Abstract
With high productivity and stress tolerance, numerous grass genera of the Andropogoneae have emerged as candidates for bioenergy production. To optimize these candidates, research examining the genetic architecture of yield, carbon partitioning, and composition is required to advance breeding objectives. Significant progress has been made developing genetic and genomic resources for Andropogoneae, and advances in comparative and computational genomics have enabled research examining the genetic basis of photosynthesis, carbon partitioning, composition, and sink strength. To provide a pivotal resource aimed at developing a comparative understanding of key bioenergy traits in the Andropogoneae, we have established and characterized an association panel of 390 racially, geographically, and phenotypically diverse Sorghum bicolor accessions with 232,303 genetic markers. Sorghum bicolor was selected because of its genomic simplicity, phenotypic diversity, significant genomic tools, and its agricultural productivity and resilience. We have demonstrated the value of sorghum as a functional model for candidate gene discovery for bioenergy Andropogoneae by performing genome-wide association analysis for two contrasting phenotypes representing key components of structural and non-structural carbohydrates. We identified potential genes, including a cellulase enzyme and a vacuolar transporter, associated with increased non-structural carbohydrates that could lead to bioenergy sorghum improvement. Although our analysis identified genes with potentially clear functions, other candidates did not have assigned functions, suggesting novel molecular mechanisms for carbon partitioning traits. These results, combined with our characterization of phenotypic and genetic diversity and the public accessibility of each accession and genomic data, demonstrate the value of this resource and provide a foundation for future improvement of sorghum and related grasses for bioenergy production.
Keywords: Bioenergy Association Panel, carbon partitioning, biomass composition, nonstructural sugars, Multiparent Advanced Generation Inter-Cross (MAGIC), multiparental populations, MPP
ALTHOUGH numerous plant species have been evaluated as potential bioenergy feedstocks, many of the most promising candidates belong to a tribe of grasses, the Andropogoneae, that includes many agriculturally important species, such as maize, sorghum, and sugarcane. The genetic improvement of bioenergy candidates within this tribe is challenging because little is understood about the genetic architecture of many of their most relevant bioenergy traits. Further complicating this improvement, the Andropogoneae have distinct phenotypic characteristics, such as a type II cell wall (Vogel 2008), C4 photosynthetic mechanisms, and various carbon partitioning patterns (Braun and Slewinski 2009), which limit the pertinence of basic research in C3 non-grass model organisms, e.g., Arabidopsis. Additionally, many of the proposed candidates, such as switchgrass (Panicum virgatum) and members of the Saccharum genus, including sugarcane, have complex genomes, which limits the generation and dissemination of genetic and genomic resources. The designation of a functional model grass species and the subsequent development of a community resource for the genetic dissection of carbon partitioning, biomass composition, and other yield-related bioenergy traits is needed to increase collaboration and accelerate bioenergy improvement.
Sorghum bicolor (L.) Moench has emerged as one of the preferred candidates for bioenergy feedstocks, and has warranted continued investment for development as a dedicated bioenergy crop due to its high productivity, widespread adaptability, and relative ease of genomic analysis (TERRA 2015). Sorghum is a drought-tolerant C4 grass with a diverse gene pool that can be exploited for a variety of traits, including those most desirable for bioenergy production. Currently, the existing sorghum germplasm contains four predominant types: grain, sweet, forage, and biomass. Each of these has a preferred ideotype with varying proportions of grain, stalks, leaves, non-structural sugars, etc. This range of phenotypic diversity not only allows sorghum to serve as a functional model to study various bioenergy and biomass-related traits in Andropogoneae but it also allows sorghum to be optimized to serve as raw material for promising conversion technologies (Calviño and Messing 2012).
Currently, grain, sweet, and biomass sorghums all serve as feedstocks for various conversion technologies. Grain sorghum, which accumulates starch in the seed, is used as a key feedstock for starch–ethanol conversion throughout the United States (Wu et al. 2010). Sweet and biomass sorghums, which are respectively characterized by the accumulation of non-structural and structural carbohydrates in the stalk, provide promise for high-yielding, sustainable bioenergy production. Biomass sorghums have recorded yields of up to 30 dry tons per hectare while sweet sorghums have shown the potential to produce 6000 liters of ethanol per hectare (Wu et al. 2010). Both sweet and cellulosic types have great potential for various bioenergy production methods already in use across locations worldwide. Understanding the genetic mechanisms underlying their differences will be key to maximizing their potential as bioenergy crops.
The distinguishing factor among the different sorghum bioenergy types, and the other bioenergy candidates in general, is how each partitions, translocates, and stores carbon, although the biochemical pathways, machinery, and their genetic controls that allocate carbon to various compositional constituents (i.e., lignin, cellulose, and hemicellulose) are not fully understood (Vogel 2008). Structural carbohydrates, including cellulose, hemicellulose, and pectin, along with the phenolic polymer lignin, are the major components of cell walls (Vogel 2008), while the primary constituents of non-structural carbohydrates in sorghum are sucrose, fructose, glucose, and starch (Saballos 2008). While variation within the structural carbohydrate profile has been documented in sorghum (Murray et al. 2008a), few studies have examined the genetic architecture and control of these traits in sorghum or other grasses.
Association studies in sorghum have revealed genetic controls of many phenotypes, including height (Brown et al. 2008; Murray et al. 2009), flowering time (Mace et al. 2013a), panicle architecture (Brown et al. 2006), seed size (Zhang et al. 2015), and various domestication traits (Morris et al. 2013a). Most of the studies have been conducted to elucidate the genetic architecture of complex traits as they relate to grain production, not bioenergy production. Because breeding for bioenergy crops with high biomass or fermentable sugars requires a conceptual adjustment from the traditional dwarfed cereals (Salas Fernandez et al. 2009), a characterized resource specifically arranged to represent critical bioenergy phenotypes not only allows for greater progress in the explication and exploitation of sorghum’s natural genetic diversity but also the diversity of the broader Andropogoneae tribe.
To facilitate the use of genomic research for improved renewable energy through enhanced biomass-related traits, we created a focused genomic resource, the Sorghum Bioenergy Association Panel (BAP). With a total of 390 accessions and 232,303 SNPs, the BAP captures sufficient diversity, yet restricts the panel to bioenergy types to allow for more efficient and informative association mapping. In this study, we introduce the BAP and demonstrate its useful diversity for understanding key bioenergy phenotypes. We also examine the relationship of carbon partitioning between structural, represented by neutral detergent fiber (NDF), and non-structural carbohydrates, represented by non-fibrous carbohydrates (NFC). Because these traits of carbon allocation are defining characteristics between sweet and biomass sorghum, understanding the genetic controls allows for more efficient improvement by enabling marker-assisted breeding and genomic selection for both types of bioenergy sorghum. Our goal in this research was not only to identify candidate genes that may be the future targets of crop improvement but also to lay a broader foundation of genetic and genomic resources for future studies that seek to maximize the potential of sorghum and other Andropogoneae as bioenergy crops.
Materials and Methods
Selection and representation of genetic resources
To ensure the accuracy and availability of this panel for future research, all of the accessions have PI inventory numbers and may be requested through the US Department of Agriculture’s Germplasm Repository Information Network (GRIN) (Supplemental Material, File S1). This panel can be divided into two subsets: sweet and biomass types (File S1), which represent to the two most important bioenergy types. Sweet lines were defined as having a Brix value of over 10% at the milk development stage or at physiological maturity. The sweet lines consist of 152 accessions, and the 238 biomass types make up the remaining accessions. Sweet accessions include cultivars from previously defined panels: the sweet sorghum association panel (Murray et al. 2009) and the US historic sweet sorghum panel (Wang et al. 2009). The additional sweet accessions and the biomass lines were chosen based on diversity of worldwide geographic distribution, racial categorization, and agronomic characteristics (File S1). The 390 lines comprise accessions from all five major sorghum races (bicolor, caudatum, durra, guinea, and kafir) with representatives from the entire African continent, Asia, and the Americas (File S1). Several important lines were also added, including lines sequenced at the Joint Genome Institute and the first source of the reference genome, BTx623 (Paterson et al. 2009).
Field design, phenotypes, and phenotyping protocols
The BAP was phenotyped in Florence, South Carolina, at the Clemson University Pee Dee Research and Education Center in 2013 and 2014. Trials were planted on 76 cm rows at a planting density of approximately 96,000 plants/ha in loamy sand soil on May 16, 2013 and May 6, 2014, and were irrigated at the time of planting and on an as-needed basis. Two complete randomized blocks or replicates of the BAP were planted in each year. Due to the extreme height of many of the accessions, which were taller than the irrigation pivot, no irrigation took place approximately 90 days after planting. Seed obtained through GRIN (http://www.ars-grin.gov) was treated with a chemical slurry of Concep II, NipIt, Apron XL, and Maxim XL. This seed treatment allowed for the application of Bicep II Magnum for weed control at a rate of 3.5 liters/ha prior to seed germination. Atrazine at a rate of 4.7 liters/ha was applied before plants had reached a height of 45 cm. Additionally, 125 kg/ha of layby N was applied approximately 30 days after planting. Besides the chemicals used as part of the seed treatment, no other insecticides or fungicides were applied.
Anthesis was determined when 50% of the plot had begun to shed pollen. Height measurements were taken at physiological maturity, or at a set harvest date of October 1, from the base of the stalk to the apex of the panicle, or, if no panicle was present, to the apex of the shoot apical meristem. When possible, each plot was harvested at physiological maturity of the genotype, with the exception of genotypes that did not flower, which were harvested as a single time point. At the time of harvest, three plants were cut at the base of the stalk, panicles were removed, and fresh weights were recorded. To remove the confounding effects of tillering on a per area basis, yield and compositional data were generated using three representative plants. Based on planting density, this represents approximately 0.5 m of row length. Biomass samples were dried at 40°. Dry weight was recorded once samples had obtained a constant weight. Dry tons per hectare were extrapolated based on the dry weight of the samples at the approximate planting density of 96,000 plants/ha. Compositional data, which included NDF, NFC, acid detergent fiber (ADF), and lignin, were generated by analyzing the dried samples with a Perten DA7250 near-infrared spectroscopy (NIR) instrument (https://www.perten.com). The custom NIR curves were developed by the Perten Applications team using wet chemistry data from 107 unique samples and ten blind technical replicates generated by Dairyland Labs (http://www.dairylandlabs.com). Lignin and ADF (a cumulative measurement of lignin and cellulose) wet chemistry data were generated using the Association of Official Agricultural Chemists (AOAC) protocol 973.18 whereas NDF (a cumulative measurement of cellulose, hemicellulose, and lignin) and NFC (a cumulative measure of non-structural carbohydrates) data were generated using AOAC protocol 2002.04.The wet chemistry samples were selected based on phenotypic and spectra diversity, a protocol recommended from the Perten Applications team. Yield and composition were compared in the BAP to the Sorghum Association Panel (SAP) (Casa et al. 2008), a previously defined sorghum panel focusing on grain sorghum. Dry weights and compositional components in the SAP were calculated based on five representative plants at a rate of 131,000 plants/ha. Compositional data for the SAP were generated using a NIR analysis provided by Chromatin, Inc. (https://www.chromatininc.com). All compositional data are presented as a percentage of dry matter (DM). GRIN provided racial and geographic origin information. To provide a control phenotype as confirmation of the genomic data, pericarp pigmentation, which is conditioned by a known gene (Ibraheem et al. 2010), was characterized from the seed provided by GRIN following previously outlined methods (Rooney 2000). Phenotypes for the BAP are located in File S2.
Genotyping, SNP calling, filtering, and imputation
For each entry, five seeds from each plant were grown for 2 weeks in a growth chamber, and DNA was extracted from whole seedlings using a DNeasy Plant Mini kit from Qiagen. Genotyping-by-sequencing (GBS) libraries were generated using an ApeKI digestion, and following previously outlined protocols (Elshire et al. 2011). Sequencing was performed on an Illumina HiSequation 2000, with 95 barcoded individuals and one negative control included in each lane. Single-end reads for the 343 individuals have been deposited in the NCBI Sequence Read Archive (SRA) under the BioProject identification number PRJNA298892.
Raw sequencing reads were filtered and processed using the TASSEL 5.0 pipeline (Bradbury et al. 2007), and BWA (Li and Durbin 2009) was used to align the filtered sequences to sorghum reference genome version 2 available from Phytozome (Paterson et al. 2009; Goodstein et al. 2012). A minimum aligned read depth of 10 was required for calling SNPs in any individual. (See File S3 and File S4 for details, sample command lines, and Perl scripts.)
After trimming and filtering raw data for quality, we retained over 350 million 64-bp sequencing reads, which corresponded to 1.8 million unique mapped tag locations in the sorghum genome, and 327,121 putative SNP sites. After filtering low-coverage SNPs, individuals with too many missing sites, and sites with a minor allele frequency below 5%, 232,303 SNPs in 343 accessions were retained. Missing genotypes were fully imputed with the software fastPHASE (Scheet and Stephens 2006), with 20 independent starts of the EM algorithm. There is a mean distance of 2–3 kb between each SNP, which is consistent with the level and density of SNP discovery in the previously published SAP (Morris et al. 2013a). The fully imputed data set was used for all association analysis and heritability.
To make comparisons between the BAP and the SAP, raw data from both panels were merged, and then filtered using similar methods. However, for these analyses, SNPs were filtered with a minor allele frequency of 1% with a coverage of at least 60% of individuals, and imputed loci with less than 80% confidence were considered missing. The final analyses of allele frequencies and expected heterozygosity were performed on 187,766 common SNPs between the BAP and the SAP.
Genetic differentiation and population structure
Levels of genetic differentiation between grain, sweet, and biomass sorghums were calculated using Wright’s FST (Wright 1969). For these estimations, we used non-imputed SNP data, and selected sites with a minimum of 80 individuals per type present, as well as a minimum minor allele frequency of 5%. To determine if mean FST values were significantly different from zero, permutation tests were performed where individual genotypes (across all polymorphic sites) were randomly permuted into groups of the same size 1000 times, and the mean FST was recalculated to determine a null distribution.
Genomic comparisons between the SAP and the BAP were calculated using R statistical software (R Development Core Team 2011). The expected heterozygosity was calculated using the R-package “pegas” (Paradis 2010). Heterozygosity was calculated on a per SNP basis and in a 20-kb sliding window with a 2-kb overlap. The 20-kb region was chosen based on the established linkage disequilbrium (LD) in sorghum (Hamblin et al. 2004; Mace et al. 2013b). To determine significance, permutation tests were performed by randomly assigning individuals into groupings of the same size as the original BAP and SAP for 100 permutations. The difference in heterozygosity between the two panels was recalculated for each permutation, and P-values were generated by counting the number of permuted values that were equal to or greater than the observed heterozygosity difference. Sites with P-values lower than 0.01 were considered significant.
Population structure was estimated using the program STRUCTURE (Pritchard et al. 2000). The genetic data were thinned to 1 SNP every 20 kb using the vcftools v0.1.13 thin function (Danecek et al. 2011). This left approximately 1 SNP per linkage group. Final structure analysis was performed with 16,476 loci from the 343 individuals with genomic data. Analysis was performed with K-values ranging from 1 to 12. Five independent replicates were generated for each K-value with a 10,000 run burn-in period followed by 200,000 sampling iterations. Principal component analysis was conducted using the EIGENSTRAT method (Patterson et al. 2006) (version 6.0.1) using the “Smart PCA” Perl command.
Genome-wide association scans
Single-SNP tests of association were performed using models implemented in the R-package GAPIT (Team 2011; Lipka et al. 2012). Association scans were performed using a general linear model (GLM), a mixed linear model (MLM) with internally calculated kinship and population structure, an MLM with kinship and an externally calculated population structure via STRUCTURE, and the compressed mixed linear model (CMLM) (Zhang et al. 2010), which internally controls for population structure and kinship among individuals and uses cluster analysis to assign individuals to groups. The MLMs and the CMLM both incorporate a kinship (K) matrix and population structure (Q-matrix), which has been shown to increase statistical power and reduce false positives (Yu et al. 2006). Before presenting genome-wide association study (GWAS) results, the model fit was compared by examining the QQ plots (File S5), and the CMLM was selected as the model with superior fit. To further reduce the chance of false positives, significance levels in these tests were determined using the Bonferroni correction method, resulting in a significance cut-off of approximately 3.0 × 10−7. Due to an earlier than expected frost in 2013, only 211 individuals were included for genomic analysis. In 2014, a total of 331 individuals were used in genomic analysis.
LD was calculated locally within a 1-mb region surrounding each significant locus. Within each region, a pairwise LD between each SNP was calculated using the R-package Genetics. The extent of LD was determined to decay when the r2 value was less than 0.1 (File S6). Genes potentially linked to any significantly associated SNP were identified by scanning version 2.1 of the S. bicolor genome (Goodstein et al. 2012). Gene function was determined using the Panther Classification System (Mi et al. 2013) and the European Bioinformatic Institute’s PFAM identification (Finn et al. 2014). Candidate genes were selected based on functional annotations provided by Phytozome, the Panther Classification System, and the PFAM database. SNP effects were predicted by the software snpEff (Cingolani et al. 2012).
Phenotypic analysis
Phenotypic analysis was conducted using R statistical software (Team 2011). Maximum, minimum, mean, and standard deviation values for the BAP were calculated using the mean values of both replicates per year. Phenotypic values in the SAP were calculated based on two replicates in 2013. Accessions that did not flower (i.e., photoperiod sensitive accessions) were not included in the anthesis analysis.
Correlations were determined using the phenotypic mean of the two replicates per year. Pearson correlations and the subsequent P-values were calculated using R statistical software with the “cor.test” function. Marker-based estimation of narrow-sense heritability was calculated with the “heritability” package (Team 2011; Kruijer et al. 2015).The phenotypic means for each year were treated as replicates in the input. Since the narrow-sense heritability calculation uses the genomic markers (Kruijer et al. 2015), a random subset of 100 individuals with complete datasets (ADF, NDF, NFC, lignin, height, and dry weight) from 2013 and 2014 were used in the calculations to avoid discrepancies based on genotypes. The centered relatedness matrix used with the marker-based heritability analysis was generated from GEMMA (Zhou et al. 2013).
To ensure that phenotypic values (and therefore genomic associations) were not confounded with the block effect, a model was developed for the phenotypic values that included effects of accession and block. Since the blocks contained up to 400 accessions, there may have been field heterogeneity that impacted the phenotypic values. Using the predicted values from the model above (basically the average of the two observations) hopefully minimized the impact of the field heterogeneity. To ensure that the phenotypic values were not confounded with field heterogeneity, an additional model was developed for the phenotypic values that also included covariates associated with the field effect. For this study, the covariates chosen were anthesis and height (see the descriptions below). Fortunately, these covariates turned out to have almost no relationship (not statistically significant) with the primary phenotypes of interest, and even after adjusting for the covariates, the phenotypic values of the accessions remained essentially unchanged (File S7). File S7 also contains the model used for the analysis and the scatterplots for the actual and predicted phenotypic values. Therefore, we concluded potential field effects were not creating a systematic bias in the phenotypic data, and used the predicted phenotypic value for each accession from the model including block effects in the subsequent association analyses. For the GWAS results, values were standardized by subtracting the mean, dividing by the standard deviation, and then averaging across replicates.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Genomic diversity and differentiation
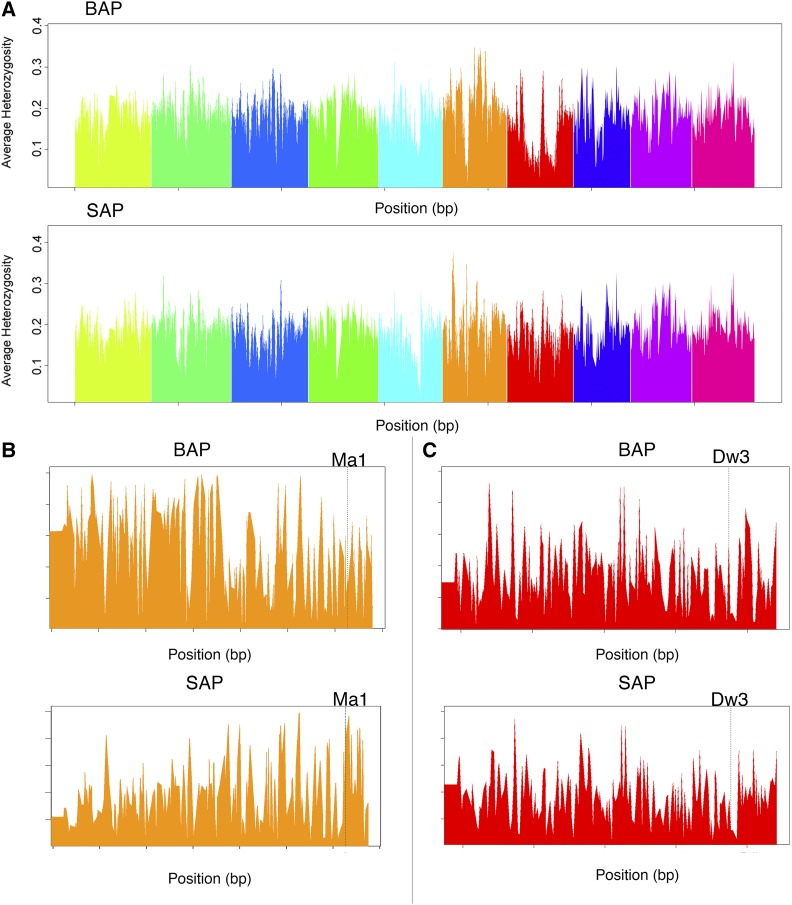
To identify genomic regions differentiated between the SAP and BAP, expected heterozygosity was calculated for individual SNPs and within a 20-kb sliding window with a 2-kb overlap. There were 187,766 common SNPs between the panels. Of these SNPs, 14,841 loci differed in expected heterozygosity by more than 25%. To look at global patterns in differentiation between the two resources, the SNPs were divided into sliding windows of 20 kb representing genomic regions within the estimated LD distance, and the mean heterozygosity for each block was compared (Figure 1). This resulted in 26,110 regions in which 525 differed in the expected heterozygosity by more than 25%. Since grain types have been selected for early maturity in temperate environments for grain maturation and bioenergy types have been selected for delayed flowering and increased biomass, it would be expected that regions surrounding major maturity genes would differ in the expected heterozygosity. To test this hypothesis, the expected heterozygosities of the 20-kb flanks surrounding known maturity genes [Ma1 (Murphy et al. 2011), Ma3 (Childs et al. 1997), and Ma6 (Murphy et al. 2014)] and a known dwarfing gene [Dw3 (Multani et al. 2003)] were compared between the two panels. The regions surrounding Ma1, Ma3, and Dw3 in the BAP and SAP were significantly different whereas Ma6 was not. There was low SNP coverage around the Ma6 locus, which may explain why the Ma6 locus was not differentiated between the two data sets. Although the SAP had a greater average heterozygosity near Ma1, regions surrounding Ma3 and Dw3 had higher average heterozygosities in the BAP than the SAP (Figure 1). These data highlight the fundamental differences in the two panels and suggests that there may be unexploited genetic diversity in the BAP due to a selective bottleneck for dwarfed, early maturing grain accessions in temperate environments.
Figure 1.
(A) Genome-wide heterozygosity calculated for the BAP (top) and SAP (bottom) with a 500-kb sliding window. (B) Average heterozygosity in 20-kb windows with a 2-kb overlap for the region on chromosome 6 containing the Ma1 gene, Sobic.006G057900, in the BAP (top) and the SAP (bottom). (C) Average heterozygosity in 20-kb windows with a 2-kb overlap for the region on chromosome 7 containing the Dw3 gene, Sobic.007G047300, in the BAP (top) and the SAP (bottom).
Because sweet and biomass sorghum are the primary types used for bioenergy production, determining how differentiated these two types are could provide insights into the genetic architecture of compositional components. However, the level of differentiation (as measured by FST) between the sweet and biomass types of sorghum was overall very low (mean FST = 0.024, where 0 is no differentiation and 1 is complete differentiation), although it was significantly greater than the null distribution (File S8). The maximum value of FST is 0.276, highlighting that there were no fixed differences between types in the data set despite significant phenotypic differences.
Population structure
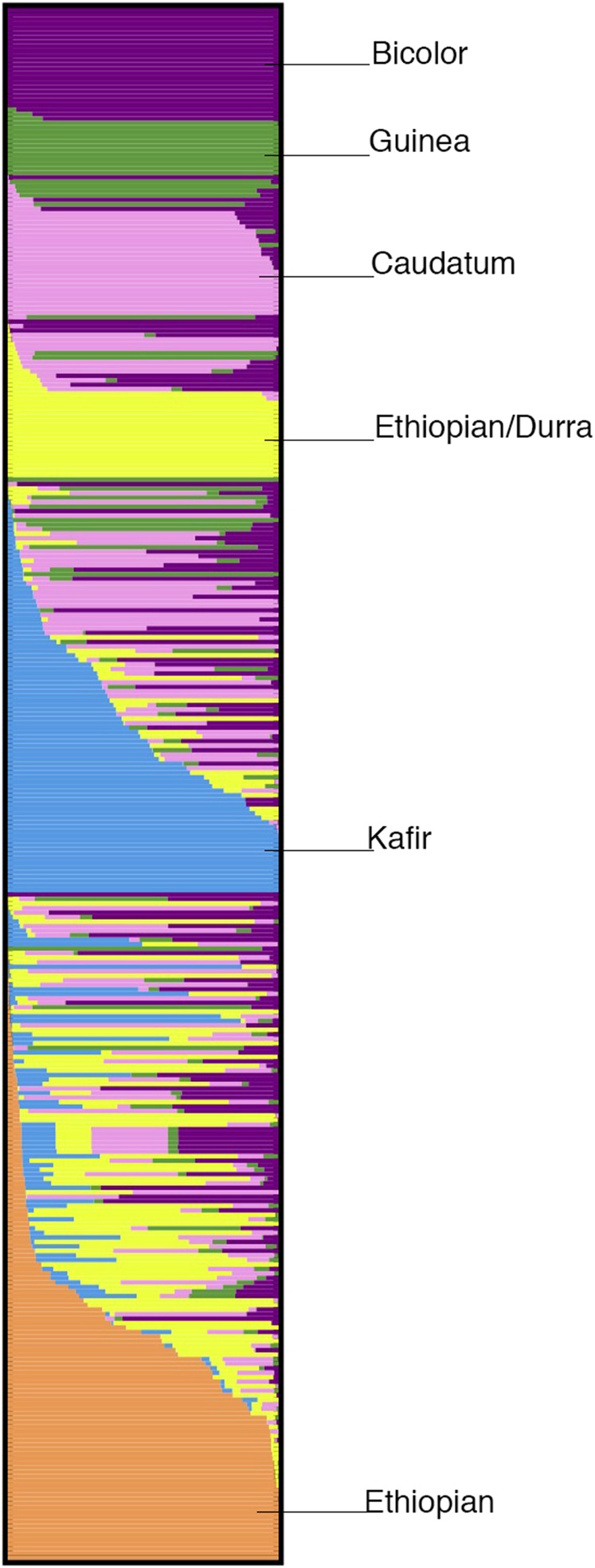
Previous work in the SAP has shown that population structure is related to the categorization of sorghum to the five botanical races and numerous geographic regions of sorghum colonization (Casa et al. 2008; Brown et al. 2011). Previous work has also demonstrated that these phenotypically based classifications are genetically supported (Brown et al. 2011). Based on these observations it would be expected that similar population patterns would appear in the BAP. Definitive patterns emerged supporting the previous findings that race and geographical origin help define subpopulation categorization (Figure 2). Figure 2 shows the STRUCTURE results from K = 6 of 343 individuals in the BAP. As expected, each of the five botanical races emerges as a subpopulation. Additionally, a sixth cluster appears that divides the Ethiopian accessions into two distinct groups. Since Ethiopia is the center of diversity for sorghum, it is not unexpected that distinct subpopulations could emerge when analyzing population structure. Racial data were not provided by GRIN for any of the accessions included in the orange cluster (Figure 2). Since racial classification is determined, at least in part, by panicle architecture and seed characteristics, it was not possible to establish racial classifications for this group due to the limited panicle emergence in the photoperiod-sensitive accessions. Interestingly, the most distinct group, the guinea population in the green cluster, cluster heavily together and have the lowest proportion of membership. Principal component analysis also showed clustering of the West African guinea types as well as the unclassified Ethiopian accessions. Additional STRUCTURE and principal component analysis results are in File S9.
Figure 2.
Population structure results with six defined subpopulations. The purple cluster represents bicolor accessions. The green cluster has the fewest number of members, and is mainly made up of guinea accessions. The pink cluster represents caudatum accessions. The yellow cluster represents durra accessions that are mainly from Ethiopia. The blue cluster includes individuals that cluster with kafir types. This grouping is usually associated with photoperiod insensitivity. The orange cluster represents accessions from Ethiopia, but no racial data were available for these lines.
Phenotypic means, distributions, correlations, and heritability
To highlight the differences between the grain-dominated SAP and the BAP, data were collected for phenotypes important for bioenergy sorghums. Comparison between the two panels revealed distinct patterns of phenotypic selection for each of the two types (Table 1). The average anthesis date in the BAP was almost 30 days longer than the SAP. This would have been even greater if photoperiod-sensitive lines had been included in the analysis. The average height was nearly 2 m greater in the bioenergy panel than in the grain panel. Also, the accumulation of above-ground biomass was significantly greater in the bioenergy panel. The composition traits as a proportion of dry matter (DM) did not differ as much between the two panels; however, when extrapolating the compositional components based on the dry weight, differences between two panels become more apparent. For example, the average accumulation of NDF/ha would be nearly 13 tons vs. 6 tons in the SAP. Not surprisingly, NFC as a percentage of DM is higher in the BAP than the SAP. Since 139 of the accessions in the BAP are classified as sweet types that have been selected to accumulate non-structural carbohydrates, it is reasonable to expect that the BAP would have a higher percentage and maximum value for the accumulation of non-structural sugars.
Table 1. Phenotypic comparisons between the SAP and BAP.
| Phenotype | BAP | SAP | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Average | Minimum | Maximum | Standard deviation | N | Average | Minimum | Maximum | Standard deviation | ||
| Anthesis (days) | 217 | 97 | 66 | 153 | 24 | 369 | 68 | 50 | 111 | 7 | |
| Height (cm) | 390 | 341.2 | 75.0 | 536.0 | 86.8 | 383 | 147.3 | 63.5 | 414.5 | 57.7 | |
| Dry weight (tons/ha) | 390 | 19.4 | 3.3 | 70.9 | 11.3 | 344 | 7.7 | 2.21 | 28.6 | 3.9 | |
| ADF (% of DM) | 387 | 41.5 | 14.0 | 54.9 | 7.9 | 379 | 37.5 | 24.8 | 61.2 | 5.5 | |
| NDF (% of DM) | 387 | 67.1 | 47.1 | 81.2 | 7.1 | 379 | 62.9 | 43.2 | 78.4 | 6.1 | |
| NFC (% of DM) | 387 | 27.6 | 13.9 | 50.0 | 8.0 | 369 | 20.3 | 10.5 | 45.5 | 6.4 | |
| Lignin (% of DM) | 387 | 6.6 | 1.6 | 10.5 | 1.6 | NA | NA | NA | NA | NA | |
Of the phenotypes collected in the BAP, the marker-assisted narrow-sense heritability estimates were generally high. Overall, the heritability of each phenotype is similar to previously published work (Table 2). However, anthesis heritability was much higher in the BAP than previously published studies (Murray et al. 2008a). This may be because many of the accessions in the BAP rely on photoperiod induction to initiate reproductive tissue formation. Since the heritability estimation used data from only one geographic location, the heritability estimate likely does not reflect the actual impact of the various latitudes and day lengths on photoperiod-sensitive lines. If anthesis values were collected in an environment with a shorter day length and the same analysis conducted to calculate heritability, these values would probably be much lower. The compositional phenotype heritabilities were similar to previously published results (Murray et al. 2008a).
Table 2. Heritability and correlations of phenotypes in the BAP.
| Phenotype | H2 calculation | h2 estimation | Anthesis | Height | Dry weight | ADF | NDF | NFC | Lignin |
|---|---|---|---|---|---|---|---|---|---|
| Anthesis | 0.86 | 0.90 | — | 0.724*** | 0.687*** | 0.530*** | 0.163* | −0.088 | 0.579*** |
| Height | 0.72 | 0.82 | 0.724*** | — | 0.549*** | 0.430*** | 0.245*** | −0.141** | 0.527*** |
| Dry weight | 0.39 | 0.32 | 0.687*** | 0.549*** | — | 0.009 | −0.088 | 0.183*** | 0.056 |
| ADF | 0.55 | 0.62 | 0.530*** | 0.430*** | 0.009 | — | 0.837*** | −0.866*** | 0.872*** |
| NDF | 0.51 | 0.54 | 0.163* | 0.245*** | −0.088 | 0.837*** | — | −0.963*** | 0.721*** |
| NFC | 0.50 | 0.56 | −0.088 | −0.141** | 0.183*** | −0.866*** | −0.963*** | — | −0.704*** |
| Lignin | 0.57 | 0.70 | 0.579*** | 0.527*** | 0.056 | 0.872*** | 0.721*** | −0.704*** | — |
Significance at 0.05 probability; **significance at 0.01; ***significance at 0.001.
Validation of the GWAS results using seed color as a control
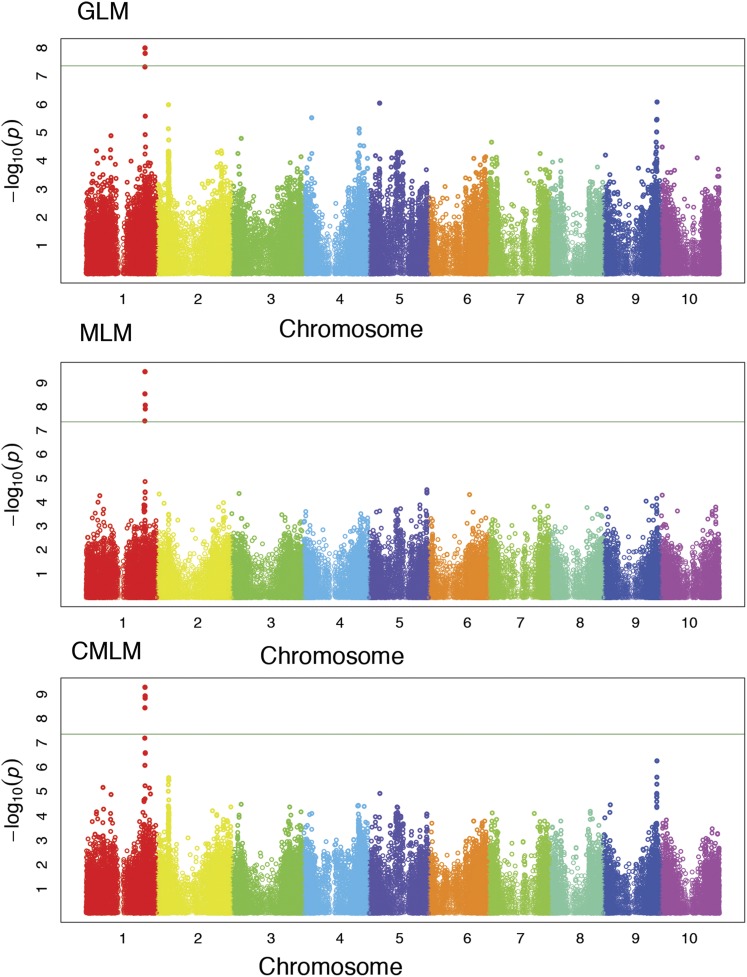
Pericarp pigmentation in sorghum seeds is a well-studied trait that is known to be controlled by an MYB transcription factor (Y1; Yellow seed1) (Rooney 2000; Ibraheem et al. 2010; Morris et al. 2013b). Since this gene has been mapped in the SAP (Morris et al. 2013b), pericarp pigmentation was used as a control in this study to validate the genetic data. As expected, all of the models in GAPIT (GLM, MLM, and CMLM to control for population structure and kinship) identified a single region within the transcript of the Y1 locus (Sobic.001G397900) that was strongly associated with seed color in the BAP (Figure 3).
Figure 3.
A single locus, the Y1 MYB transcription factor, was identified in all three models as expected. This phenotype represents a control to validate correct SNP calling, imputation, and GWAS methodology.
Association mapping for structural and non-structural carbohydrates
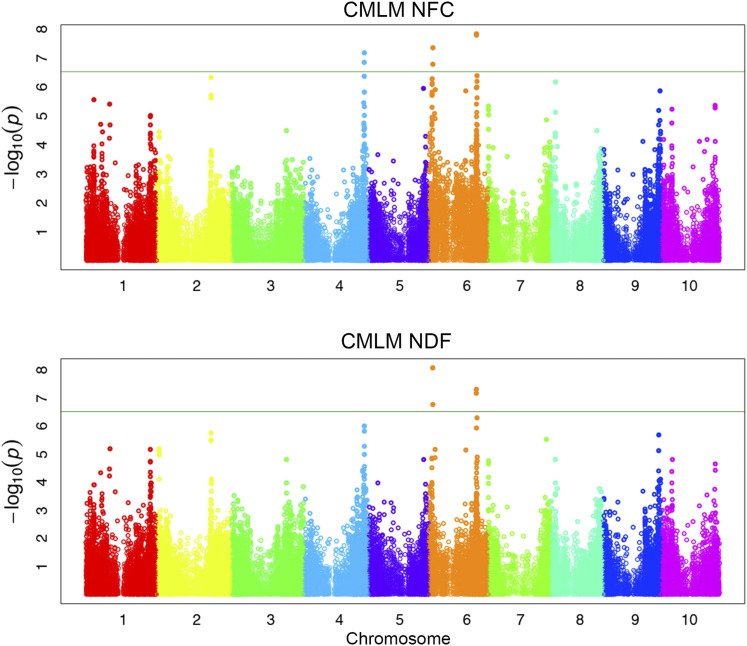
Association mapping revealed genomic regions strongly associated with NDF and NFC. Since these phenotypes are inversely related to one another, it would be expected that many of the same significant loci identified for one phenotype were also present in the other phenotype. The association scans from NDF and NFC demonstrate this relationship (Figure 4).
Figure 4.
A total of eight unique SNPs, five loci, and 22 genes were identified using the CMLM for NFC and NDF. SNPs with a P-value of less than 3.00 × 10−7 were considered significant.
Using the CMLM from GAPIT, the association scans revealed a total of eight significant SNPs, representing five loci and 22 genes (File S10). LD was calculated locally for each significant SNP (File S6). Significantly associated SNPs within the distance of LD decay of on another were considered a single locus; also, any gene within the LD estimate was considered linked, and plausibly implicated in the determination of the phenotype. Of the eight significant SNPs, two are intragenic missense variants, indicating the higher likelihood that specific genes contribute to the phenotype.
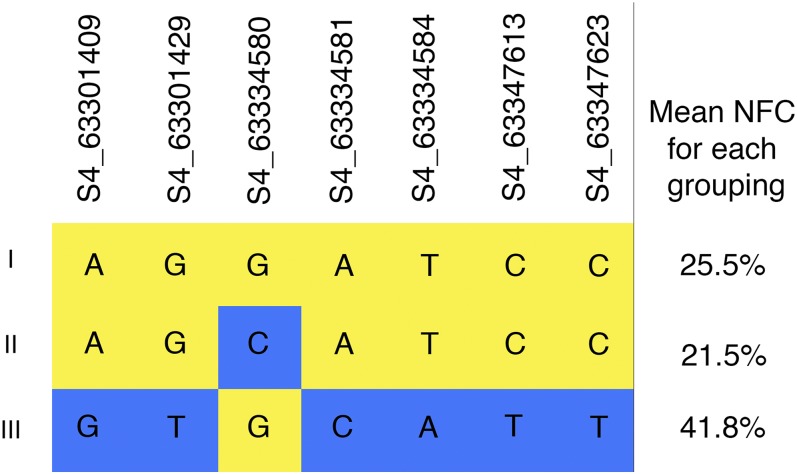
A total of five regions were identified through the association methods: two loci were located on chromosome 4, and three on chromosome 6. Although most loci identified had plausible explanations of their impact on biomass compositional components, one of the regions on chromosome 4 is particularly interesting. An SNP in this region causes an amino acid change to a vacuolar iron transporter. The SNPs in this linkage group appear to create a distinctive haplotype structure. There were three haplotypes in this region (Figure 5). The mean NFC of haplotype III was 41.8% while the mean value of haplotype I was only 25.5% NFC. Haplotype II, which only differed from haplotype I by a single base pair, also had a low NFC value (21.5%). Of the individuals with an NFC of over 40% DM (29 individuals), 11 individuals have haplotype III. The top five individuals all have haplotype III at this location. Historically important sweet lines such as Rio, Wray, Leoti, and Sugar Drip each possessed haplotype III at the specified locus (Figure 5). The strong association with NFC coupled with the clustering of historically important accessions provides evidence that this region impacts the accumulation of nonstructural carbohydrates in Sorghum bicolor, and could be important for bioenergy sorghum improvement.
Figure 5.
Three haplotypes on chromosome 4. This region was significantly associated with NFC in the CMLM in 2014. Yellow indicates the more frequent allele, and blue indicates the less frequent allele. Haplotypes I and II correspond to low values of NFC while haplotype III corresponds to high levels of NFC.
Due to the potentially confounding effects of height and maturity on accumulation of structural and non-structural carbohydrates, the candidate genes were compared to the locations of known maturity genes (Ma1 – Ma6) (Mace and Jordan 2010) and known dwarfing genes (Dw1 – Dw4) (Mace and Jordan 2010) . There was no co-localization among any of the maturity genes or dwarfing genes with any of the significantly associated regions. Furthermore, there was no overlap among the nearly 221 candidate genes identified for maturity (Mace et al. 2013a) and the candidate genes for structural or non-structural carbohydrates identified in this study. In addition, GWASs were conducted on height and flowering time from the data in the BAP; no significant SNPs co-localized with the results from NFC and NDF (File S11).
Candidate gene identification
Each region identified through the GWASs has plausible candidates for biomass composition (Table 3). Most notably, SNP S4_63347613, shown in haplotype III (Figure 5), causes an amino acid change from an alanine to a valine in a vacuolar iron transporter family protein. Previous studies have shown that sucrose accumulation in plants regulates an iron-deficient response (Lin et al. 2016). Furthermore, in a previous comparison of divergence between sweet and grain types, this region underwent a segmental duplication from their most recent common ancestor, suggesting possible neofunctionalization of the two vacuolar iron transporter between sweet and grain sorghum (Jiang et al. 2013). Additionally, a vacuolar-processing enzyme was identified in this region. Vacuoles serve a major role in sucrose accumulation and mobilization in plants (Leigh 1984). The other region on chromosome 4 contains four genes, one of which, a B-box zinc finger protein, shares homology with a salt tolerance homolog. Sugar accumulation has been shown to be a molecular response to salt stress in sorghum (Sui et al. 2015).
Table 3. Significant SNPs, candidate genes, and number of genes within LD of significant SNP.
| SNP | P-value | Local LD (kb) | No. of genes in the region | Candidate gene | Distance to the candidate gene (bp) |
|---|---|---|---|---|---|
| S4_63301409 | 6.85 × 10−8 | 23 | 4 | Salt-tolerance homolog | 18,095 downstream |
| S4_63301429 | 6.85 × 10−8 | 23 | 4 | Salt-tolerance homolog | 18,105 downstream |
| S4_63347613 | 1.41 × 10−7 | 23 | 8 | Vacuolar iron transporter | Intragenic |
| S4_63347623 | 1.41 × 10−7 | 23 | 8 | Vacuolar iron transporter | Intragenic |
| S6_4320818 | 4.40 × 10−8 | 1 | 0 | NA | NA |
| S6_4330906 | 1.64 × 10−7 | 1 | 0 | NA | NA |
| S6_49773083 | 1.68 × 10−8 | 16 | 9 | Cellulase (glycosyl hydrolase) | 13,666 downstream |
| S6_49784457 | 1.48 × 10−8 | 16 | 4 | Transducin/WD40 homolog | 773 upstream |
The region identified on chromosome 6 had two genes coding for cellulase enzymes, Sobic.006G122200 and Sobic.006G122300. These genes hydrolyze glycosidic bonds in complex carbohydrates, such as cellulose, which is the major component of NDF. These SNPs were associated with increased levels of non-structural carbohydrates and decreased levels of structural carbohydrates. These glycoside hydrolase family 5 proteins could be involved in the degradation of structural components of the cell wall. These were the only two genes to have GO terms associated with carbohydrate metabolic process (GO:0005975). Additionally, a transducin/WD40 family protein was identified from a significantly associated SNP 773 bp upstream. Transducin/WD40 proteins have been shown to increase biomass accumulation (Gachomo et al. 2014). Although the genes identified in this study are plausible candidates for biomass compositional components, further evidence will be needed to dissect the true effect of these allelic variants.
Discussion
Sorghum as a functional model for bioenergy and the value of the BAP
Of the potential bioenergy Andropogoneae candidates, sorghum has emerged as one of the preferred species for direct commercialization as a bioenergy crop and as a functional model for other Andropogoneae. Sorghum has natural advantages as a model for this family of grasses because of its relatively small diploid genome (∼730 Mb), significant breeding history, and substantial natural diversity. This extensive genetic and phenotypic diversity provides the foundation for gene discovery and crop improvement. It also allows sorghum to serve as a model for other bioenergy Andropogoneae because of its adaptability to various bioenergy conversion technologies. Due to its high levels of sugar accumulation and its close evolutionary history, it can also serve as a relevant reference for the Saccharum genus. Since there are no reported genomic incompatibilities among the four types of sorghum, genes identified that improve bioenergy sorghum performance in the BAP could be incorporated into grain and forage types as well.
The BAP was constructed by using publicly available racial and geographic as well as agronomic data from field evaluations. Since previous studies have shown that the racial classifications are genetically supported (Brown et al. 2011), the hypothesis was that by selecting lines incorporating the major botanical races, we would be able to capture a sufficient amount of genetic diversity. The botanical races are correlated with geographic regions. After we selected individuals based on racial distribution, we supplemented under-represented regions with accessions with known geographic origins. Phenotypically, we restricted accessions to tall, photoperiod-sensitive, late-maturing accessions. We also chose accessions screened for resistance to a major sorghum disease, anthracnose. This was an attempt to remove the confounding effects of varying resistances and susceptibilities, since the presence of the disease could alter the carbon composition profile of the individual accession. Although we tightly constrained the amount of diversity for flowering time, height, and disease susceptibility, we captured an appropriate amount of genomic diversity compared with other panels. Finally, historically important lines used in breeding and lines that were sequenced at the Joint Genome Institute were included. All accessions are available for public distribution through the GRIN system.
The development of a genetic and genomic resource specifically designed to capture the natural genetic and phenotypic diversity of sorghum for carbon partitioning and biomass composition increases the efficiency and efficacy of association genetics and incorporation of favorable alleles into a breeding pipeline. Although nested association mapping (NAM) populations and multi-parent advanced-generation inter-cross (MAGIC) populations have been shown to improve the detection of small effect loci and reduce the false-discovery rate (Cavanagh et al. 2008; Yu et al. 2008), these populations severely restrict the diversity and thus the detection of novel gene candidates or rare, favorable alleles. In addition, diversity panels developed for conservation of genetic resources and analysis of genetic diversity impede many efforts to identify causal genes either because of the confounding effects as a consequence of the diversity or the lack of statistical power from a low phenotypic frequency. The BAP’s construction limits the confounding effects associated with flowering time and height (Flint-Garcia et al. 2005) by limiting the panel to tall, late-flowering, photoperiod-sensitive accessions. Furthermore, the selection of accessions with known phenotypic diversity increases the likelihood that variants are at higher frequencies in the mapping population, which increases the probability of a true positive association (Myles et al. 2009). The creation, evaluation, and characterization of a diversity panel with the public dissemination of data provides insights to create better constructed NAM, MAGIC, recombinant inbred lines (RILs), or candidates for whole-genome resequencing. Overall, the BAP was created to overcome the limitations with other genomic resources, and the effective mapping of two key phenotypes show the advantages of using the BAP for critical bioenergy traits, but future studies should implement better field designs for improved statistical analysis. An important insight from this study is that the large number of accessions allowed a thorough analysis of the associations, but resulted in a design with very large block size. Even though we corrected for possible field heterogeneity from the large block size, additional studies using this resource should use superior designs such as an incomplete block design with multiple row plots. This allows for adjustment due to competition effects and other field variants. With more appropriate design, the BAP has the potential to serve as a critical resource for the continued advancement of sorghum as a preferred bioenergy feedstock.
Conclusions
The objective in this study was to expand the existing foundation of genetic and genomic resources for bioenergy research in non-model Andropogoneae. By creating the sorghum BAP, we provide a genetic and genomic resource that not only provides a foundational knowledge for determining the genetic architecture of traits important for bioenergy but also expands the current germplasm in the sorghum community. Although this panel limits phenotypic variance of the included accessions to bioenergy-like ideotypes, genetic and phenotypic diversity of the overall species was maintained. The strong heritabilites and the low correlations of the compositional phenotypes to dry weight suggested that composition can be improved without affecting the total yield (Murray et al. 2008b). The association analysis identified regions of the genome that could be targeted to improve biomass quality. However, others have suggested that increasing total yield is more important than improving composition quality for maximizing extractible energy per unit input (Murray et al. 2008a). Since increasing sink strength has been shown to advantageously affect yield (Bihmidine et al. 2013), understanding the genetic controls of the compositional components could allow for improved sink strength with a positive yield outcome. By identifying genomic regions independently affecting yield and composition, researchers could simultaneously select for both yield and increased quality instead of selecting for one or the other. This would allow researchers to increase yield and compositional quality concurrently, promoting an increase in breeding efficiency and bioenergy optimization. Furthermore, determining the genetic controls of carbon allocation in sorghum may be useful in elucidating the genetic mechanisms controlling biomass yield, sugar accumulation, and other compositional constituents in other C4 grasses.
By analyzing phenotypic and genomic data from the BAP, researchers can better design experiments to study the genetics of bioenergy sorghum. Providing corroborating evidence on how sorghum populations are structured not only reinforces previous studies (Casa et al. 2008; Morris et al. 2013a) but also provides valuable information pertaining to how certain botanical races of sorghum may perform in a bioenergy context. The establishment, characterization, and subsequential genomic analysis of this resource have highlighted regions of the genome and possible candidate genes for targeted improvement in bioenergy sorghum. These candidate genes need further validation, such as analysis of segregating populations, targeted gene sequencing, and functional tests. The need for the grass community to develop appropriate resources for gene identification with functional annotations is imperative for the continued improvement of bioenergy feedstocks. The creation and analysis of this foundational resource provides researchers with valuable tools and essential knowledge for continued experimentation with bioenergy sorghum and other Andropogoneae. Providing easily accessible accessions with genomic information allows for greater efficiency of research by encouraging collaboration and the dissemination of information. The establishment, characterization, and analysis of the BAP facilitate the advancement of sorghum for bioenergy production and optimization worldwide, and provide a foundational resource for the development of renewable energy.
Acknowledgments
The authors thank Lauren McIntyre of the University of Florida for insights and perspective that strengthened the content and message of this manuscript. We also thank Matthew Lennon and Alexander Cox for dedication to the project and their tireless efforts in collecting accurate phenotypic measurements. We acknowledge the Clemson University Genomics and Bioinformatics Laboratory for aiding in the generation of genomic data. Analysis was conducted on Clemson’s high-performance computing resource, the Palmetto Cluster. This material is based upon work that is supported by the National Institute of Food and Agriculture, the US Department of Agriculture under award number 2011-67009-23490, and the US Department of Energy under award number DE-AR0000595. This work is also supported by the United Sorghum Checkoff Program and the Robert and Lois Coker Endowment.
Footnotes
Communicating editor: F. van Eeuwijk
Supplemental Material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.183947/-/DC1
Literature Cited
- Bihmidine S., Hunter C. T. I., Johns C. E., Koch K. E., Braun D. M., 2013. Regulation of assimilate import into sink organs: update on molecular drivers of sink strength. Front. Plant Sci. 4: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury P. J., Zhang Z., Kroon D. E., Casstevens T. M., Ramdoss Y., et al. , 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. [DOI] [PubMed] [Google Scholar]
- Braun D. M., Slewinski T. L., 2009. Genetic control of carbon partitioning in grasses: roles of sucrose transporters and tie-dyed loci in phloem loading. Plant Physiol. 149: 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P. J., Klein P. E., Bortiri E., Acharya C. B., Rooney W. L., et al. , 2006. Inheritance of inflorescence architecture in sorghum. Theor. Appl. Genet. 113: 931–942. [DOI] [PubMed] [Google Scholar]
- Brown P. J., Rooney W. L., Franks C., Kresovich S., 2008. Efficient mapping of plant height quantitative trait loci in a sorghum association population with introgressed dwarfing genes. Genetics 180: 629–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P. J., Myles S., Kresovich S., 2011. Genetic support for phenotype-based racial classification in sorghum. Crop Sci. 51: 224–230. [Google Scholar]
- Calviño M., Messing J., 2012. Sweet sorghum as a model system for bioenergy crops. Curr. Opin. Biotechnol. 23: 323–329. [DOI] [PubMed] [Google Scholar]
- Casa A. M., Pressoir G., Brown P. J., Mitchell S. E., Rooney W. L., et al. , 2008. Community resources and strategies for association mapping in sorghum. Crop Sci. 48: 30–40. [Google Scholar]
- Cavanagh C., Morell M., Mackay I., Powell W., 2008. From mutations to magic: resources for gene discovery, validation and delivery in crop plants. Curr. Opin. Plant Biol. 11: 215–221. [DOI] [PubMed] [Google Scholar]
- Childs K. L., Miller F. R., Cordonnier-Pratt M. M., Pratt L. H., Morgan P. W., et al. , 1997. The sorghum photoperiod sensitivity gene, ma3, encodes a phytochrome b. Plant Physiol. 113: 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, snpeff: Snps in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire R. J., Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K., et al. , 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6: e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Miller B. L., Clements J., Bateman A., 2014. iPfam: a database of protein family and domain interactions found in the Protein Data Bank. Nucleic Acids Res. 42: D364–D373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint-Garcia S. A., Thuillet A.-C., Yu J., Pressoir G., Romero S. M., et al. , 2005. Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J. 44: 1054–1064. [DOI] [PubMed] [Google Scholar]
- Gachomo E. W., Jimenez-Lopez J. C., Baptiste L. J., Kotchoni S. O., 2014. GIGANTUS1 (GTS1), a member of Transducin/WD40 protein superfamily, controls seed germination, growth and biomass accumulation through ribosome-biogenesis protein interactions in Arabidopsis thaliana. BMC Plant Biol. 14: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein D. M., Shu S., Howson R., Neupane R., Hayes R. D., et al. , 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40: D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin M. T., Mitchell S. E., White G. M., Gallego W., Kukatla R., et al. , 2004. Comparative population genetics of the panicoid grasses: sequence polymorphism, linkage disequilibrium and selection in a diverse sample of Sorghum bicolor. Genetics 167: 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibraheem F., Gaffoor I., Chopra S., 2010. Flavonoid phytoalexin-dependent resistance to anthracnose leaf blight requires a functional yellow seed1 in Sorghum bicolor. Genetics 184: 915–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S.-Y., Ma Z., Vanitha J., Ramachandran S., 2013. Genetic variation and expression diversity between grain and sweet sorghum lines. BMC Genomics 14: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijer W., Boer M. P., Malosetti M., Flood P. J., Engel B., et al. , 2015. Marker-based estimation of heritability in immortal populations. Genetics 199: 379–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh R., 1984. The role of the vacuole in the accumulation and mobilization of sucrose. Plant Growth Regul. 2: 339–346. [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Ye Y., Fan S., Jin C., Zheng S., 2016. Increased sucrose accumulation regulates iron-deficiency responses by promoting auxin signaling in Arabidopsis plants. Plant Physiol. 170: 907–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka A. E., Tian F., Wang Q., Peiffer J., Li M., et al. , 2012. GAPIT: genome association and prediction integrated tool. Bioinformatics 28: 2397–2399. [DOI] [PubMed] [Google Scholar]
- Mace E. S., Jordan D. R., 2010. Location of major effect genes in sorghum (Sorghum bicolor (L.) Moench). Theor. Appl. Genet. 121: 1339–1356. [DOI] [PubMed] [Google Scholar]
- Mace E. S., Hunt C. H., Jordan D. R., 2013a Supermodels: sorghum and maize provide mutual insight into the genetics of flowering time. Theor. Appl. Genet. 126: 1377–1395. [DOI] [PubMed] [Google Scholar]
- Mace E. S., Tai S., Gilding E. K., Li Y., Prentis P. J., et al. , 2013b Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat. Commun. 4: 2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Thomas P. D., 2013. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41: D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. P., Ramu P., Deshpande S. P., Hash C. T., Shah T., et al. , 2013a Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc. Natl. Acad. Sci. USA 110: 453–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. P., Rhodes D. H., Brenton Z., Ramu P., Thayil V. M., Deshpande S., et al. , 2013b Dissecting genome-wide association signals for loss-of-function phenotypes in sorghum flavonoid pigmentation traits. G3 Genes Genomes Genet. 3: 2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multani D. S., Briggs S. P., Chamberlin M. A., Blakeslee J. J., 2003. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302: 81–84. [DOI] [PubMed] [Google Scholar]
- Murphy R. L., Klein R. R., Morishige D. T., Brady J. A., Rooney W. L., et al. , 2011. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc. Natl. Acad. Sci. USA 108: 16469–16474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R. L., Morishige D. T., Brady J. A., Rooney W. L., Yang S., et al. , 2014. Ghd7 (Ma6) represses sorghum flowering in long days: Ghd7 alleles enhance biomass accumulation and grain production. Plant Genome 7: 1–10. [Google Scholar]
- Murray S. C., Rooney W. L., Mitchell S. E., Sharma A., Klein P. E., et al. , 2008a Genetic improvement of sorghum as a biofuel feedstock: II. QTL for stem and leaf structural carbohydrates. Crop Sci. 48: 2180–2193. [Google Scholar]
- Murray S. C., Sharma A., Rooney W. L., Klein P. E., Mullet J. E., et al. , 2008b Genetic improvement of sorghum as a biofuel feedstock: I. QTL for stem sugar and grain nonstructural carbohydrates. Crop Sci. 48: 2165–2179. [Google Scholar]
- Murray S. C., Rooney W. L., Hamblin M. T., Mitchell S. E., Kresovich S., 2009. Sweet sorghum genetic diversity and association mapping for brix and height. Plant Genome 2: 48–62. [Google Scholar]
- Myles S., Peiffer J., Brown P. J., Ersoz E. S., Zhang Z., et al. , 2009. Association mapping: critical considerations shift from genotyping to experimental design. The Plant Cell Online 21: 2194–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., 2010. Pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26: 419–420. [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Bowers J. E., Bruggmann R., Dubchak I., Grimwood J., Gundlach H., Haberer G., et al. , 2009. The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. [DOI] [PubMed] [Google Scholar]
- Patterson N., Price A. L., Reich D., 2006. Population structure and eigenanalysis. PLoS Genet. 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team , 2011. R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Rooney W., 2000. Sorghum: Origin, History, Technology, and Production. John Wiley, New York. [Google Scholar]
- Saballos A., 2008. Development and Utilization of Sorghum as a Bioenergy Crop. Springer, New York. [Google Scholar]
- Salas Fernandez M. G., Becraft P. W., Yin Y., Lübberstedt T., 2009. From dwarves to giants? Plant height manipulation for biomass yield. Trends Plant Sci. 14: 454–461. [DOI] [PubMed] [Google Scholar]
- Scheet P., Stephens M., 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78: 629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui N., Yang Z., Liu M., Wang B., 2015. Identification and transcriptomic profiling of genes involved in increasing sugar content during salt stress in sweet sorghum leaves. BMC Genomics 16: 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TERRA, 2015 Financial Assistance Funding Opportunity Announcement No. DE-FOA-0001211. Technical report. Advanced Research Projects Agency—Energy, Washington, DC. [Google Scholar]
- Vogel J., 2008. Unique aspects of the grass cell wall. Curr. Opin. Plant Biol. 11: 301–307. [DOI] [PubMed] [Google Scholar]
- Wang M. L., Zhu C., Barkley N. A., Chen Z., Erpelding J. E., et al. , 2009. Genetic diversity and population structure analysis of accessions in the US historic sweet sorghum collection. Theor. Appl. Genet. 120: 13–23. [DOI] [PubMed] [Google Scholar]
- Wright S., 1969. Evolution and Genetics of Populations. The Theory of Gene Frequencies, Vol. 2 University of Chicago Press, Chicago. [Google Scholar]
- Wu X., Staggenborg S., Propheter J. L., Rooney W. L., Yu J., et al. , 2010. Features of sweet sorghum juice and their performance in ethanol fermentation. Ind. Crops Prod. 31: 164–170. [Google Scholar]
- Yu J., Pressoir G., Briggs W. H., Bi I. V., Yamasaki M., et al. , 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208. [DOI] [PubMed] [Google Scholar]
- Yu J., Holland J. B., McMullen M. D., Buckler E., 2008. Genetic design and statistical power of nested association mapping in maize. Genetics 178: 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Li J., Compton R. O., Robertson J., Goff V. H., Epps E., et al. , 2015. Comparative genetics of seed size traits in divergent cereal lineages represented by sorghum (Panicoidae) and rice (Oryzoidae). G3 Genes Genomes Genet. 5: 1117–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Ersoz E., Lai C.-Q., Todhunter R. J., Tiwari H. K., et al. , 2010. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42: 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Carbonetto P., Stephens M., 2013. Polygenic modeling with Bayesian sparse linear mixed models. PLoS Genet. 9: e1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.