Abstract
Chromosomal inversions are widespread among taxa, and have been implicated in a number of biological processes including adaptation, sex chromosome evolution, and segregation distortion. Consistent with selection favoring linkage between loci, it is well established that length is a selected trait of inversions. However, the factors that affect the distribution of inversion breakpoints remain poorly understood. “Sensitive sites” have been mapped on all euchromatic chromosome arms in Drosophila melanogaster, and may be a source of natural selection on inversion breakpoint positions. Briefly, sensitive sites are genomic regions wherein proximal structural rearrangements result in large reductions in local recombination rates in heterozygotes. Here, I show that breakpoints of common inversions are significantly more likely to lie within a cytological band containing a sensitive site than are breakpoints of rare inversions. Furthermore, common inversions for which neither breakpoint intersects a sensitive site are significantly longer than rare inversions, but common inversions whose breakpoints intersect a sensitive site show no evidence for increased length. I interpret these results to mean that selection favors inversions whose breakpoints disrupt synteny near to sensitive sites, possibly because these inversions suppress recombination in large genomic regions. To my knowledge this is the first evidence consistent with positive selection acting on inversion breakpoint positions.
Keywords: chromosomal inversions, Drosophila melanogaster, sensitive sites, structural variation, breakpoints
SUCCESSFUL chromosomal inversions are generally thought to be favorable due to reduced recombination rates between arrangements (Krimbas and Powell 1992; Hoffmann and Rieseberg 2008; Kirkpatrick 2010). In the majority of theoretical treatments, it is believed that inversions are favored because they suppress recombination between genetically distant alleles that are favored in similar contexts. For example, this can result from an array of biological processes including local adaptation (Kirkpatrick and Barton 2006), sex chromosome evolution (Charlesworth et al. 2005), and maintenance of segregation distortion complexes (Lyon 2003). Consistent with this idea, longer chromosomal inversions are expected to be favored by selection relative to shorter inversions because they can suppress recombination between a larger number of genetically distant loci (Crumpacker and Kastritsis 1967; Krimbas and Powell 1992). Indeed, numerous studies in Drosophila have shown that high frequency and fixed inversions are significantly longer than rare and low frequency inversions (Olvera et al. 1979; Ruiz et al. 1984; Cáceres et al. 1997). Furthermore, common inversions are significantly longer than theoretical predictions of the length distribution that should be generated through neutral processes (Van Valen and Levins 1968; Brehm and Krimbas 1991; Krimbas and Powell 1992). Although some evidence indicates that intermediate length inversions are favored relative to short and very long inversions (Cáceres et al. 1997, 1999), a general consensus has emerged that evolutionarily successful chromosomal inversions tend to be longer than rare inversions. These observations are consistent with the idea that chromosomal inversions that are long relative to the average newly-formed inversion are favored by natural selection because they tend to suppress recombination in larger genomic intervals and can therefore maintain linkage between a large number of alleles that are favorable when in linkage disequilibrium (Brehm and Krimbas 1991; Krimbas and Powell 1992; Cáceres et al. 1997, 1999).
While the factors that affect inversion lengths are well characterized, the selective and neutral processes that influence inversion breakpoint positions remain largely obscure. Many authors have found that chromosomal inversion breakpoint positions tend to cluster more closely than would be expected if breaks occur at random (Tonzetich et al. 1988; Krimbas and Powell 1992; Cáceres et al. 1997; Ranz et al. 2007; but see Olvera et al. 1979). These observations may result from neutral processes if certain regions are more prone to breakage. Alternatively, they may result from selective processes if breakpoints in certain genomic regions are selectively favorable. Hence, at present it is not clear if inversion breakpoint positions constitute an important source of natural selection affecting the evolution of chromosomal inversions (although see Tadin-Strapps et al. 2004; Castermans et al. 2007; and Corbett-Detig and Hartl 2012 for examples of negative selection on inversion breakpoint effects).
Several studies have shown that the Drosophila melanogaster genome contains numerous “sensitive sites.” These sites are believed to be necessary to produce normal crossover frequencies in the surrounding genomic regions. When synteny is disrupted near a sensitive site, e.g., due to translocations or inversions, recombination is suppressed in large genomic regions surrounding that sensitive site (Roberts 1970, 1972; Hawley 1980; Coyne et al. 1993; Navarro and Ruiz 1997). In his pioneering work, Roberts (1970, 1972) conducted a survey for X-ray-induced recombination suppressors in D. melanogaster. He found that translocations and inversions whose breakpoints were near to specific cytological bands on the autosomal chromosomes strongly suppressed recombination across much of the chromosome arm. In a related work, Coyne et al. (1993) studied the fertility effects of heterozygosity for pericentric inversions and found that chromosomal inversions for which one or both breakpoints was near to a pairing, sensitive sites would partially restore fertility in heterozygous females. This indicates that inversion breakpoints near to sensitive sites result in suppressed recombination over the majority of chromosome arms. Finally, Hawley (1980) performed similar experiments to those of Roberts (1970, 1972), and used X-4 translocations to map sensitive sites on the X chromosome. In total, Hawley identified and fine mapped four sensitive sites across the euchromatic arm of the X chromosome. The D. melanogaster genome therefore contains a minimum of eight sensitive sites, many of which are relatively finely mapped.
Because of the sensitive sites’ powerful effects in promoting normal recombination during meiosis, it is plausible that proximity to these sites is an important selective force affecting inversion breakpoint positions. In this work, I compare the breakpoints of common and rare naturally-occurring chromosomal inversions in D. melanogaster. I show that the breakpoints of common inversions are significantly more likely than those of rare inversions to be found within cytological bands containing a sensitive site. In addition, common inversions that do not intersect a cytological band that contains a sensitive site are significantly longer than rare inversions. These data suggest that inversions whose breakpoints are near to sensitive sites have higher fitness than the average newly-formed inversions. This may indicate that inversions are favored by natural selection either because they are long or because they affect chromosome synteny nearby to a sensitive site. Finally, I discuss possible interpretations of these results.
Materials and Methods
Inversions analyzed
Owing to decades of study, >500 naturally-occurring chromosomal inversions have been reported in D. melanogaster (Krimbas and Powell 1992; Capy and Gibert 2012). For the purposes of this analysis, I focused on the exceptionally well-curated data set presented by Krimbas and Powell (1992). Traditionally, inversions in D. melanogaster are placed in one of four categories: common cosmopolitan, rare cosmopolitan, recurrent endemic, and unique endemic (Mettler et al. 1977; Krimbas and Powell 1992). Inversions in the first three categories achieve moderate to high frequencies either locally or globally in D. melanogaster populations, and due to modest sample sizes, I grouped inversions from these three categories into one: common inversions. Importantly, all inversions in this group have reached moderate frequencies suggesting that they experience higher relative fitness than rare inversions. I contrasted this common set with the unique endemic inversions, which are sufficiently rare polymorphisms that they should more closely reflect the neutral mutational processes that generate inversions (Messer 2009). I favored comparisons between common and rare inversions, rather than the more common framework contrasting fixed and polymorphic mutations (Hudson et al. 1987; McDonald and Kreitman 1991), because the D. melanogaster lineage has fixed only a single large paracentric inversion since its common ancestry with D. yakuba (Krimbas and Powell 1992; Ranz et al. 2007), therefore the sample size for fixed inversions in the D. melanogaster lineage is insufficient for quantitative analyses. Nonetheless, it is interesting to note that the distal breakpoint of the inversion that fixed on the D. melanogaster lineage is directly adjacent to the cytological band that contains the pairing sensitive site on chromosome arm 3R. In order to exclude their potentially confounding effects on fertility, I also excluded all pericentric inversions from these analyses.
Sensitive sites
Although the sensitive sites on the X and chromosome 3 are well mapped, and two independent studies identified strikingly-similar sensitive sites on chromosome 3; mapping positions of sensitive sites on chromosome 2 are discrepant. Rather than a single site, chromosome arm 2L appears to have a large sensitive region spanning many cytological bands (Roberts 1972), and consequentially recombination on this chromosome arm is particularly susceptible to disruptions. Furthermore, although Roberts (1972) identified cytological band 53 as the location of the sensitive site on chromosome arm 2R, the analysis of Coyne et al. (1993) instead identified band 49 as the location of the sensitive site on this chromosome arm. This disagreement presumably reflects the much smaller sample sizes for inversions and translocation breakpoints on chromosome 2R relative to the arms of chromosome 3 in both studies (7 and 10 breakpoints total are on 2R in Coyne et al. 1993 and Roberts 1972, respectively), and the consequentially poorer mapping resolution for this chromosome arm. For these reasons, I excluded chromosome 2 from all subsequent analyses. In total, I considered sensitive sites at cytological bands 3, 7, 11, 18, 68, and 92. Finally, I excluded all inversions that occurred exclusively on a chromosome arm with another inversion as interpreting the effects of breakpoint positions in these complex rearrangements is challenging. Breakpoint data and types for all inversions considered in this work are presented in Supplemental Material, Table S1.
Permutation tests
To determine if the number of common inversion breakpoints that intersect cytological bands containing sensitive sites exceeds what we would expect based on neutral mutational processes, I randomly sampled an equal number of rare inversions from each chromosome arm that contained a common inversion. In other words, the permuted sets sample the same number of inversions from each chromosome arm as the common inversions. This approach therefore controls for mutational heterogeneity between chromosome arms as well as among cytological bands, and is feasible here because of the large number of rare inversions that have been documented in natural populations of D. melanogaster. Within this set, I then counted the number of times a sampled inversion’s breakpoints intersected a cytological band containing a sensitive site and asked if this number was equal to or greater than the number of common inversion breakpoints that intersect cytological bands containing sensitive sites. I repeated this procedure 100,000 times, and the P-value is then the proportion of resampled rare inversion sets whose breakpoints intersect as many or more sensitive sites as the common inversions (this test is therefore one-tailed).
Breakpoint positions and length depend on a complex and largely unknown joint distribution. It is infeasible within a permutation framework to simultaneously account for all of these factors. Nonetheless, I employed two tests that control for length. First, to control for inversion lengths, regardless of breakpoint positions, I recorded the lengths of all common inversions, and then placed these inversions at random along their respective chromosome arms. In instances where the inversion would extend past the end of the chromosome arm, I drew a new starting breakpoint at random. I then counted the number of randomly-distributed inversions having a breakpoint that intersects a cytological band containing a sensitive site. I repeated this procedure 100,000 times and recorded the proportion that resulted in as many or more breakpoints intersecting sensitive sites as in the common inversions. Second, to control for inversion length and the position of one breakpoint, I recorded the lengths of all common inversions, and then sampled single inversion breakpoints at random from the rare inversions. I then inferred the corresponding breakpoint of this inversion based on the inversion’s length and the randomly sampled breakpoint. In instances where the other breakpoint is beyond the end of the chromosome arm, I drew a new breakpoint from the rare inversions. I then repeated the procedure above using this set of resampled inversions. For consistency with the previous works studying inversion breakpoint positions and sensitive sites, I measured inversion length in a number of cytological bands. I also obtained similar results using lengths measured in centimorgans and using lengths measured in base pairs (Figure S1). All permutation tests were conducted using custom PERL scripts. These are available as File S1 and File S2.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
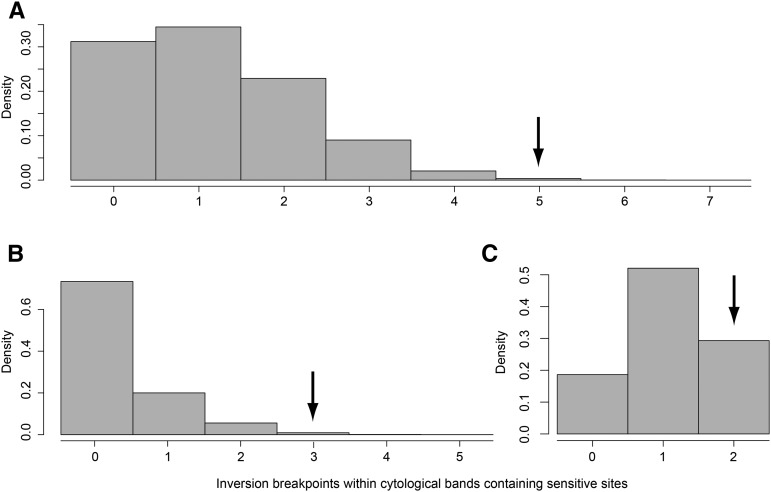
If the proximity of breakpoints to sensitive sites affects the fitness of chromosomal inversions, inversions for which one breakpoint intersects a cytological band containing a sensitive site will tend to have higher fitness than a randomly-selected inversion. Therefore, under this hypothesis, breakpoints of common inversions (n = 12) are expected to intersect these cytological bands at a higher rate than the breakpoints of rare inversions (n = 192, Table S1). Among the 12 common inversions in this data set, five have one breakpoint within a cytological band containing a sensitive site. Despite the relatively small sample size of common inversions, the probability of intersecting five or more sensitive sites is small (P = 0.028, 100,000 permutations; Figure 1). Furthermore, the difference between common and rare inversion breakpoints on chromosome 3 is marginally significant when this chromosome is analyzed in isolation; while the X chromosome, for which there are only two common inversions in D. melanogaster populations, does not show a significant enrichment for breakpoints in sensitive sites, but does trend in the expected direction (P = 0.065 and P = 0.29 for chromosome 3 and X, respectively; 100,000 permutations; Figure 1). Hence, these data are consistent with the idea that the proximity of breakpoints to sensitive sites affects the fitness of inversions in D. melanogaster populations.
Figure 1.
Distribution of the number of inversion breakpoints within permuted sets of rare inversions that intersect cytological bands containing sensitive sites. Arrows indicate the number of sensitive sites intersected by common inversion breakpoints in D. melanogaster populations. (A) For all inversions considered, (B) for inversions on chromosome 3, and (C) for inversions on chromosome X.
An important consideration for interpreting these results is that inversion breakpoint positions are not independent of length (Cáceres et al. 1997). Given that a substantial body of work indicates that inversion length is a selected trait (Olvera et al. 1979; Ruiz et al. 1984; Krimbas and Powell 1992; Cáceres et al. 1997), we must consider whether selection on inversion length alone may be a sufficient explanation for the enrichment of cooccurrence of common inversion breakpoints and sensitive sites. Importantly, longer inversions will tend to have breakpoints toward the center of chromosome arms (Cáceres et al. 1997), which is precisely where the sensitive sites tend to be located. However, when I randomly placed inversions of identical lengths to the set of common inversions, I found that the common inversions’ breakpoints are more likely to intersect cytological bands containing sensitive sites than is expected by chance (P = 0.023, 100,000 permutations). Furthermore, when I resampled a single breakpoint from the rare inversions and selected the other breakpoint at a distance equal to the inversion length to the common inversion, I obtained a similar result; indicating a significant excess of common inversion breakpoints are located within cytological bands containing sensitive sites (P = 0.028, 100,000 permutations). Collectively, these results suggest that selection on inversion lengths is not the primary selective force causing common inversions’ breakpoints to intersect cytological bands that contain sensitive sites at a higher rate than is expected by chance.
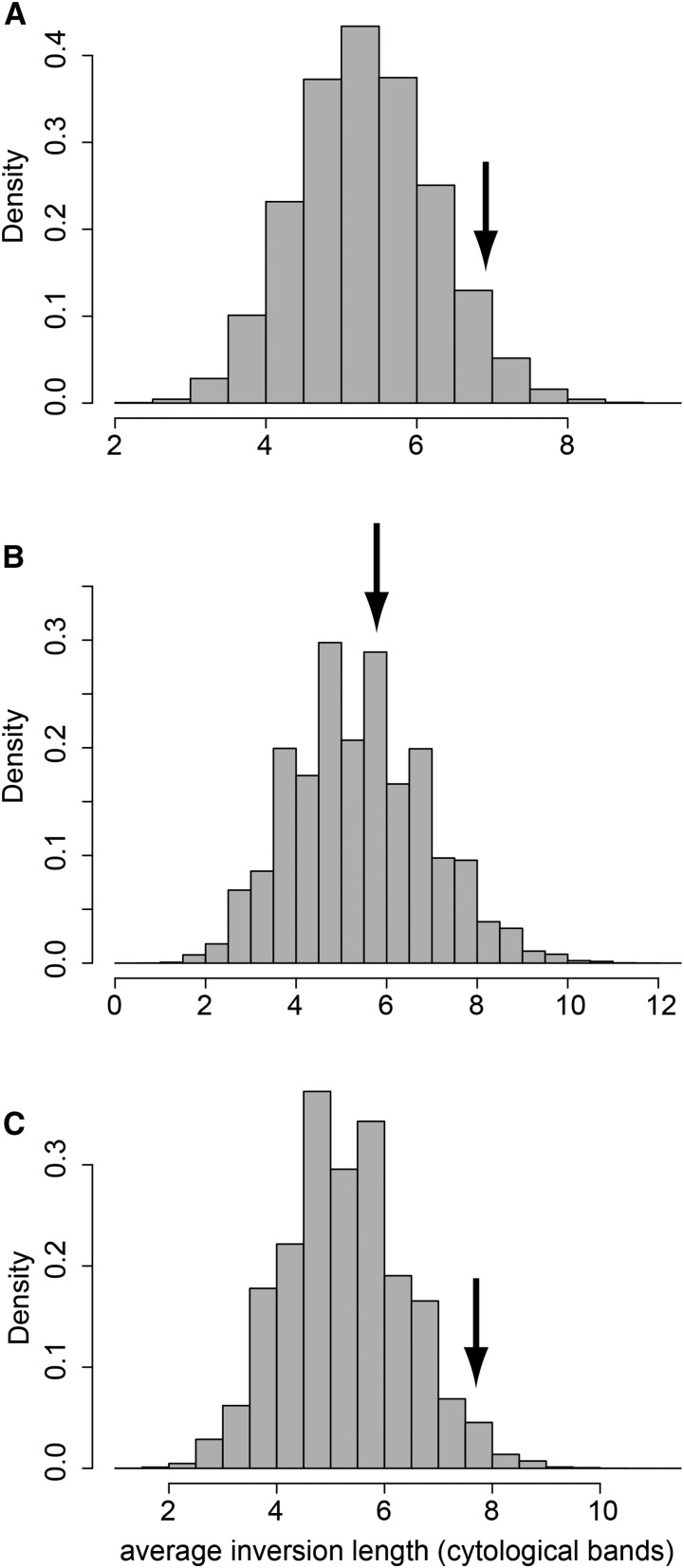
Nonetheless, given the strength of evidence demonstrating that natural selection acts on chromosomal inversion lengths, it is useful to consider how selection might impact inversion lengths in conjunction with breakpoint positions. I first asked if common inversions are longer on average than rare inversions, and I obtained marginally-significant results supporting this relationship (P = 0.052, 100,000 permutations; Figure 2). I then divided the common inversions into two groups. One group contained the five common inversions for which one breakpoint intersects a cytological band containing a sensitive site, and the other group contained the seven common inversions for which neither breakpoint intersects a cytological band that contains a sensitive site. When I repeated the above test, I found that inversions in the latter group, for which neither breakpoint intersects a sensitive site, are significantly longer than the rare inversions (P = 0.028, 100,000 permutations; Figure 2), while the common inversions that do intersect a sensitive site are much closer in length to that of rare inversions (P = 0.42, 100,000 permutations; Figure 2). Thus, for those chromosomal inversions whose breakpoints do not intersect a cytological band that contains a sensitive site, it appears that natural selection may favor increased inversion lengths.
Figure 2.
Distribution of the mean lengths of inversions within permuted sets of rare inversions. Arrows indicate the mean length of common inversions in D. melanogaster. (A) For all inversions considered, (B) for inversions for which one breakpoint intersects a cytological band containing a sensitive site, and (C) for inversions for which neither breakpoint intersects a cytological band containing a sensitive site.
Discussion
It is widely believed that chromosomal inversions are selected for their effects on suppressing recombination between alleles that are favorable in combination with one another (Krimbas and Powell 1992; Hoffmann and Rieseberg 2008; Kirkpatrick 2010). Consistent with these models, it is well established that longer inversions, which suppress recombination over larger genomic regions, are favorable relative to smaller inversions (Krimbas and Powell 1992; Cáceres et al. 1997). By comparison, the selective forces that influence breakpoint positions of chromosomal inversions are poorly understood, and inversion breakpoint positions have been comparatively understudied relative to inversion lengths. Based on the analyses presented here, it appears that in D. melanogaster, breakpoints of common inversions are significantly more likely to intersect a cytological band containing a sensitive site than are rare inversions. However, the common inversions whose breakpoints do not intersect a sensitive site are significantly longer than rare inversions, while the lengths of common inversions whose breakpoints do intersect a sensitive site show no evidence for increased lengths. Thus although length is an important aspect of inversion evolution, the forces that result in the colocalization of inversion breakpoints and sensitive sites may be largely independent from those that affect inversion lengths.
Although these data are consistent with differences in fitness between inversions whose breakpoints are near to sensitive sites and those whose breakpoints are not, there are numerous important complexities that should be considered. First, although the analysis presented here assumes that inversions can be treated independently of one another, inversion breakpoints are not necessarily strictly independent. For example, if suppressed recombination is favorable on many genetic backgrounds within a small genomic window, and that window is near to a sensitive site, it could contribute to this pattern observed here even without direct selective effects associated with sensitive sites. Alternatively, this explanation might instead predict that inversions would overlap the genomic window and would not necessarily require breakpoints to be in close proximity. Second, the sample sizes of common inversions and sensitive site locations are inherently limited. Given the breadth of cytological sampling of D. melanogaster populations, it is very unlikely that novel common inversions will be identified in this species. Furthermore, because the species has fixed only a single inversion since its common ancestry with D. yakuba, it is not feasible to investigate the relationship between fixed inversions and sensitive sites. I therefore caution that although these results are consistent with fitness differences associated with breakpoint proximity to sensitive sites, other factors may also explain the observations presented here and a conclusive link between sensitive sites and the relative fitness of chromosomal inversions will require additional studies.
Nonetheless, these data are consistent with the idea that inversions whose breakpoints are near to sensitive sites have higher fitness than the average newly-formed inversion. This could be consistent with at least two potential causes. First, it is possible that most novel inversions have deleterious fitness effects. If disrupting synteny near to sensitive sites alleviates those fitness costs, this group of inversions would have higher fitness than many newly-formed inversions. For example, many authors have reported that heterokaryotypic females display higher rates of nondisjunction than homokaryotypes (e.g., Roberts 1962; Forbes 1962). If disrupting synteny near to a pairing sensitive site decreases rates of nondisjunction, these inversions would tend to have higher fitness relative to those inversions that do not intersect sensitive sites. This or similar processes may enable these inversions to drift to polymorphic frequencies more readily than the majority of newly-formed inversions.
Alternatively, as in the majority of theoretical models that aim to explain why chromosomal inversions are sometimes favored by natural selection, it is possible that inversions that disrupt synteny near a sensitive site suppress recombination over large genetic distances. If these chromosomal inversions capture or acquire alleles that are favorable on the same haplotype that is sheltered from recombination, they may experience selective pressures similar to those that have been proposed in theoretical models of inversion evolution. This could explain why inversions for which one breakpoint is near to a sensitive site disproportionately achieve polymorphic frequencies in natural populations. These explanations are not mutually exclusive, and, undoubtedly, additional factors not specifically considered here will impact the evolution of polymorphic inversions in natural populations.
For two D. melanogaster inversions whose breakpoints are located in cytological bands containing sensitive sites, population genetic evidence is consistent with positive selection acting due to suppressed recombination between distinct alleles. In(X)Be’s proximal breakpoint is located within the cytological band containing the centromere-proximal sensitive site on the X chromosome. This inversion shows moderate sex-ratio distortion in laboratory crosses and despite its recent origin (estimated to be ∼600 generations), In(X)Be is present at ∼8% frequency across many populations in Sub-Saharan Africa (Corbett-Detig and Hartl 2012). It is likely that this inversion is favored by selection because it has captured a distorter locus and its modifiers, or several independent distorter loci (see Jaenike 2001 for a comprehensive review of sex-ratio inversions). In(3R)Mo also displays compelling evidence that natural selection is a primary factor influencing the evolution of this inversion and the centromere-proximal breakpoint of In(3R)Mo is located within the cytological band that contains the pairing sensitive site on this arm. In Raleigh, North Carolina, In(3R)Mo has increased dramatically in frequency from nearly undetectable levels in 1977 (Mettler et al. 1977) to ∼20% frequency in 2003 (Langley et al. 2012). Similar frequency increases have been observed for In(3R)Mo in many other populations as well (Kapun et al. 2014, 2016). Furthermore, In(3R)Mo maintains linkage between large haplotypes (∼1 Mb) that are distant from the inversion breakpoints both inside and outside of the inverted region (Corbett-Detig and Hartl 2012; Langley et al. 2012), potentially consistent with strongly-suppressed recombination across much of chromosome arm 3R and selection to maintain linkage between distant alleles. Nonetheless, further work is required to conclusively demonstrate that natural selection has favored these inversions because the proximity of their breakpoints to a sensitive site has resulted in suppressed recombination between mutations that are favored when in linkage disequilibrium.
Regardless of the specific factors that affect the relative fitness of inversions whose breakpoints are located near to sensitive sites, these results may carry a number of important implications for our understanding and interpretations of inversion breakpoint clustering that have been reported in a variety of species (Tonzetich et al. 1988; Krimbas and Powell 1992; Cáceres et al. 1997; Ranz et al. 2007). First, selection affecting inversion breakpoint positions due to proximity to sensitive sites may provide an explanation for observations of breakpoint clustering, as successful inversions will tend to contain breakpoints near to sensitive sites and will therefore necessarily colocalize with the breakpoints of other successful inversions. Second, if similar sensitive sites are present in the center of chromosome arms in the D. buzzatti subgroup species and other Drosophila species, selection for breakpoint proximity to those sites may partially explain the observation that intermediate length inversions appear to be favored by natural selection, though it is not immediately obvious how this effect could explain the inverse correlation between total map length and inversion lengths (Cáceres et al. 1997, 1999). Third, these data suggest that observed excess inversion breakpoint clustering have the potential to aid in the identification of sensitive sites that are important for recombination during meiosis in other species. Consistent with this idea, it is interesting to note that two of three common inversions on chromosome arm 2R in D. melanogaster have one breakpoint within band 49, which is the band that Coyne et al. (1993) identified as containing a sensitive site in their analysis; suggesting that this may be the location of the sensitive site on this chromosome arm. Breakpoint clustering in evolutionarily-successful inversions therefore offers a complementary approach to direct mapping experiments for identifying genomic features that are important for crossing over during meiosis. Confirming that band 49 and other potential sensitive sites affect recombination in heterokaryotypic individuals will be a target of future research in my group.
Although these data suggest a potential fundamental evolutionary process affecting genome structure evolution, there are several unanswered questions that limit our current understanding of sensitive sites. Perhaps the most important is that the basic nature of the sensitive sites is not well understood. Despite the fact that independent studies have demonstrated the existence of sensitive sites on different genetic backgrounds in D. melanogaster (Roberts 1970, 1972; Coyne et al. 1993), we lack a specific understanding of the mechanism through which these sites impact the distribution of chiasmata in this species. Furthermore, although Hawley (1980) identified intercalary heterochromatin in the regions associated with sensitive sites on the X chromosome, other intercalary heterochromatic regions do not act as sensitive sites. Advancing our understanding of the genetics of sensitive sites is essential to understanding these evolutionary processes. Finally, it is not known how many species require pairing at sensitive sites to produce normal crossover frequencies, and the available evidence is limited to a few well-characterized model organisms. At a minimum, the D. melanogaster (Roberts 1970, 1972; Hawley 1980; Coyne et al. 1993) and Caenorhabditis elegans genomes (McKim et al. 1988, 1993) contain numerous sensitive sites, suggesting that selection on breakpoint proximity to sensitive sites may affect the evolution of inversion polymorphism in a wider array of species. Future work is necessary to fully explore the selective and functional effects of proximity to sensitive sites in chromosomal inversions and other types of structural rearrangements in D. melanogaster as well as in other species.
Acknowledgments
I thank Rebekah Rogers, Shelbi Russell, Stefan Prost, Rasmus Nielsen, John Pool, David Begun, Peter Andolfatto, and four anonymous reviewers for helpful comments on this manuscript. This work was supported by a University of California Berkeley Chancellor’s Postdoctoral Fellowship and by a National Science Foundation dissertation defense improvement grant to R.B.C.-D. I have no conflicts of interest to declare.
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.190389/-/DC1.
Communicating editor: D. J. Begun
Literature Cited
- Brehm A., Krimbas C. B., 1991. Inversion Polymorphism in Drosophila obscura. J. Hered. 82: 110–117. [DOI] [PubMed] [Google Scholar]
- Capy, P., and P. Gibert, 2012 Drosophila melanogaster, Drosophila simulans: So Similar, So Different, pp. 5–16 in Contemporary Issues in Genetics and Evolution, Vol. 11, edited by P. Capy, P. Gibert, and I. Boussy. Springer Science & Business Media, Dordrecht, The Netherlands. [Google Scholar]
- Castermans D., Vermeesch J. R., Fryns J.-P., Steyaert J. G., Van de Ven W. J. M., et al. , 2007. Identification and characterization of the TRIP8 and REEP3 genes on chromosome 10q21.3 as novel candidate genes for autism. Eur. J. Hum. Genet. 15: 422–431. [DOI] [PubMed] [Google Scholar]
- Cáceres M., Barbadilla A., Ruiz A., 1997. Inversion length and breakpoint distribution in the Drosophila buzzatii species complex: is inversion length a selected trait? Evolution 51: 1149–1155. [DOI] [PubMed] [Google Scholar]
- Cáceres M., Barbadilla A., Ruiz A., 1999. Recombination rate predicts inversion size in Diptera. Genetics 153: 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., Marais G., 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128. [DOI] [PubMed] [Google Scholar]
- Corbett-Detig, R. B., and D. L. Hartl, 2012 Population Genomics of Inversion Polymorphisms in Drosophila melanogaster. PLoS Genet. 8: e1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., Meyers W., Crittenden A. P., Sniegowski P., 1993. The fertility effects of pericentric inversions in Drosophila melanogaster. Genetics 134: 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crumpacker D. W., Kastritsis C. D., 1967. A New Gene Arrangement in the Third Chromosome of Drosophila Pseudoobscura. J. Hered. 58: 3–6. [PubMed] [Google Scholar]
- Forbes C., 1962. The Effect of Heterozygous Inversions on Primary Nondisjunction in Drosophila Melanogaster. Genetics 47: 1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley R. S., 1980. Chromosomal sites necessary for normal levels of meiotic recombination in Drosophila melanogaster. I. Evidence for and mapping of the sites. Genetics 94: 625–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Rieseberg L. H., 2008. Revisiting the Impact of Inversions in Evolution: From Population Genetic Markers to Drivers of Adaptive Shifts and Speciation? Annu. Rev. Ecol. Evol. Syst. 39: 21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Kreitman M., Aguadé M., 1987. A Test of Neutral Molecular Evolution Based on Nucleotide Data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike J., 2001. Sex chromosome meiotic drive. Annu. Rev. Ecol. Syst. 32: 25–49. [Google Scholar]
- Kapun M., Schalkwyk H., McAllister B., Flatt T., Schlotterer C., 2014. Inference of chromosomal inversion dynamics from Pool‐Seq data in natural and laboratory populations of Drosophila melanogaster. Mol. Ecol. 23: 1813–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapun M., Fabian D. K., Goudet J., Flatt T., 2016. Genomic Evidence for Adaptive Inversion Clines in Drosophila melanogaster. Mol. Biol. Evol. 33: 1317–1336. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M., 2010. How and Why Chromosome Inversions Evolve. PLoS Biol. 8: e1000501–e1000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., Barton N. H., 2006. Chromosome Inversions, Local Adaptation and Speciation. Genetics 173: 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimbas, C. B., and J. R. Powell, 1992 Drosophila Inversion Polymorphism, CRC Press, Boca Raton, FL. [Google Scholar]
- Langley, C. H., K. Stevens, C. Cardeno, Y. C. G. Lee, D. R. Schrider et al., 2012 Genomic Variation in Natural Populations of Drosophila melanogaster. 192: 533–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M. F., 2003. Transmission ratio distortion in mice. Annu. Rev. Genet. 37: 393–408. [DOI] [PubMed] [Google Scholar]
- McDonald J. H., Kreitman M., 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- McKim K. S., Howell A. M., Rose A. M., 1988. The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics 120: 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K. S., Peters K., Rose A. M., 1993. Two types of sites required for meiotic chromosome pairing in Caenorhabditis elegans. Genetics 134: 749–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer P. W., 2009. Measuring the Rates of Spontaneous Mutation From Deep and Large-Scale Polymorphism Data. Genetics 182: 1219–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettler L. E., Voelker R. A., Mukai T., 1977. Inversion clines in populations of Drosophila melanogaster. Genetics 87: 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A., Ruiz A., 1997. On the Fertility Effects of Pericentric Inversions. Genetics 147: 931–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olvera O., Powell J. R., De La Rosa M. E., Salceda V. M., 1979. Population genetics of Mexican Drosophila VI. cytogenetic aspects of the inversion polymorphism in Drosophila pseudoobscura. Evolution 33: 381–395. [DOI] [PubMed] [Google Scholar]
- Ranz J. M., Maurin D., Chan Y. S., von Grotthuss M., Hillier L. W., et al. , 2007. Principles of Genome Evolution in the Drosophila melanogaster Species Group. PLoS Biol. 5: e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P. A., 1962. Interchromosomal effects and the relation between crossing-over and nondisjunction. Genetics 47: 1691–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P. A., 1970. Screening for X-Ray-Induced Crossover Suppressors in Drosophila melanogaster: Prevalence and Effectiveness of Translocations. Genetics 65: 429–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P. A., 1972. Differences in synaptic affinity of chromosome arms of Drosophila melanogaster revealed by differential sensitivity to translocation heterozygosity. Genetics 71: 401–415. [DOI] [PubMed] [Google Scholar]
- Ruiz A., Naveira H., Fontdevila A., 1984. La historia evolutiva de Drosophila buzzatii. IV. Aspectos citogenéticos de su polimorfismo cromosómico. Genet. Iber. 36: 13–35. [Google Scholar]
- Tadin-Strapps M., Warburton D., Baumeister F. A. M., Fischer S. G., Yonan J., et al. , 2004. Cloning of the breakpoints of a de novo inversion of chromosome 8, inv (8)(p11.2q23.1) in a patient with Ambras syndrome. Cytogenet. Genome Res. 107: 68–76. [DOI] [PubMed] [Google Scholar]
- Tonzetich J., Lyttle T. W., Carson H. L., 1988. Induced and natural break sites in the chromosomes of Hawaiian Drosophila. Proc. Natl. Acad. Sci. USA 85: 1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen L., Levins R., 1968. The origins of inversion polymorphisms. Am. Nat. 102: 5–24. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.