altered Chi activity
Mutations altering the tunnel of RecC were produced on plasmid pSA607 using the QuikChange Multi Site-Directed Mutagenesis kit (Agilent Technologies) with the oligonucleotides (Integrated DNA Technologies) listed in Table S2. Oligonucleotides were designed with the desired mutation(s) using the web-based QuikChange Primer Design Program (http://www.genomics.agilent.com/primerDesignProgram.jsp).
For patch mutagenesis, the base composition of the mutagenic oligonucleotide was mixed so that each targeted codon could be efficiently changed to any other. Accordingly, at each position the wild-type base was present at 94% of the total; the remaining 3 bases were present at 2% each (see Table S2). For single-codon random mutagenesis, all 4 bases were mixed (25% each) at each of three contiguous positions in the oligonucleotide. Specific site-directed mutations were made with the QuikChange kit and oligonucleotides designed to introduce the desired point mutation (e.g., W70A or D136A).
Following mutant-strand synthesis with 100 ng of pSA607 and 125 ng of the appropriate oligonucleotide, DNA was digested with DpnI, and 1.5–5 µl of the reaction mix (50 µl) was used for transformation of XL10 Gold ultracompetent cells (Agilent Technologies). Ampicillin-resistant (AmpR) colonies were selected on LBA plates. For preliminary characterization of the mutagenesis reaction, 5–10 isolated colonies were grown overnight in 5 ml of LB. Plasmid DNA was prepared (Pure Link Miniprep Kit; Invitrogen, Carlsbad, CA), and a portion of recC was amplified by a DNA polymerase chain reaction (PCR) using oligonucleotides listed in Table S2. The PCR products were purified (QIAquik PCR purification kit; QIAGEN, Valencia, CA) and sequenced using the appropriate oligonucleotide primers to determine the frequency and distribution of mutations. Sequence data included ∼500 bp, centered on the position of the mutation target.
For screening recC phenotypes following mutagenesis, ∼500 AmpR colonies were pooled into 5 ml of LB and incubated at 37° for 2 hr. Plasmid DNA was isolated as described above and used to transform the hsdR17 hsdM+ modification-proficient strain DH5α. A total of ∼500 AmpR colonies were pooled; DNA was prepared as before and used to transform strain V2831 (ΔrecBCD2731 < kan> recF143) to AmpR. The chromosomal recBCD deletion allowed us to assess the plasmid-borne RecBCD phenotype, and the recF mutation limited recombination to the RecBCD pathway. Unusual Chi-related phenotypes were observed in ∼5 and 65% of the colonies screened following patch and single-codon mutagenesis, respectively (see below).
Chi activity in mutant candidates was first estimated by comparing the plaque sizes of λ 872 (χ°; i.e., lacking Chi) and λ 873 (χ+76; i.e., containing Chi) phage; both contain b1453, which deletes red and most of gam. For large-scale screening, a 50 µl aliquot of each phage stock (8 × 106 PFU/ml) was applied as a line (∼5 × 85 mm) to a BBL YE plate. Isolated colonies of transformants and recBCD+ control strains were picked from LBA plates and cross-streaked first against λ χ° and then against λ χ+76 phage. Plaque size was examined after overnight incubation at 37°. For isolates producing apparently equal-size plaques, more accurate plaque sizes were determined on lawns of bacteria as described below.
Chi alleles
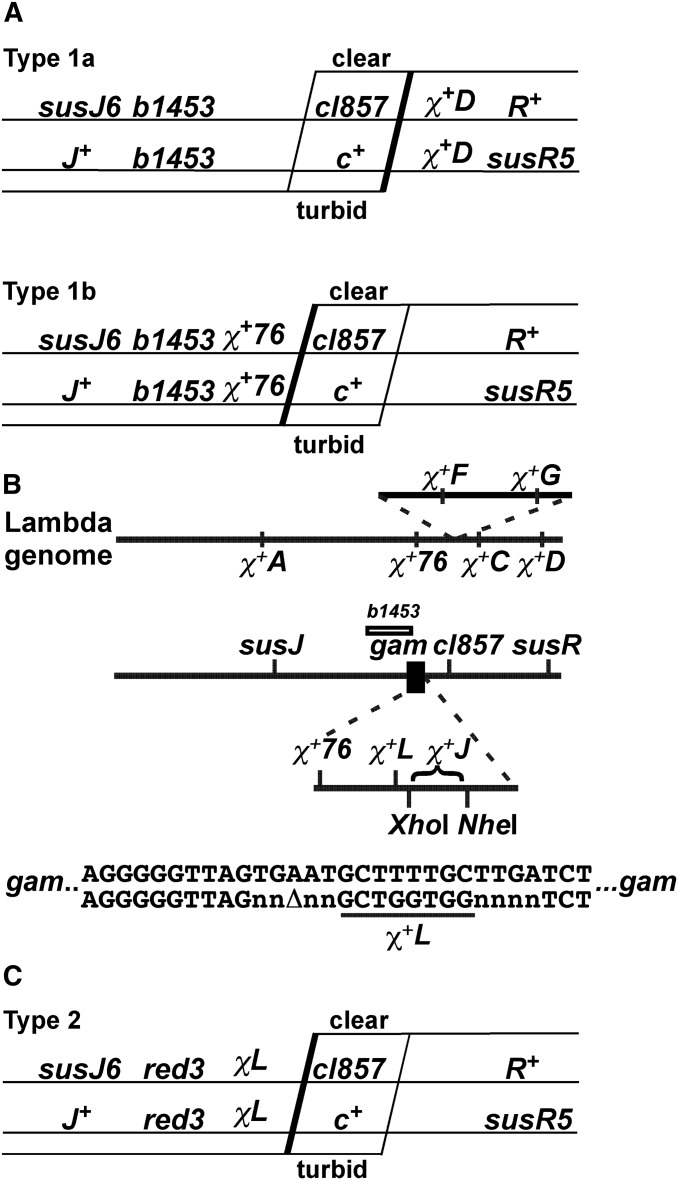
To assess Chi activity in genetic assays, we used phage with Chi alleles at eight locations in the genome (Figure 3B). χ+A, χ+76, χ+C, χ+D, χ+F, and χ+G have been described previously (see references in Table 2). We created Chi at two new loci, χL in the gam gene and χJ to its right. χJ alleles were identified in a phage library containing insertions of random 25-mers (see File S1). For χ+L251, the gam gene was inactivated by a 1-bp frameshift near the N terminus of the gene, and the Chi octamer (5′ GCTGGTGG 3′) was created by substitutions at another 3 bp nearby (Figure 3B; Smith et al. 1981a). Four nucleotides flanking the Chi octamer on each side were randomized to generate χ+L252, 253, and 254. The designation χ+ indicates the allele contains the Chi octamer, 5′ GCTGGTGG 3′; χ− indicates the allele contains a C to T change in the octamer (5′ GTTGGTGG 3′), which inactivates Chi (Schultz et al. 1981). The flanking nucleotides are identical for χ+ and χ− alleles with the same numerical designation.
Figure 3.
Physical and genetic map of λ and crosses to assess Chi activity. (A) λ crosses to determine Chi hotspot activity of wild type and RecC tunnel mutants. Single horizontal lines represent the ds DNA of each parent in the cross with the indicated genetic markers. Diagonal lines represent the position of exchanges among J+ R+ recombinants; a thick line indicates exchange stimulated by an active Chi in each cross. The plaque morphology (clear or turbid) generated by the exchange is indicated. Crosses of type 1a and 1b measure Chi hotspot activity (Stahl and Stahl 1977). (B) Physical maps of λ and of the gam region. Markers on each line, which represents the λ genome or an insertion in which χ+F and χ+G arose, are placed to scale. The b1453 deletion extends from bp 27,728–33,032; red (bp 31,348–32,810) with the red-3 mutation (T to A at bp 31,966, creating exo D21E, and G to A at bp 32,382, creating bet W143*; unpublished data) is to the left of gam (bp 32,816–33,232). χ+76 is an A to G mutation at bp 33,058 (unpublished data), and the χ+L octamer is at bp 33,196–33,203. Restriction enzyme cleavage sites for XhoI (bp 33,498) and NheI (bp 34,679) were used to clone 25-mers creating χ+J. Part of the nucleotide sequence of gam is shown with the 1-bp deletion to create χ+L (underlined) flanked in this case by four random nucleotides (n). (C) Type 2 crosses to measure χ+L activity and its context-dependence.
Table 2. Chi alleles and their nucleotide sequence contexts.
| Chi allele | Sequence (5′ → 3′)a | Description (refs)b |
|---|---|---|
| χ+76c | GGCGAGCTGCTGGTGGTGACGCGCCC | Chi+ mutant of λ (Stahl and Stahl 1977; Kobayashi et al. 1982) |
| χ+D123 | TCGTGAAAGCTGGTGGCAGGAGGTCG | Chi+ mutant of λ (Smith et al. 1981a) |
| χ+C157 | GCAGATCAGCTGGTGGAAGAGGGACT | Chi+ mutant of λ (Sprague et al. 1978) |
| χ−C206 | GCAGATCAGTTGGTGGAAGAGGGACT | Octamer mutation of χ+C157 (Schultz et al. 1981) |
| χ+F225 | CCGGCTAGGCTGGTGGGGTTGCCTTA | Chi+ mutant of λ (Smith et al. 1981b) |
| χ+G218 | AAACCACCGCTGGTGGCGGTGGTTTT | Chi+ mutant of λ (Smith et al. 1981b) |
| χ+J270 | TGGGTGTGCTCTGCTGGTGGGCGGG | Random 25-bp insertion in gam |
| χ+J271 | TGGTTGTTTTTGGCTGGTGGGTGGT | “ |
| χ+J272 | TGGGGTGTGCTCTGCTGGTGGGCGG | “ |
| χ+J273 | GATCGGGTCAGGGGAGAGCTGGTGG | “ |
| χ+J274 | GGAGGTGCTGGTGGCCTTTGGTACG | “ |
| χ+J275 | CACCAGCTGGTGGTCCTAGGTGTTC | “ |
| χ+J276 | GCTGGTGGAGGCAGGTAATGCCATT | “ |
| χ+L251 | TTAGTGATGCTGGTGGTTGATCTCA | Chi octamer in gam; wt flanks |
| χ−L251 | TTAGTGATGTTGGTGGTTGATCTCA | Octamer mutation of χ+L251 |
| χ+L252 | TTAGCCATGCTGGTGGACGGTCTCA | Chi octamer in gam; mutant flanks |
| χ−L252 | TTAGCCATGTTGGTGGACGGTCTCA | Octamer mutation of χ+L252 |
| χ+L253 | TTAGCAAAGCTGGTGGCGACTCTCA | Chi octamer in gam; mutant flanks |
| χ+L254 | TTAGCATAGCTGGTGGAGACTCTCA | Chi octamer in gam; mutant flanks |
| χ+L255 (2-3) | TTAGCCATGCTGGTGGCGACTCTCA | Swap of 5′ and 3′ flanks |
| χ+L256 (2-4) | TTAGCCATGCTGGTGGAGACTCTCA | “ |
| χ+L257 (3-2) | TTAGCAAAGCTGGTGGACGGTCTCA | “ |
| χ+L258 (4-2) | TTAGCATAGCTGGTGGACGGTCTCA | “ |
| χ+L259 | TTAGTGATGCTGGTGGTCGAAAACA | “Hottest” flank cut by RecBCD |
| χ−L259 | TTAGTGATGTTGGTGGTCGAAAACA | Octamer mutation of χ+L259 |
| χ+L262 | TTAGTGATGCTGGTGGGGGCTTCCA | “Colder” flank cut by RecBCD |
refs, references; wt, wild type.
Sequence surrounding the Chi octamer (underlined) in the indicated allele in λ. The χ− octamer mutation (C → T) is in boldface type.
See Materials and Methods for sources of nonreferenced alleles.
χ+76 is A → G mutation at nucleotide 33,058 in the gam gene, changing gctggtAg to gctggtGg (unpublished data).
Screen for phage sensitivity and Chi activity by spot test
Spot tests for phage growth were done as described previously (Schultz et al. 1983). Cells were grown in TB supplemented with maltose (0.1%), thiamine (0.5 µg/ml), and ampicillin to ∼1 × 108 CFU/ml. Phage stocks (∼3 × 103 PFU/ml in SM) were applied as 10 µl aliquots to the lawn of bacteria in TB top agar on TBA plates. To eliminate null mutants from further analysis, candidates were tested for the retention of RecBCD nuclease activity using a phage T4 gene 2 triple-nonsense mutant (Amundsen et al. 2012), which forms plaques only on cells lacking RecBCD nuclease activity (Oliver and Goldberg 1977). About 1% of the mutant candidates tested allowed T4 gene 2 mutant plaque-formation; most of these contained plasmids with recC nonsense mutations.
We next assayed λ plaque size by spot test of λ 872 (χ°), λ 873 (χ+76), and λ 801 (red+ gam+). As expected, wild-type (red+ gam+) λ phage formed large plaques on all strains. Phage containing the χ+76 allele formed large plaques on recBCD+ cells, but not on the RecC tunnel mutants reported here (see Results). Phage with other Chi alleles (i.e., χ+A, χ+C, χ+D, χ+F, χ+G, χ+J, or χ+L) formed plaques larger than isogenic χ° phage on many tunnel mutants (see Results). While the absolute plaque size of a given phage on a given strain varied slightly from day to day, the relative sizes of plaques were highly reproducible.
λ recombination and Chi activity assays
Recombination proficiency and Chi activity in λ crosses were measured as described (Schultz et al. 1983). Chi activity was quantified in two types of crosses (Figure 3). (i) Chi hotspot activity was determined from two normalized crosses between λ 1081 and λ 1082 (χ+D123; cross 1a) and between λ 1083 and λ 1084 (χ+76; cross 1b) (Stahl and Stahl 1977). The frequency of J+ R+ recombinant phage in both crosses was determined by plating on strain C600 (supE44) or strain V3477 (recD2741 supE44) for total phage, and on strain 594 (sup+) or strain V222 (recD1013 sup+) for recombinants. The number of clear and turbid plaques on strain 594 was determined by visual inspection; at least 50, and usually >100, plaques of each type were counted to determine the turbid/clear (t/c) ratio. These crosses are normalized by an interval lacking Chi in each cross; Chi hotspot activity is expressed as √(t/c)1a / (t/c)1b, where t/c is the ratio of turbid to clear plaques on strain 594 or V222 from crosses 1a and 1b as indicated (Stahl and Stahl 1977). (ii) Chi activity was determined from a nonnormalized cross between susJ6 cI857 and cI+ susR5 phage with various homozygous χL alleles. The frequency of J+ R+ recombinant phage and their plaque morphology were determined as described above. Chi activity is expressed as the ratio of clear to turbid plaques (c/t) on strain 594 or V222. Data are mean ± SEM, or individual values.
Hfr recombination assays
Recombination proficiency was measured by selecting His+ StrR (streptomycin-resistant) exconjugants following mating between Hfr donor strain V1306 (Hfr PO44 his+ rpsL+) and plasmid transformants of recipient strain V2831 (ΔrecBCD2731 hisG4 rpsL31). The ratio of donor to recipient cells was ∼1:10.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article and Supplemental Material.
Experimental design to identify RecBCD mutants having altered interaction with Chi
To rapidly screen RecC tunnel mutant candidates and subsequently to obtain quantitative measures for Chi activity and recombination, we used several phage-based assays to measure intracellular nuclease and Chi activity of the RecBCD enzyme. RecBCD influences the growth, and consequently plaque formation, of E. coli phages in several ways. RecBCD exonuclease activity blocks the growth and plaque formation of phage T4 mutants lacking the protective gene 2 protein that binds to the ends of infecting DNA. These protein caps are thought to block RecBCD binding and subsequent exonucleolytic degradation (Oliver and Goldberg 1977), so that T4 2+ but not T4 2− phage form plaques on RecBCD nuclease-proficient strains. Wild type and all RecC tunnel mutants reported here were RecBCD nuclease-proficient by this assay; indicating that RecBCD enzyme was functionally intact, at least for nuclease activity, which requires the helicase and ATPase activities (Hsieh and Julin 1992; Yu et al. 1998).
Chi activity and recombination proficiency were measured using λ red gam mutant phage. (Hereafter, “λ” means “λ red gam mutant” except where noted.) We used the λ plaque-size assay noted in the Introduction to qualitatively assess Chi activity in wild type and RecC tunnel mutants. We quantified intracellular Chi activity and recombination proficiency in λ vegetative crosses by determining the position of genetic exchange among selected recombinants (Figure 3, A and C). Chi acts primarily to its left as shown here (Faulds et al. 1979; Kobayashi et al. 1982; Taylor et al. 1985), resulting in an excess of exchanges in the Chi-containing interval compared to another without Chi. The first measure of Chi hotspot activity used a pair of crosses, each with a Chi-free interval that acted as a control for the other, thereby normalizing Chi activity (Figure 3A, type 1a and 1b) (Stahl and Stahl 1977). Each parental phage had a mutation (susJ6 or susR5) flanking the region containing a Chi site located to the right (χ+D123) or left (χ+76) of cI+ (or the alternative cI857 allele), a plaque morphology marker. J+ R+ recombinants were selected on nonsuppressing (sup+) E. coli host cells, and plaques were scored as turbid (c+) or clear (cI857). When Chi is active, there is an excess of turbid plaques in cross 1a and an excess of clear plaques in cross 1b. Chi hotspot activity is expressed as √(t/c)1a / (t/c)1b (Stahl and Stahl 1977), where t/c is the ratio of turbid to clear plaques in cross 1a or 1b, as indicated. In these crosses the Chi hotspot activity of recBCD+ strains is ∼5; in recBCD null strains, it is essentially 1, as expected from the normalization and from Chi hotspot activity requiring RecBCD.
To determine the role of nucleotide context in the Chi response, as indicated by our results below, we created Chi at a new locus (χL) within the gam gene, ∼150 bp to the right of the χ+76 locus (Figure 3B). As a control, we introduced into certain χ+L alleles the corresponding χ−L mutation, a C to T change in the Chi octamer that inactivates Chi at the well-studied χC locus (χ−C206; 5′GTTGGTGG 3′; Sprague et al. 1978; Schultz et al. 1981). We expressed Chi activity as the ratio of clear (cI857) to turbid (cI+) plaques (c/t) among J+ R+ phages from such crosses (Figure 3C, type 2). As shown below, alteration of the bp flanking χ+L significantly raised or lowered the c/t ratio in wild type and RecC tunnel mutants by as much as fivefold, demonstrating remarkable DNA context-dependence of Chi activity that is as strong as the stimulation of recombination by Chi itself. [We use “Chi hotspot activity” for normalized crosses and “Chi activity” for the c/t ratio in single (i.e., not normalized) crosses.]
To measure overall recombination proficiency, we used standard assays of Hfr conjugational and λ vegetative recombination (Stahl and Stahl 1977; Schultz et al. 1983). In these assays, the recombinant frequencies in recBC null mutants are lower than those in wild type (recBCD+) by factors of ∼500 and ∼10, respectively.
Results
RecC tunnel mutants that lack Chi activity but retain nuclease and recombination-promoting activities
To identify amino acids involved in detecting or signaling the presence of Chi on ss DNA, we mutagenized four patches of codons encoding amino acids that line the RecC tunnel, as indicated by the crystal structure (Table S3) (Singleton et al. 2004). In the crystal structure these patches cover a significant portion of the tunnel surface (Figure 2) and are largely composed of amino acids that extend into the tunnel interior, possibly allowing interaction with ss DNA. Although there is no direct assay for Chi recognition, such as stable binding of RecBCD enzyme to Chi-containing DNA (see Discussion), we quickly assessed the consequence of Chi recognition by comparing the plaque sizes of λ with a Chi site (χ+76) and without (χ°) as noted above.
Mutation of one to three codons within codon patches 37–44, 58–67, 643–652, and 687–691 of recC resulted in many mutants (∼5% of the isolates tested) on which λ χ° and χ+76 made plaques equal in size but ranging from tiny to large, depending on the recC mutant (Table S3; wild-type λ red+ gam+ formed large plaques on all the mutants); in some cases no plaques were observed on the recC mutant but were observed on the wild-type control strain. This spectrum of plaque sizes is expected, as mutation of recC could lead to loss of Chi activity (no or small-plaque formation by both phage) or to the ability to respond to a sequence present in both χ° and χ+76 (large-plaque formation by both phage). Most mutants retained nearly full recombination proficiency as measured following Hfr conjugation (Table S3). Chi hotspot activity in the mutants, using the normalized crosses type 1a and 1b with χ+76 and χ+D123 (Figure 3A), ranged from 1.0 to 3.9 compared to 5.0 in the wild-type control (Table S3). These results confirm the loss or reduction of Chi activity initially detected by the plaque-size assay using χ+76.
We used two criteria for selecting individual codons for further analysis. First, codon changes that arose frequently in the “patch” mutants were identified. For example, 7 of 14 isolates from mutagenesis of codon patch 37–44 in recC contained a change in codon G41. Chi activity was reduced whether this mutation was present alone or in combination with additional mutations. Second, we identified additional codons for mutagenesis following alignment of the RecC polypeptides from bacterial species with or without known or suspected Chi activity (Schultz and Smith 1986; Smith et al. 1986; McKittrick and Smith 1989). We chose amino acids with 100% identity within the RecC tunnel of species with Chi activity but not in species without suspected Chi activity. Based on these criteria, we mutagenized the eight codons noted below.
Single amino acid changes in the RecC tunnel apparently abolishing Chi activity
To test the requirement for individual amino acids in Chi activity, we mutagenized single codons of plasmid-borne recC using DNA oligonucleotides with all three randomized nucleotides at codon Q38, S39, G41, L64, K88, R644, Q652, or R708, whose encoded amino acids protrude into the tunnel (Figure 2). This randomization allowed us to screen, at each position, for any amino acid change that alters Chi activity. From each mutagenesis, we transformed a recBCD deletion strain and analyzed ∼300 transformants that were T4 2− resistant and therefore RecBCD nuclease-proficient; ∼50–65% of these isolates produced equal plaque sizes of λ χ° and χ+76 phage and were characterized further (Table 1 and Table S4). Both phage made equally large plaques on some mutants (Q38D, G, and Y; S39A, R, L, P, and W; L64I and Y; K88D; and R644D, Q, E, G, I, S, W, and V), suggesting the presence of a DNA sequence in both χ° and χ+76 phage that stimulates recombination in these RecC tunnel mutants. All of these mutants were recombination-proficient, and many had hotspot activity in normalized crosses (unpublished data; see Discussion). Other mutants, including the 25 thoroughly analyzed below, had reduced λ plaque size in the presence or absence of a Chi site, suggesting the loss of Chi activity (S39, G41, L64, K88, Q652, and R708; Table 1). This conclusion was supported by strongly reduced hotspot activity in normalized crosses (Table 1 and Table S4; Figure 3A). Recombination proficiency of these mutants in Hfr and λ crosses was only partially reduced (Table S5).
Table 1. Recombination proficiency and Chi activity of RecC tunnel mutants.
| recC codon changea | χ°/χ+76/ χ+L252 plaque sizeb | Phage λ recombination | |||
|---|---|---|---|---|---|
| χ+L crosses | |||||
| χ+76, χ+D hotspot crosses | Chi activity (c/t ratio)d | ||||
| Chi hotspot activityc | χ+L251 | χ+L252 | χ−L252 | ||
| + | S/L/L | 5.1 ± 0.2 | 3.9 ± 0.3 | 4.9 ± 0.2 | 1.9 ± 0.1 |
| recBCD null | L/L/L | 1.1 ± 0.1 | 1.4 ± 0.2 | 1.3 ± 0.3 | 1.1 ± 0.1 |
| S39E | T/T/T | 1.0 ± 0.1 | 1.5 ± 0.2 | 1.9 ± 0.2 | 1.3 ± 0.1 |
| S39V | T/T/M | 1.5 ± 0.1 | 2.3 ± 0.4 | 4.6 ± 0.4 | 1.6, 1.5 |
| G41Q | N/N/S | 0.9, 1.4 | 2.1 ± 0.3 | 2.3 ± 0.1 | 1.2, 1.4 |
| L64A | T/T/M | 1.5 ± 0.1 | 2.1 ± 0.2 | 3.3 ± 0.3 | 1.4 ± 0.2 |
| L64S | N/N/S | 1.0 ± 0.1 | 2.5 ± 0.2 | 2.9 ± 0.2 | 1.5 ± 0.3 |
| L64V | N/N/S | 1.3 ± 0.1 | 2.7 ± 0.3 | 3.4 ± 0.1 | ND |
| K88I | N/N/M | 1.6 ± 0.3 | 3.3 ± 0.1 | 4.3 ± 0.2 | 1.5 ± 0.2 |
| Q652L | T/T/T | 1.2 ± 0.1 | 2.4 ± 0.2 | 3.7 ± 0.9 | 1.3, 1.2 |
| R708D | N/N/M | 1.4 ± 0.1 | 2.9 ± 0.2 | 3.8 ± 0.2 | 1.3 ± 0.3 |
| R708V | T/T/M | 1.4 ± 0.2 | 2.8 ± 0.2 | 3.2 ± 0.2 | 1.5, 1.3 |
S, small; L, large; T, tiny; M, medium; N, no visible plaques.
Strains are transformants of strain V2831 (ΔrecBCD2371) with the indicated recC codon change on derivatives of plasmid pSA607 (recBC2773D). recBCD null is recB21 (IS186 insertion).
Relative size of isolated plaques on the indicated strain.
Chi hotspot activity for each set of crosses (λ 1081 × 1082 and λ 1083 × 1084) was determined as described (Stahl and Stahl 1977). Chi hotspot activity = √(t/c1a) / (t/c1b) where t/c1a is the ratio of turbid (c+) to clear (cI857) plaques from the λ 1081 × 1082 cross and t/c1b is the ratio of turbid to clear plaques from the λ 1083 × 1084 cross among J+ R+ recombinants in type 1 crosses (Figure 3A). Data are from two independent experiments (listed separately) or the mean ± SEM from 4–10 experiments.
Chi activity for crosses with λ χ+L phage is the ratio of clear (cI857) to turbid (c+) recombinant plaques (c/t) among J+ R+ recombinants (determined as described above) in type 2 crosses (Figure 3C).
RecC tunnel mutants reveal context-dependence of Chi hotspot activity
The results above defined several RecC tunnel amino acids involved in Chi recognition or signal transduction. We tested the hypothesis that those RecC tunnel mutants on which both χ° and χ+76 phage made no visible plaques or only tiny plaques recognize a different “Chi-like” DNA sequence not found in λ. A hotspot sequence active in these mutants could stimulate recombination and thus formation of large λ plaques. We prepared a λ library with insertions of random 25-mers at the newly-defined χJ locus in the gam gene (Figure 3B). We expected any octamer (like Chi) to occur once per ∼4000 phage, given 48 (65,536) octamers and 18 possible positions of an octamer within the insert.
A total of ∼10,000 phage from the library were plated on recBCD+ cells or four RecC tunnel mutants (L64A, Q652L, R708D, and R708V), and a few plaques (1 per ∼3000) larger than the vast majority were observed. Seven phage of independent origin found on recBCD+ cells contained Chi, as expected. Remarkably, however, all of the six phage of independent origin found on the tunnel mutants also contained Chi (Table 2). (Because of limited diversity in the library, these six were apparently sisters of the first seven found on recBCD+ cells.) This unexpected finding indicates that Chi activity is possible in some of the tunnel mutants if the Chi sequence is present at a different position in λ, possibly because of local sequence context. We did not observe larger plaques on nine additional RecC mutants tested (S39E and F; G41E, Q, and W; L64S, T, and V; Q652E), perhaps because of the rarity of such large-plaque phage and the limited number of phage plated. In addition, although we rarely observed a plaque-size increase by Chi in any context with these mutants, most do have detectable hotspot activity (Table S6), indicating that even these mutants recognize Chi (see Discussion).
To test the inference that Chi at a different position is active in certain RecC tunnel mutants, we determined plaque-formation by λ containing Chi at five other loci (χ+A, χ+C, χ+D, χ+F, and χ+G) on wild type and 20 RecC tunnel mutants. χ+C, χ+D, and χ+F phage formed significantly larger plaques on 7 of 20 tunnel mutants that allowed no or only tiny plaque-formation by χ+76 phage; as expected, all of these χ+ phage formed large plaques on wild-type recBCD+ cells. This test of Chi in additional contexts demonstrated that Chi activity in the tunnel mutants was dictated by more than the Chi octamer (5′ GCTGGTGG 3′), which was present in all phage tested.
Context-dependence of χ+L activity in wild type and RecC tunnel mutants
In order to assess more thoroughly the role of context on Chi activity in the mutant strains, we created Chi at another newly-defined locus, designated χL in gam (Figure 3B). As we reported recently (Taylor et al. 2016), the new Chi site, χ+L251, was active in recBCD+ cells with a c/t ratio of 3.9 in type 2 crosses (Table 1; Figure 3C), indicating an excess of exchanges in the interval containing χ+L. In the standard recBCD null mutant recB21, the c/t ratio was 1.4 ± 0.2, reflecting low-level RecBCD-independent recombinants about equally frequent in the two intervals left and right of χL. With χ−L251 (5′ GTTGGTGG 3′), the c/t ratio was 1.3 ± 0.1 in recBCD+ cells and 1.1 ± 0.1 in recB21, indicating little if any activity of this Chi-minus allele. Remarkably, most of the RecC tunnel mutants had c/t ratios with χ+L251 that were noticeably higher (2.1–3.3) than that of recB21 (1.4; Table 1 and Table S6), indicating that in these RecC tunnel mutants χ+L251 was active, albeit less than in recBCD+ cells, and RecBCD dependent.
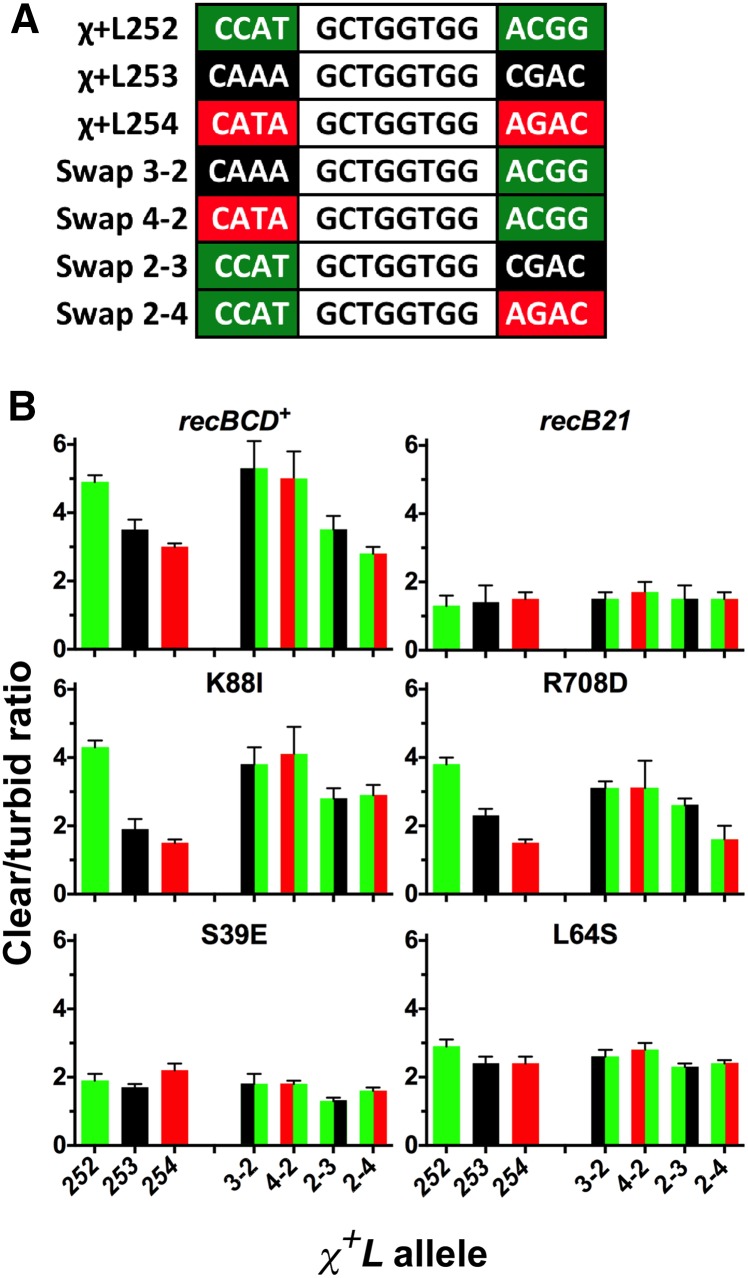
We tested the idea that the immediately surrounding nucleotide context affects Chi hotspot activity of χ+L251 by randomizing 4 bp on each side of the Chi octamer. For this analysis, we chose three representative χL alleles with different nucleotide sequences flanking the Chi octamer (χ+L252, χ+L253, and χ+L254; Table 2). On wild type and on the recC tunnel mutants tested (L64A, K88I, and R708D), χ+L252 formed the largest plaques, χ+L253 formed intermediate-sized plaques, and χ+L254 formed the smallest plaques. All three phages made plaques much larger than those of χ−L252. These results indicate that plaque size and presumably Chi activity are influenced by the nucleotides surrounding Chi, since all phage had Chi at the same position in λ but differed only in the 4 bp flanking each side.
To quantify these results, we determined the Chi activity (c/t ratio) from type 2 crosses (Figure 3C) with these four Chi alleles in recBCD+ cells and the RecC tunnel mutants (Figure 4; Table 1 and Table S6). In recBCD+ cells the c/t ratio of χ+L252 was 4.9, higher than that for χ+L251 (3.9); for χ+L253 and χ+L254 the c/t ratios were 3.5 and 3.0, respectively, corresponding to the order of their plaque sizes. χ+L252 gave the highest Chi activity in most of the RecC tunnel mutants; the c/t ratios decreased for the other Chi alleles in the same order as in recBCD+ cells (χ+L252 > χ+L253 > χ+L254), again corresponding to the order of plaque sizes. High Chi activities (c/t ratios ≳ 3) were frequent and observed in 16 of the 25 mutants tested with codon changes in S39, G41, L64, W70, K88, D136, Q652, and R708 (Table S6 and Figure 4). The Chi activity of three tunnel mutants (S39V, K88I, and R708D) was similar to that in wild type for χ+L252 despite the very low Chi hotspot activity in normalized crosses with χ+76 and χ+D123 in these mutants (Table 1), confirming the locus (context) dependence of Chi activity in these RecC tunnel mutants. Of the tunnel mutants analyzed, S39E had the weakest Chi activity: the c/t ratios from crosses in this mutant with seven of eight Chi alleles were not significantly different from those of two alleles with a mutation in the Chi octamer (χ−L252 and χ−L259; Figure 4; Table 1, Table S6, and Table S7; see below).
Figure 4.
Context-dependence of Chi activity in wild type (recBCD+), RecBCD null (recB21), and RecC tunnel mutants. (A) Diagrammatic representation of χ+L alleles and swaps of flanking bp tested for context-dependence. The Chi octamer, common to all alleles, is shown in black font. The origin of the 4 bp 5′ and 3′ of the octamer is indicated by the last digit of the allele number separated by a hyphen and the bar colors. For example, χ+L252 (green) with the 3′ context of χ+L254 (red) is indicated as 2-4 (green/red). (B) Chi activity of the indicated strains. Strains are transformants of V2831 (ΔrecBCD::kan) with the indicated rec alleles or amino acid changes in RecC carried on derivatives of plasmid pSA607 (RecBC2773D). Chi activity, measured as the ratio of clear to turbid plaques from type 2 crosses (Figure 3C), is shown for the indicated χ+L alleles. Bar colors represent the source of the 4 bp flanking Chi as described above. Data represent the mean ± SEM for four to eight crosses. Data for recBCD+ and recB21 are from Taylor et al. (2016).
Nucleotide context 3′ of χ+L influences Chi activity in wild type and RecC tunnel mutants
Our results above suggest that some or all of the 8 bp flanking the Chi octamer strongly influence Chi activity. We therefore randomized the 4 bp adjacent to either the 5′ side or the 3′ side of χ+L252 (largest plaques on many mutants) and χ+L254 (smallest plaques on many mutants) and tested the resultant phage for Chi activity by plaque size. A total of 10–25 phage for each of the four randomizations were sequenced, and their plaque sizes, with parental χ+L252 and χ+L254 controls, were determined on wild type and RecC tunnel mutants (unpublished data). Alteration of the 5′ context of χ+L252 or χ+L254 had no significant effect on plaque size on recBCD+ and three recC tunnel mutants (L64A, K88I, and R708D). Alteration of the 3′ context of χ+L252 and χ+L254, however, had a dramatic effect on plaque size. For example, the plaques of χ+L254 phage were tiny to small on R708D and K88I, but one-third (4 of 12) of the nucleotide changes 3′ of the Chi octamer resulted in formation of larger plaques. Conversely, χ+L252 phage formed larger plaques on L64A, K88I, and R708D, and about one-third (9 of 25) of the 3′ bp changes resulted in formation of much smaller plaques. These data indicate that sequences flanking Chi on the 3′ side influence the activity of χ+L, although no correlations between nucleotide sequences and plaque size were evident from this limited analysis (see below).
We extended this analysis in a recent report (Taylor et al. 2016) and found that swapping the 3′, but not 5′, flanking 4 bp of the three χ+L alleles studied above changed Chi activity in wild-type recBCD+ cells. For example, putting the 3′ flank of χ+L252 (more active) at χ+L254 (less active) increased the activity of χ+L254 and, conversely, putting the 3′ flank of χ+L254 (less active) at χ+L252 (more active) reduced the activity of χ+L252. These results were found in both recBCD+ cells (Figure 4) and most tunnel mutants (Figure 4; Table S6). For example, in K88I the c/t ratio increased from 1.9 in χ+L253 to 3.8 in χ+L253 with the 3′ flank of χ+L252; similarly, the c/t ratio increased from 1.5 in χ+L254 to 4.1 in χ+L254 with the 3′ flank of χ+L252. Conversely, in K88I the c/t ratio was reduced from 4.3 in χ+L252 to 2.8 in χ+L252 with the 3′ flank of χ+L253 and to 2.9 in χ+L252 with the 3′ flank of χ+L254. For all of these comparisons, P < 0.05 (unpaired t-test; Table S7). Thus, K88I behaves similarly to wild type for this set of Chi sites, even though K88I has little if any activity with χ+76 and χ+D153 (Table 1). Similar results were seen with tunnel mutants altered at S39V, L64V, and R708D (Figure 4; Table S6).
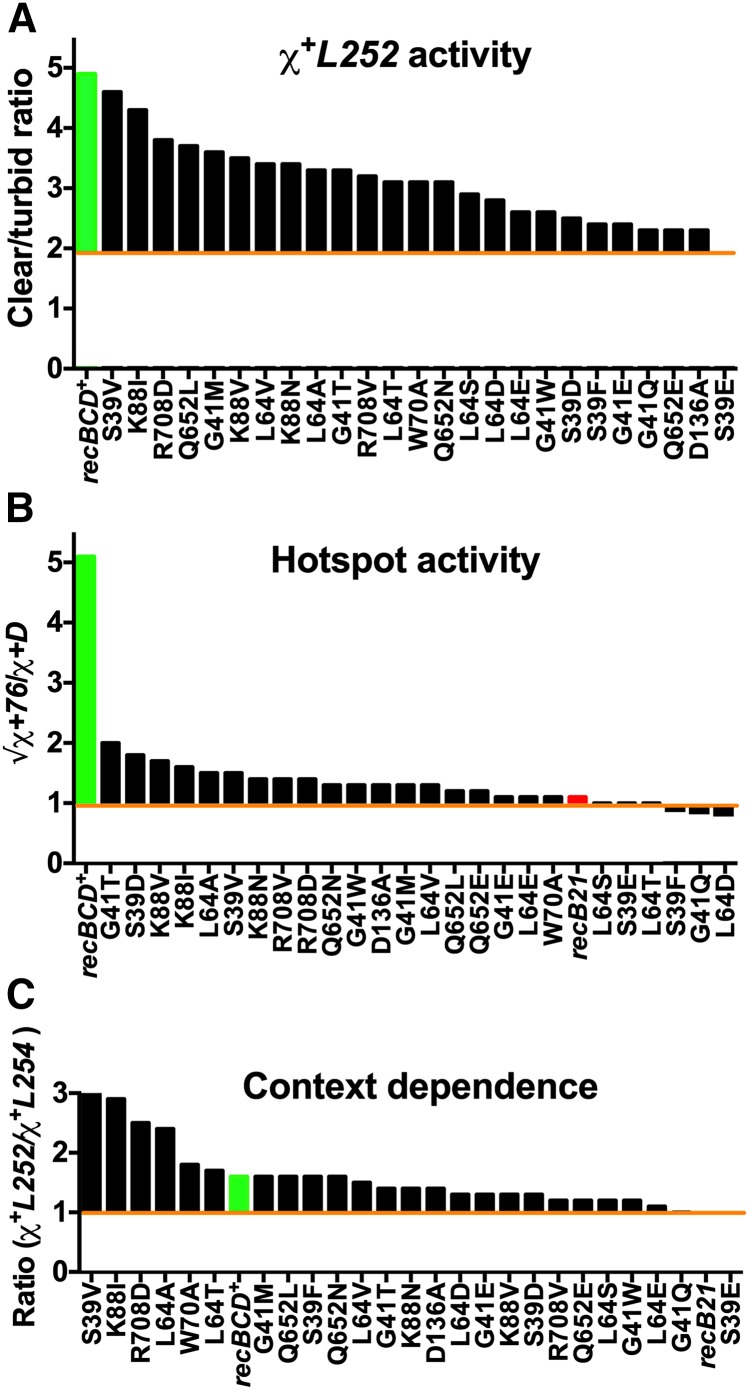
A continuous spectrum of Chi activity and Chi context-dependence in RecC tunnel mutants
To determine if the RecC tunnel mutants fell into distinct classes, perhaps from alteration of distinct, separable ways of interrupting the RecBCD-Chi interaction, we evaluated two parameters for each mutant and wild-type RecBCD. First is Chi activity (the ability of each enzyme to respond to Chi), assayed here as the c/t ratio of eight χ+L alleles in crosses of type 2, and as the hotspot activity from normalized crosses of types 1a and 1b with χ+76 and χ+D123 (Figure 3A). For normalized crosses (type 1), 1 indicates no Chi hotspot activity (Stahl and Stahl 1977); for nonnormalized (type 2) crosses, the c/t ratios in the recBCD null mutant recB21 were ∼1.3, indicating no Chi activity. In both types of crosses the highest values were ∼5–5.5, indicating strong Chi activity (Table 1 and Table S6). Second is Chi context-dependence (the effect of flanking sequence context on Chi activity), assayed here as the ratio of the activities (c/t) of two χ+L alleles in crosses of type 2; values ranged from ∼3, indicating strong context-dependence, to 1, indicating no context-dependence (Table S8).
When we ranked the 25 recC mutants according to their activity with the most active χ+L allele used above, χ+L252, we found that the Chi activities fell into a continuum with no apparent break points that would suggest classes of mutants (Figure 5A). The highest Chi activity (4.9) was observed with recBCD+ cells, not significantly different from that of the recC S39V mutant (4.6) (P = 0.23; unpaired t-test). The lowest Chi activity (1.9) was observed with the S39E mutant, not significantly different from that of the recB21 null mutant (1.3) (P = 0.22), as noted above. Between these two extremes there were recC mutants with nearly every activity. The Chi hotspot activity (in normalized crosses) of these mutants also formed a continuum (Figure 5B), although the hotspot activity of the most active mutant G41T (2.0) was much less than that of wild type (5.1). This behavior likely reflects the low activity of χ+76 in the RecC tunnel mutants, which was used to isolate the mutants, and perhaps of χ+D123 as well (see Discussion). The hotspot activities of several mutants, such as S39E, were not significantly different from that of the recB21 null mutant, which has no Chi activity (Stahl and Stahl 1977). Ranked according to activity of other χ+L alleles, the mutants also formed a continuum (Table S6).
Figure 5.
Chi activity and its context dependence in RecC tunnel mutants vary along a continuum. Data are from Table 1, Table S4, and Table S6. (A) The c/t ratios (Chi activity) from type 2 crosses (Figure 3C) with χ+L252 are shown for the indicated RecC tunnel mutants. The c/t ratios in recBCD+ for χ+L252 (4.9; green bar) and for χ−L252 (1.9; orange line) are shown for comparison. (B) Weak Chi hotspot activity of the RecC tunnel mutants in normalized χ+D123 and χ+76 hotspot crosses (Figure 3A, type 1 crosses; Table 1). Orange line at 1.0 indicates no Chi hotspot activity. (C) Context dependence (ratio of the c/t ratios from crosses with χ+L252 vs. χ+L254) varies from strong (∼3 in some tunnel mutants) to medium (∼1.5) in recBCD+ and other tunnel mutants, to low (∼1) in recBCD null and other tunnel mutants. Orange line at 1.0 indicates no Chi context-dependence. Data for recBCD+ and recB21 are from Taylor et al. (2016).
We observed similar continua when we ranked the mutants according to their context-dependence. For example, the c/t ratio for the most active χ+L allele (χ+L252) divided by that of the least active (χ+L254) ranged from 3.1 for S39V (strong context-dependence) to 0.9 for S39E, not significantly different from that of the recB21 null mutant (no apparent context-dependence) (Figure 5C). (We note that there can be no context-dependence of an inactive Chi site; see Discussion.) There was a suggestion of two classes in Figure 5C defined by the difference between L64A (2.4) and W70A (1.8), but this difference was not statistically significant (P = 0.28). Ranked according to the ratios of other pairs of χ+L alleles, the mutants also formed continua, with maximal context-dependence as high as or higher than that of wild-type RecBCD (∼3) and minimal values near that of recB21 (∼1; Table S6).
We noted that several mutants ranked high by both Chi hotspot activity and context-dependence and some low by both criteria. For example, S39V, K88I, and R708D had the highest Chi activity assayed by the c/t ratio with χ+L252, and S39E and G41Q had about the lowest (Figure 5A). These two sets of RecC tunnel mutants also had the highest and lowest context-dependence assayed by the ratio of activities of χ+L252 and χ+L254 (Figure 5C). Some mutants, however, had moderately strong Chi activity but little context-dependence. For example, L64S, L64D, and G41W were in the middle of the c/t ratio rankings from crosses with χ+L252 and χ+L253 but had low context-dependence ranking (Chi activity ratios ranging from ∼1 to ∼1.4).
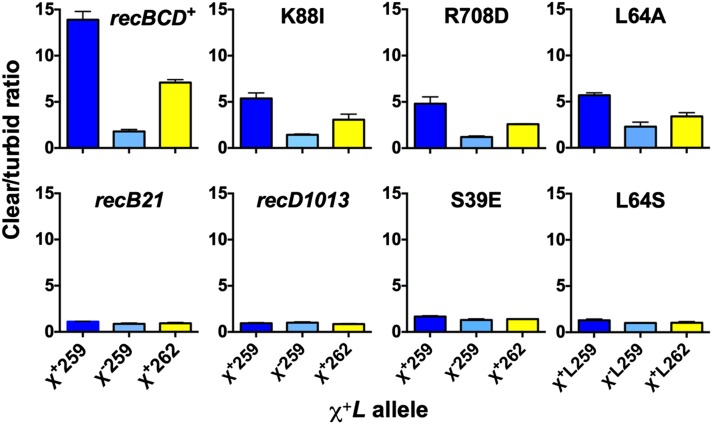
RecC tunnel mutants manifest a range of activities with exceptionally strong Chi alleles
We recently reported that the Chi octamer flanked by certain 3′ flanking sequences has Chi activity in recBCD+ cells even greater than that of the Chi alleles used above (Taylor et al. 2016). We tested two of these Chi alleles, χ+L259 and χ+L262, in five RecC tunnel mutants that manifested high or low Chi activity with the χ+L252 allele used above and found qualitatively similar responses. For example, with χ+L259, tunnel mutants K88I and R708D had c/t ratios in type 2 crosses of 5.4 and 4.8, respectively (Figure 6; Table S9), even higher than they had with χ+L252 (4.3 and 3.8, respectively; Table 1 and Table S6; Figure 4). L64A behaved similarly. These high Chi activities depend on the Chi octamer, because the χ−L259 mutation gave a low c/t ratio in all cases. Similarly, two RecC tunnel mutants, S39E and L64S, which had low activity with χ+L252, had negligible activity with χ+L259 (1.7 and 1.3, respectively), only slightly greater than with the χ−L259 Chi octamer mutant (1.3 and 1.0, respectively). Another Chi allele, χ+L262, with high activity in recBCD+ cells also had strong activity in the K88I, R708D, and L64A mutants but essentially no activity in S39E and L64S. Thus, these last two mutants had little or no Chi activity in any case tested, whereas the others had activity in all cases tested except with χ+76, used to isolate the mutants, or (for L64A) with the translocated χ+C locus (Handa et al. 2012).
Figure 6.
Activity of strong Chi alleles in RecC tunnel mutants. Shown are results of type 2 crosses (Figure 3C) with two χ+L alleles that have the same 5′ flanking sequence as χ+L251 but different 3′ flanks (Table 2), which confer high Chi activity in recBCD+ cells (Taylor et al. 2016). The Chi octamer mutant χ−L259 was used as a control, as were the recBCD null mutant recB21 and the recD1013 mutant, in both of which Chi is inactive (Stahl and Stahl 1977; Amundsen et al. 1986). Data for recBCD+ and recB21 are from Taylor et al. (2016).
Discussion
To test the hypothesis that the recombination hotspot Chi is recognized by a tunnel in the RecC subunit (Figure 2), we sought recC tunnel missense mutants that do not respond to Chi. Unexpectedly, during this study we found that the activity of Chi strongly depends on the nucleotide sequences flanking the Chi octamer 5′ GCTGGTGG 3′. Changing these flanking nucleotides changed Chi activity by factors as great as the stimulation of recombination by Chi itself, about a factor of five. Our study of recC tunnel mutants and of Chi’s flanking sequences demonstrates a complex interaction between these two parts of Chi and, we propose, two domains of RecBCD (the RecC tunnel and the RecB nuclease domain) that are critical for proper DNA break repair and genetic recombination. Here, we discuss the implications of our results for understanding how RecBCD enzyme is regulated by Chi hotspots.
Analysis of RecBCD mutants reveals an additional feature of Chi recombination hotspots: context-dependence of the Chi octamer
As noted in the Introduction, Chi was equated with 5′ GCTGGTGG 3′ based on seven thoroughly analyzed Chi sites having only this octamer in common and mutations creating or inactivating Chi falling exclusively in this octamer (Smith et al. 1984). Synthetic octamer inserted into DNA gives a substrate that purified RecBCD enzyme readily cuts at Chi and onto which it loads RecA protein (Dixon and Kowalczykowski 1991; Anderson and Kowalczykowski 1997). We are unaware, however, of such synthetic Chi sites being previously tested for recombination hotspot activity in living cells, although synthetic Chi on a linear DNA fragment stimulates transformation of E. coli cells, and on a circular plasmid it stimulates formation of high molecular weight DNA (Dabert et al. 1992; Dabert and Smith 1997). It remained possible, however, that Chi in a given context is active with purified proteins but not as a recombination hotspot in cells. For example, a Chi-related sequence (5′ GCTGGTGCTCG 3′), denoted χ*, weakly stimulates cutting of DNA and loading of RecA protein by the purified RecBC1004D pseudorevertant mutant and wild-type RecBCD enzymes, but χ* has no detectable recombination hotspot activity in cells, either recBCD+ or recC1004, a frameshift pseudorevertant with eight amino acid changes in the RecC tunnel (Schultz et al. 1983; Handa et al. 1997; Arnold et al. 2000). With this exception, the published data were consistent with both the genetic and biochemical activities of Chi being determined simply by 5′ GCTGGTGG 3′, but our observations reported here force a revision of this view.
The precise effect of the sequence context on Chi activity remains to be determined, but our data show that the flanking 3′ bp are more influential than the flanking 5′ bp. This conclusion is supported both by our randomization of bp on each side of the octamer and by our swapping of flanking bp between more active χ+L alleles and less active χ+L alleles (Figure 4). As predicted, swapping the 3′, but not the 5′, flanks of active alleles increased the activity of less active alleles and vice versa, and randomization of the 3′ flanks revealed more changes affecting Chi activity than randomization of the 5′ flanks. Roughly one-third of the 80 randomizations examined had detectable effects, demonstrating that multiple flanking sequences influence Chi activity, but we were unable to discern a pattern in these sequences indicating the essential features. Our recent analysis of vast numbers of sequences flanking the octamer revealed the importance of bp 4–7 on the 3′ flank for activation of RecBCD’s nicking activity at Chi (Taylor et al. 2016). At these positions, purines appear to be favored over pyrimidines. We used one of these sequences in χ+L259 (5′ GCTGGTGGTCGAAAA 3′), an exceptionally “hot” Chi allele (Figure 6), to demonstrate that some RecC tunnel mutants, but perhaps not others, retain Chi recognition, as discussed below. The complete rules of how the sequences flanking Chi determine Chi’s activity, however, remain to be elucidated.
Previously reported RecC Chi-recognition mutants also retain Chi activity
We began this study with the goal of determining the amino acids in the RecC tunnel that are required for Chi recognition. Some of the recC mutants we generated allowed large-plaque formation by λ without Chi, and we surmised that these RecC mutant enzymes recognize a new hotspot with a sequence other than 5′ GCTGGTGG 3′ and present in the 48.5 kb of λ DNA. Handa et al. (2012) drew the same conclusion from a study of other RecC tunnel mutants with amino acids changed to alanine and their interaction with Chi. The possibility that their and our mutants of this type recognize a new hotspot sequence remains to be tested because of the lack of an easy assay for novel Chi sites with activity in these recC mutants. Some other recC mutants, however, allowed only small or no plaque formation, which provided a simple way to screen or select for Chi-like sequences active in these recC mutants. To our surprise, all of the six independent sequences isolated contained Chi (5′ GCTGGTGG 3′) but obviously at a locus different from those used previously (χ+76 and χ+D123), which were only weakly active in these mutants. This result implied a context-dependence of Chi’s activity, which was demonstrated by our subsequent experiments reported here and by Taylor et al. (2016).
But these observations also showed that the recC mutants we had isolated were not bona fide Chi recognition mutants. Instead, they were Chi context-dependence mutants: their phenotype depended on the Chi site used for assay. After we came to this conclusion, Handa et al. (2012) reported 11 recC mutants with reduced Chi activity, each with alanine in place of the wild-type amino acid, which they concluded lacked Chi recognition or had relaxed Chi specificity. Their study, however, used Chi at only one locus: transplacement of χ+C157 and its surrounding DNA into the λ int gene. We tested three of the recC mutants they reported (L64A, W70A, and D136A) and found that each in fact has Chi activity with χ+L252 (Table 1 and Table S6). Although we have not tested all of the mutants they reported, the high frequency of Chi context-dependence mutants isolated in our screen, which is similar to that of Handa et al. (2012), suggests that many, and perhaps most or all, of their mutants are Chi context-dependent mutants. Until a putative Chi recognition recBCD mutant is tested with a large number of sequences flanking Chi, it will remain unclear if the mutant genuinely lacks Chi activity.
Some of the mutants we isolated had in fact little or no activity with most of the Chi sites in the 25 different contexts that we tested. For example, S39E was not significantly different from the recBCD null mutant recB21 in its response to Chi in seven of eight contexts tested (Table S6). This mutant may in fact lack Chi recognition, but it may also have Chi activity that depends on a context that we did not test. This possibility complicates the interpretation of mutants that are thought to lack Chi recognition, as noted above.
An additional complication is the lack of a direct assay for Chi recognition. The available assays measure the response to Chi, which involves both recognition of Chi and transduction of the Chi-generated signal to the relevant active site in RecBCD enzyme (Figure 1B; Amundsen et al. 2007). Because RecBCD responds to Chi only as it is unwinding DNA rapidly (∼1 kb/sec) (Ponticelli et al. 1985; Taylor et al. 1985), no direct assay, such as binding to Chi, is available. It might, however, be possible to incorporate a moiety at Chi that would emit a signal, such as fluorescence, when productively encountered by RecBCD enzyme without the necessity of further changes to RecBCD.
Several other RecC tunnel mutants were particularly illuminating. The R708D mutant blocked plaque-formation by λ red gam χ+76 phage but allowed plaque-formation by λ red gam χ+L252 phage (Table 1). Consistent with these observations, R708D had moderately high Chi activity with χ+L252 (c/t = 3.8 compared to 4.9 with wild-type recBCD+) but low hotspot activity in normalized hotspot crosses using χ+76 and χ+D123 (1.4 compared to 5.1 with wild type). R708D also had high context-dependence as assayed by the ratio of activity with χ+L252 to that with χ+L254 (2.5), even higher than that of wild-type RecBCD (1.6) (Table S4, Table S6, and Table S8; Figure 4 and Figure 5). S39V and K88I behaved similarly. Thus, we might not have detected the R708D and R708V mutants if we had used χ+L252 rather than χ+76 in our screen. This outcome emphasizes the need to expand the spectrum of Chi sites used to deduce the parts of RecBCD required for Chi recognition.
The L64V and K88V mutants had moderately high Chi activity with χ+L252 (3.4 and 3.5, respectively) and with χ+L254 (2.2 and 2.8, respectively), and consequently low context-dependences by the ratio of these two activities (1.5 and 1.3, respectively) (Table S6 and Table S8; Figure 5). Yet in normalized crosses with χ+76 and χ+D123 these mutants had much lower hotspot activities (1.3 and 1.7, respectively) than that in recBCD+ (5.1) (Table 1 and Table S4; Figure 5B). These mutants also illustrate differential activity of Chi in different contexts, i.e., Chi context-dependence.
Molecular basis of RecBCD’s recognition of Chi and its context-dependence
Current evidence indicates that Chi is recognized as the ss DNA with 5′ GCTGGTGG 3′ during its passage through the RecC tunnel (Figure 2; Smith 2012; Wigley 2013). Amino acids lining the tunnel are thought to contact specific bases of the DNA; the correct fit presumably sends a signal to the other RecBCD subunits to alter RecBCD’s activity (Figure 1B). We report here that this signaling requires more than 5′ GCTGGTGG 3′: additional nucleotides on the 3′ side of this octamer strongly influence the activity of Chi. The context-dependence of wild-type RecBCD is as high as a factor of 5.0 in genetic crosses (Figure 4 and Figure 6; Table 1, Table S6, and Table S9) and 7.4 in enzymatic nicking of DNA (Taylor et al. 2016), i.e., as high as or higher than Chi hotspot activity itself (Stahl and Stahl 1977). These nucleotides may extend into the RecB nuclease domain, which is at the exit of the RecC tunnel in the crystal structure (Figure 1 and Figure 2; Singleton et al. 2004); if so, Chi may be recognized by more than the RecC tunnel. For example, the RecB nuclease domain may recognize the 3′ flank but have a less stringent sequence requirement than the RecC tunnel has for the Chi octamer. Our recent report supports this interpretation (Taylor et al. 2016).
It is interesting that a previously reported RecBCD-Chi mutant combination also requires additional nucleotides on the 3′ side for alteration of RecBCD enzyme activities. The first reported mutants that blocked Chi activation, but retained RecBCD enzymatic activities and recombination proficiency, were pseudorevertants of the recC73 null allele, as noted in the Introduction (Schultz et al. 1983). One of these mutants, recC1004, has no detectable activity with standard Chi sites in λ (χ+76, χ+C157, and χ+D123) (Schultz et al. 1983) but does have partial activity with the χ* sequence 5′ GCTGGTGCTCG 3′ noted above (Handa et al. 1997; Arnold et al. 2000). This sequence confers large-plaque formation and increased burst size of λ red gam phage in both recC1004 mutant and recBCD+ cells, but it does not produce recombination hotspot activity detectable in either strain. It alters the nuclease activity of RecBCD in a way that may allow rolling-circle replication and greater phage yield, and it increases the RecA-loading activity of RecBCD, which is thought to be a function of the RecB nuclease domain (Churchill and Kowalczykowski 2000; Spies and Kowalczykowski 2006); the absence of hotspot activity in χ* mutant cells remains a mystery, however. The putative effects on RecB are consistent with our suggestion above that the 3′ flank of Chi is recognized by RecB. In nearly all of the recC mutants reported here, Chi in the correct context does have hotspot activity (Figure 4; Table S6).
Previous reports of mutants with an altered RecC tunnel of RecBCD, or the corresponding AddB tunnel in AddAB of Bacillus subtilis, have interpreted the alterations as interfering with the recognition of Chi (Handa et al. 2012; Saikrishnan et al. 2012; Yang et al. 2012; Wilkinson and Wigley 2014). For RecBCD, the crystal structure of the enzyme with ss DNA in the presumed Chi recognition tunnel has not been reported, but the RecC tunnel mutants were interpreted in terms of specific contacts between amino acid side chains and bases in the Chi octamer. Our results with three of these RecC tunnel mutants (L64A, W70A, and D136A) show that they still recognize Chi but respond to it only in the right context (Table 1, Table S4, and Table S6). Thus, these mutants warrant reinterpretation. Some or all of the other previously reported mutants may also be Chi context-dependence, not Chi recognition, mutants. As noted above, studies employing Chi in additional contexts are required to distinguish these possibilities.
Acknowledgments
We are grateful to Nishka Mittal for isolation of the recC R708 mutants; Knox Young for generating new Chi alleles; Phil Bradley, Kyle Fowler, Randy Hyppa, and Sarah Zanders for helpful discussions; and Meriem El Karoui, David Leach, Mridula Nambiar, Walt Steiner, Andrew Taylor, Jeetu Thakur, and anonymous reviewers for helpful comments on the manuscript. This research was supported by grant GM-031693 from the National Institute of General Medical Sciences to G.R.S.
Footnotes
Communicating editor: S. J. Sandler
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.191056/-/DC1.
Literature Cited
- Amundsen S. K., Taylor A. F., Chaudhury A. M., Smith G. R., 1986. recD: The gene for an essential third subunit of exonuclease V. Proc. Natl. Acad. Sci. USA 83: 5558–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundsen S. K., Taylor A. F., Reddy M., Smith G. R., 2007. Intersubunit signaling in RecBCD enzyme, a complex protein machine regulated by Chi hot spots. Genes Dev. 21: 3296–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundsen S. K., Spicer T., Karabulut A. C., Londono L. M., Eberhardt C., et al. , 2012. Small-molecule inhibitors of bacterial AddAB and RecBCD helicase-nuclease DNA repair enzymes. ACS Chem. Biol. 7: 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. G., Kowalczykowski S. C., 1997. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ regulated manner. Cell 90: 77–86. [DOI] [PubMed] [Google Scholar]
- Arnold D. A., Handa N., Kobayashi I., Kowalczykowski S. C., 2000. A novel, 11 nucleotide variant of χ, χ*: One of a class of sequences defining the Escherichia coli recombination hotspot χ. J. Mol. Biol. 300: 469–479. [DOI] [PubMed] [Google Scholar]
- Bianco P. R., Kowalczykowski S. C., 1997. The recombination hotspot χ is recognized by the translocating RecBCD enzyme as the single strand of DNA containing the sequence 5′-GCTGGTGG-3′. Proc. Natl. Acad. Sci. USA 94: 6706–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K. C., Smith G. R., 1989. Distribution of Chi-stimulated recombinational exchanges and heteroduplex endpoints in phage lambda. Genetics 123: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill J. J., Kowalczykowski S. C., 2000. Identification of the RecA protein-loading domain of RecBCD enzyme. J. Mol. Biol. 297: 537–542. [DOI] [PubMed] [Google Scholar]
- Dabert P., Smith G. R., 1997. Gene replacement in wild-type Escherichia coli: enhancement by Chi sites. Genetics 145: 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabert P., Ehrlich S. D., Gruss A., 1992. χ sequence protects against RecBCD degradation of DNA in vivo. Proc. Natl. Acad. Sci. USA 89: 12073–12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon D. A., Kowalczykowski S. C., 1991. Homologous pairing in vitro stimulated by the recombination hotspot, Chi. Cell 66: 361–371. [DOI] [PubMed] [Google Scholar]
- Faulds D., Dower N., Stahl M. M., Stahl F. W., 1979. Orientation-dependent recombination hotspot activity in bacteriophage λ. J. Mol. Biol. 131: 681–695. [DOI] [PubMed] [Google Scholar]
- Gillen J. R., Clark A. J., 1974. The RecE pathway of bacterial recombination, pp. 123–136 in Mechanisms in Recombination, edited by Grell R. F. Plenum, New York. [Google Scholar]
- Handa N., Ohashi S., Kusano K., Kobayashi I., 1997. χ*, a χ-related 11-mer sequence partially active in an E. coli recC* strain. Genes Cells 2: 525–536. [DOI] [PubMed] [Google Scholar]
- Handa N., Yang L., Dillingham M. S., Kobayashi I., Wigley D. B., et al. , 2012. Molecular determinants responsible for recognition of the single-stranded DNA regulatory sequence, χ, by RecBCD enzyme. Proc. Natl. Acad. Sci. USA 109: 8901–8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson D., Weil J., 1975. Recombination-deficient deletions in bacteriophage lambda and their interaction with Chi mutations. Genetics 79: 143–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh S., Julin D. A., 1992. Alteration by site-directed mutagenesis of the conserved lysine residue in the consensus ATP-binding sequence of the RecB protein of Escherichia coli. Nucleic Acids Res. 20: 5647–5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi I., Murialdo H., Crasemann J. M., Stahl M. M., Stahl F. W., 1982. Orientation of cohesive end site cos determines the active orientation of χ sequence in stimulating recA•recBC-mediated recombination in phage λ lytic infections. Proc. Natl. Acad. Sci. USA 79: 5981–5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S. T., Stahl M. M., McMilin K. D., Stahl F. W., 1974. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics 77: 425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKittrick N. H., Smith G. R., 1989. Activation of Chi recombinational hotspots by RecBCD-like enzymes from enteric bacteria. J. Mol. Biol. 210: 485–495. [DOI] [PubMed] [Google Scholar]
- McMilin K. D., Stahl M. M., Stahl F. W., 1974. Rec-mediated recombinational hot spot activity in bacteriophage lambda. I. Hot spot activity associated with spi- deletions and bio substitutions. Genetics 77: 409–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D. B., Goldberg E. B., 1977. Protection of parental T4 DNA from a restriction exonuclease by the product of gene 2. J. Mol. Biol. 116: 877–881. [DOI] [PubMed] [Google Scholar]
- Ponticelli A. S., Schultz D. W., Taylor A. F., Smith G. R., 1985. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell 41: 145–151. [DOI] [PubMed] [Google Scholar]
- Saikrishnan K., Griffiths S. P., Cook N., Court R., Wigley D. B., 2008. DNA binding to RecD: role of the 1B domain in SF1B helicase activity. EMBO J. 27: 2222–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikrishnan K., Yeeles J. T., Gilhooly N. S., Krajewski W. W., Dillingham M. S., et al. , 2012. Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex. EMBO J. 31: 1568–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D. W., Smith G. R., 1986. Conservation of Chi cutting activity in terrestrial and marine enteric bacteria. J. Mol. Biol. 189: 585–595. [DOI] [PubMed] [Google Scholar]
- Schultz D. W., Swindle J., Smith G. R., 1981. Clustering of mutations inactivating a Chi recombinational hotspot. J. Mol. Biol. 146: 275–286. [DOI] [PubMed] [Google Scholar]
- Schultz D. W., Taylor A. F., Smith G. R., 1983. Escherichia coli RecBC pseudorevertants lacking Chi recombinational hotspot activity. J. Bacteriol. 155: 664–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton M. R., Dillingham M. S., Gaudier M., Kowalczykowski S. C., Wigley D. B., 2004. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432: 187–193. [DOI] [PubMed] [Google Scholar]
- Smith G. R., 1991. Conjugational recombination in E. coli: Myths and mechanisms. Cell 64: 19–27. [DOI] [PubMed] [Google Scholar]
- Smith G. R., 2012. How RecBCD and Chi promote DNA break repair and recombination: a molecular biologist’s view. Microbiol. Mol. Biol. Rev. 76: 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R., Kunes S. M., Schultz D. W., Taylor A., Triman K. L., 1981a Structure of Chi hotspots of generalized recombination. Cell 24: 429–436. [DOI] [PubMed] [Google Scholar]
- Smith, G. R., D. W. Schultz, A. F. Taylor, and K. Triman, 1981b Chi sites, RecBC enzyme, and generalized recombination. Stadler Genetics Symposium 13: 25–37. [Google Scholar]
- Smith G. R., Amundsen S. K., Chaudhury A. M., Cheng K. C., Ponticelli A. S., et al. , 1984. Roles of RecBC enzyme and Chi sites in homologous recombination. Cold Spring Harb. Symp. Quant. Biol. 49: 485–495. [DOI] [PubMed] [Google Scholar]
- Smith G. R., Roberts C. M., Schultz D. W., 1986. Activity of Chi recombinational hotspots in Salmonella typhimurium. Genetics 112: 429–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies M., Kowalczykowski S. C., 2006. The RecA binding locus of RecBCD is a general domain for recruitment of DNA strand exchange proteins. Mol. Cell 21: 573–580. [DOI] [PubMed] [Google Scholar]
- Sprague K. U., Faulds D. H., Smith G. R., 1978. A single base-pair change creates a Chi recombinational hotspot in bacteriophage lambda. Proc. Natl. Acad. Sci. USA 75: 6182–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl F. W., Stahl M. M., 1977. Recombination pathway specificity of Chi. Genetics 86: 715–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl F. W., Crasemann J. M., Stahl M. M., 1975. Rec-mediated recombinational hot spot activity in bacteriophage lambda. III. Chi mutations are site-mutations stimulating Rec-mediated recombination. J. Mol. Biol. 94: 203–212. [DOI] [PubMed] [Google Scholar]
- Taylor A. F., Schultz D. W., Ponticelli A. S., Smith G. R., 1985. RecBC enzyme nicking at Chi sites during DNA unwinding: Location and orientation-dependence of the cutting. Cell 41: 153–163. [DOI] [PubMed] [Google Scholar]
- Taylor A. F., Amundsen S. K., Guttman M., Lee K. K., Luo J., et al. , 2014. Control of RecBCD enzyme activity by DNA binding- and Chi hotspot-dependent conformational changes. J. Mol. Biol. 426: 3479–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A. F., Amundsen S. K., Smith G. R., 2016. Unexpected DNA context-dependence identifies a new determinant of Chi recombination hotspots. Nucleic Acids Res. DOI: 10.1093/nar/gkw541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason L. C., Oppenheim A. B., Court D. L., 2009. Modifying bacteriophage lambda with recombineering. Methods Mol. Biol. 501: 239–251. [DOI] [PubMed] [Google Scholar]
- Wigley D. B., 2013. Bacterial DNA repair: recent insights into the mechanism of RecBCD, AddAB and AdnAB. Nat. Rev. Microbiol. 11: 9–13. [DOI] [PubMed] [Google Scholar]
- Wilkinson M., Wigley D. B., 2014. Structural features of Chi recognition in AddAB with implications for RecBCD. Cell Cycle 13: 2812–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Handa N., Liu B., Dillingham M. S., Wigley D. B., et al. , 2012. Alteration of χ recognition by RecBCD reveals a regulated molecular latch and suggests a channel-bypass mechanism for biological control. Proc. Natl. Acad. Sci. USA 109: 8907–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Souaya J., Julin D. A., 1998. The 30-kDa C-terminal domain of the RecB protein is critical for the nuclease activity, but not the helicase activity, of the RecBCD enzyme from Escherichia coli. Proc. Natl. Acad. Sci. USA 95: 981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article and Supplemental Material.