Abstract
We identified loci responsible for natural variation in Arabidopsis thaliana (Arabidopsis) responses to a bacterial pathogen virulence factor, HopAM1. HopAM1 is a type III effector protein secreted by the virulent Pseudomonas syringae strain Pto DC3000. Delivery of HopAM1 from disarmed Pseudomonas strains leads to local cell death, meristem chlorosis, or both, with varying intensities in different Arabidopsis accessions. These phenotypes are not associated with differences in bacterial growth restriction. We treated the two phenotypes as quantitative traits to identify host loci controlling responses to HopAM1. Genome-wide association (GWA) of 64 Arabidopsis accessions identified independent variants highly correlated with response to each phenotype. Quantitative trait locus (QTL) mapping in a recombinant inbred population between Bur-0 and Col-0 accessions revealed genetic linkage to regions distinct from the top GWA hits. Two major QTL associated with HopAM1-induced cell death were also associated with HopAM1-induced chlorosis. HopAM1-induced changes in Arabidopsis gene expression showed that rapid HopAM1-dependent cell death in Bur-0 is correlated with effector-triggered immune responses. Studies of the effect of mutations in known plant immune system genes showed, surprisingly, that both cell death and chlorosis phenotypes are enhanced by loss of EDS1, a regulatory hub in the plant immune-signaling network. Our results reveal complex genetic architecture for response to this particular type III virulence effector, in contrast to the typical monogenic control of cell death and disease resistance triggered by most type III effectors.
Keywords: Arabidopsis thaliana, Pseudomonas syringae, type III effectors, HopAM1, QTL, genetics of immunity
PLANT pathogens have evolved complex strategies to circumvent the host innate immune response and enhance virulence, including the use of effector proteins to disrupt host immune signaling (Macho and Zipfel 2015). Plant immunity functions through extra- and intracellular receptor systems. Cell surface pattern-recognition receptors respond to pathogen molecules such as flagellin by activation of a complex defense signaling cascade called microbe (pathogen)-triggered immunity (MTI). MTI leads to production of reactive oxygen, secretion of pathotoxins, and reinforcement of cell walls at the infection site, which collectively protect the plant from infection. Successful pathogens introduce into host cells effector proteins that interact with components of the MTI response to dampen it, allowing the pathogen to evade host immunity. In response, the plant immune system evolved an intracellular class of highly polymorphic immune receptors, termed nucleotide binding leucine-rich repeat (NLR) proteins (Bonardi and Dangl 2012). There are two classes distinguished by their N-terminal domains: Toll/interleukin-1 receptor domain (TIR)-NLRs that require the function of the defense protein EDS1, and coiled-coil (CC)-NLRs that are generally independent of EDS1 (Feys et al. 2001; Wiermer et al. 2005). These proteins can associate with targets of pathogen effectors that are presumably components of the MTI response, or decoys thereof (Dangl and Jones 2001; van der Hoorn and Kamoun 2008; Dodds and Rathjen 2010; Kroj et al. 2016). When the targets are perturbed by a pathogen effector, an associated NLR can be activated, leading to an effector-triggered immune response (ETI). ETI is often thought of as an amplified MTI response (Tao et al. 2003; Jones and Dangl 2006), commonly resulting in rapid cellular desiccation and death at the site of attempted infection (Wright and Beattie 2004). This hypersensitive cell death response can isolate microbial pathogens and halt their proliferation. Because effectors are required to suppress MTI, but can trigger ETI, successful pathogens are under evolutionary pressure to continuously “re-sort” their effector collections to suppress MTI while avoiding NLR receptor activation. This evolutionary tug of war has led to bacterial pathogens with diverse collections of effectors that often function through unusual chemistries or via convergent evolution to mimic eukaryotic enzymatic functions (Fu et al. 2007; Zhang et al. 2007; Cheong et al. 2014).
MTI and ETI are accompanied by dramatic changes in host hormone signaling and altered expression of thousands of host genes (Zipfel et al. 2004, 2006; Howard et al. 2013; Lewis et al. 2015). Pathogen effector proteins interact with specific host targets that potentially influence only a sector or branch of this host immune output. Identification of host targets of pathogen effectors has been a useful method to define and study subsets of the immune system and to understand how they contribute to immunity (Deslandes and Rivas 2012; Macho and Zipfel 2015). Here, we investigated the genetics of natural variation in plant responses to toxic effects of the Pseudomonas syringae type III secretion-system effector HopAM1. P. syringae pv. tomato DC3000 (Pto DC3000), a Gram-negative bacterium virulent on tomato and on Arabidopsis thaliana (Arabidopsis), is an extensively studied model pathogen for plant-microbe interactions (Chang et al. 2005; Schechter et al. 2006; Cunnac et al. 2011; Xin and He 2013; Lewis et al. 2015). Pto DC3000 has at least 30 type III effector genes including two nucleotide sequence-identical copies of hopAM1, one in the genome (PSPTO_1022) and one in the plasmid (PSPTO_A0005) (Buell et al. 2003). The hopAM1 gene is sporadically dispersed among phylogenetically diverse P. syringae isolates (Arnold et al. 2001; Baltrus et al. 2011) and other Gram-negative plant pathogen groups, such as Ralstonia, Xanthomonas, and Pantoea (Integrated Microbial Genomes and Microbiomes, http://img.jgi.doe.gov/) (Nordberg et al. 2014). HopAM1 was originally identified from P. syingae pv. pisi via its ability to trigger a presumably NLR-mediated ETI response on some pea cultivars (Cournoyer et al. 1995). Addition of hopAM1 to the effector complement of the weakly pathogenic strain P. syringae pv. maculicola (Pma M6CdE) conferred a growth advantage on Arabidopsis (Goel et al. 2008). Furthermore, hopAM1 was one of eight type III effector genes that collectively restored nearly full virulence when added back to a completely growth-defective derivative of Pto DC3000, Pto DC3000D28E, from which the 28 most strongly expressed type III effector genes had been removed by sequential deletion (Cunnac et al. 2011). Like many other type III effectors, HopAM1 has toxic effects when overexpressed from a transgene in plants (Goel et al. 2008) and even in yeast (Munkvold et al. 2008). This latter phenotype has hampered approaches to define the biochemical function of HopAM1.
HopAM1 induces two unusual responses on Arabidopsis which are both variable across Arabidopsis accessions. HopAM1 can induce meristem chlorosis on many accessions; noted as emergence of chlorotic leaves 5–10 days after onset of HopAM1 expression when expressed from a conditionally induced transgene in Arabidopsis, or following inoculation of plant leaves with disarmed Pseudomonas strains designed to deliver HopAM1 by type III secretion. The effect appears to be systemic since it was not possible to recover any of the inoculated bacterial strain from the chlorotic leaves that emerged following hand inoculation of more mature leaves (Goel et al. 2008). Disarmed Pseudomonas strains expressing hopAM1 can also induce local cell death in leaves at the infiltration site, and the timing of onset is variable across inbred Arabidopsis accessions. We exploited this natural variation in Arabidopsis responses to HopAM1 to define host loci that control them. Using linkage analyses in both experimental crosses and the global population to describe the genetic architecture of the response to HopAM1, we identified multiple loci associated with differential response to HopAM1 for both cell death and meristem chlorosis.
Materials and Methods
A. thaliana germplasm, propagation, and growth conditions
Seeds were germinated on a mix of potting soil (13:6:3 of Metro-Mix 360, baked sand, and Perlite; 1.4 g of Peters 20-20-20 All Purpose Fertilizer; and 0.85 ml of Marathon per liter of soil mix); stratified for 5 days at 4°; transferred to growth chambers; and grown for 9 hr under light (5900 lx), 15 hr in dark at 21° (day) and 18° (night) at 45–50% relative humidity. For propagation, plants were transferred to a long day (15 hr light) greenhouse with similar conditions. Plants for HopAM1-induced meristem chlorosis were prepared differently as described below. Col-0 × Bur-0 recombinant inbred line (RIL) population was obtained from Christine Camilleri (Institut National de la Recherche Agronomique, France) (Simon et al. 2008). A. thaliana accessions, mutant, and transgenic CS_ and SALK_ lines were obtained from the Arabidopsis Biological Resource Center (ABRC) (Ohio State University, Columbus, OH). The eds1-2 line is a null mutant introgressed from a mutant in the accession Ler-0 into the Col-0 background (Bartsch et al. 2006). Mutant lines axr2-1, abi1-1, and aba2 were obtained from Jason Reed (University of North Carolina, Chapel Hill, NC) (Nagpal et al. 2000); ein2-5 and ein2-50 from Joseph Kieber (University of North Carolina, Chapel Hill, NC) (Wang et al. 2007); the global DELLA quintuple mutant from Salomé Prat (Centro Nacional de Biotecnologia, Madrid, Spain) (Cheng et al. 2004); and the triple TOC64 mutant from Úrsula Flores Pérez (Oxford University, Oxford, United Kingdom) (Aronsson et al. 2007).
Bacterial strains and growth assay
A list with strains and phenotypes assayed can be found in Supplemental Material, Table S1. Bacterial strains were maintained and grown as described (Chung et al. 2011) and in planta growth assays were done as described (Chang et al. 2005). Bacterial strains were grown in King’s B liquid media at 28° with shaking overnight. Antibiotic concentrations used for P. syringae, P. fluorescens, and P. maculicola were: 50 μg/ml rifampicin, 50 μg/ml kanamycin, 10 μg/ml spectinomycin, and 5 μg/ml tetracyclin. Weighted ANOVA was used to compare bacterial growth differences between strains from multiple experiments in Col-0 and Bur-0. Comparisons among the groups were done using Tukey’s post hoc test.
DNA extraction and PCR conditions for genotyping
We modified the protocol from Klimyuk et al. (1993) as follows. Plant material (one to two young leaves) was harvested and placed in a well (Deepwell plate 96/2000 μl; Eppendorf) along with a 4-mm borosilicate glass ball and 400 μl 0.25 M NaOH. Samples were then homogenized at 1200 strokes/min for 1 min (2000 GENO/GRINDER; Spex-CentriPrep). Plates were centrifuged for 1 min at 1000 rpm to precipitate plant tissue. A 50 μl volume of supernatant was transferred into a 96-well PCR plate (nonskirted; MultiMax) and heated at 96° for 30 sec in a Peltier thermal cycler (DNA Engine; Bio-Rad, Hercules, CA) with open lid. A 50 μl volume of 0.25 M HCl was added to neutralize the sample, and 25 μl of alkaline lysis buffer (0.5 M Tris-HCl, pH 8.0, 0.25% v/v IGEPAL CA-630) was added immediately after, mixing the samples by pipetting. The PCR plate was heated again at 96° for 120 sec and left to cool at room temperature for 5 min. A 1–2 μl volume of DNA sample was used for PCR for genotyping. PCRs were set for a 7 min “hot-start” step at 95°, followed by 35 cycles of 30 sec 95° denaturation, 30 sec X (primer-specific)° annealing, 60 sec 72° extension, and concluding with a 5 min 72° final extension. Optimized 10× PCR buffer [0.11 M KCl, 0.11 M (NH4)2SO4, 0.24 M Tris-HCl (pH 8.3), 0.022 M MgCl2, 0.6% v/v Triton X-100, 1.6% v/v Tween 20, 1.2% v/v NP-40, 1.5% v/v formamide, 0.1 mg/ml BSA, 10% v/v glycerol, and 0.02 M TMAC] was used for genotyping.
HopAM1-induced cell death assay
Cell death symptoms induced by bacteria were scored following trypan blue staining (Boyes et al. 1998) at various time intervals. Plants that were 5- to 6-weeks old were hand inoculated with bacteria. Inocula with Pto-derived strains were adjusted to an OD600 of 0.1 (∼5 × 107), and with Pfo or Pma strains to an OD600 of 0.2 (∼1 × 108 CFU/ml) in 10 mM MgCl2. For phenotyping, 3–4 leaves of 5-week-old plants were marked with a Sharpie felt pen on the axial side and the abaxial side was hand inoculated with Pto DC3000D28E(hopAM1) (see Table S1), and scored for cell death response at 24, 26, 32, 48, 72, and 96 hpi (Figure 1C). Quantitative scoring of cell death symptoms ranged from 5 (apparent by 24 hpi) to 0 (not apparent by 96 hpi) based on the time point that symptoms were first observed.
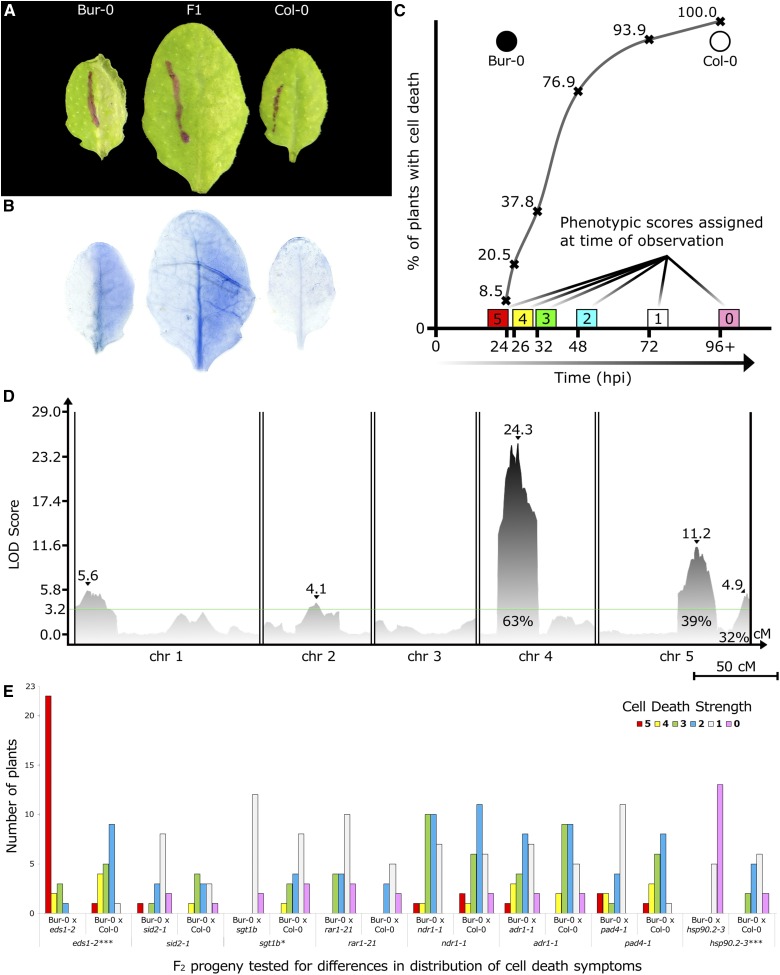
Figure 1.
HopAM1 induces variable cell death symptoms of multigenic inheritance in A. thaliana accessions. (A) HopAM1-induced cell death in Bur-0, Col-0, and F1 progeny at 48 hpi after hand inoculation with Pto DC3000D28E(hopAM1). OD600 = 0.1 (∼5 × 107 CFU/ml). Bur-0 exhibits one of the strongest onsets of symptoms as early as 23 hpi, while Col-0 exhibits only chlorosis starting ∼96 hpi. F1 progeny become symptomatic ∼48 hpi. (B) Cell death shown with trypan blue staining on the same leaves as above (48 hpi). (C) Sigmoid distribution of cell death symptoms representative of quantitative traits. The quantification of cell death at specific time points postinoculation is shown. Curve shows the percentage of plants in a Bur-0 × Col-0 RIL population that exhibit cell death symptoms at a given time point. ● = Bur-0; ○ = Col-0. (D) CIM on 342 reannotated RILs from a Col-0 × Bur-0 collection. The x-axis displays chromosomes 1 through 5 with map distances in cM. Peak LOD scores are shown by ▾ along with their values. Effects on phenotype variation are shown as percentages for QTL4 and QTL5A. The global permutation level of significance was set on LOD 3.2. Chr, chromosome. Bar, 50 cM. (E) Distribution of HopAM1-induced cell death scores among different F2 progeny between Bur-0 and mutant lines. The y-axis shows number of plants selected from each population with each phenotype score for HopAM1-induced cell death (5 = Bur-0, 0 = Col-0). Statistical significance between distributions was based on two-way ANOVA tests for each pair (statistical significance: *** P ≤ 0.001, * P ≤ 0.05).
HopAM1-induced meristem chlorosis assay
Tissue culture plates (six well, #353046; Falcon, Lincoln Park, NJ) were filled with sifted soil mix (prepared as described in Boyes et al. 1998; Holt et al. 2002). Each plate was then covered with nylon mesh and fastened with four wide rubber bands. Four plastic head pins were placed inside the corners of each plate to provide aeration during germination and support during vacuum infiltration. A total of 5–10 seeds were sown on each soil mount, plate lids were then placed on the pins, and plates were moved to 4° for 4–5 days. Seedlings were thinned to a single plant in the middle of each mount. Three 4-week-old plants per line were vacuum infiltrated with Pto DC3000D28E(hopAM1) and scored for chlorotic rosette phenotype at 10 days post inoculation (dpi): Plates were placed inverted in a rectangular plastic container filled with bacterial inoculum (OD600 of 0.05 (∼2.5 × 107 CFU/ml), 10 μM MgCl2, 60 μl/liter Silwet L-77 surfactant), 4-cm deep, and vacuum was applied for 100–120 sec. Plants were transferred into growth rooms. Phenotypic scores were assigned based on visible chlorosis in rosettes at 10 dpi. The phenotype was recorded as “any response” (1) or “no response” (0).
QTL analysis
Cell death:
Three F8 plants each of 342 lines from a Col-0 × Bur-0 RIL mapping population (Simon et al. 2008) were scored for HopAM1-induced cell death, following hand inoculation with Pto DC3000D28E(hopAM1). Initial QTL analysis on the 89 markers spanning five chromosomes of A. thaliana was performed by composite interval mapping (CIM) using WinQTL Cartographer 2.5.011 (Wang et al. 2012). After the detection of several QTL, the entire RIL population was reannotated using 60 additional markers to provide extra resolution for CIM. New markers were selected proximally and distally for each marker within the QTL regions detected. Mapping distances were recalculated using MapMaker 3.0 (Lincoln et al. 1992). For detecting QTL, a LOD score of 3.2 was chosen based on 1000 permutations at P ≤ 0.01 for CIM. Walking speed was 0.5 cM and window size was 10 cM. Epistatic interactions between the QTL identified in CIM were investigated in detail using a general linear model structure (Brady et al. 2015), which provides more power than traditional pairwise t-tests in factorial interaction studies, by using the ANOVA framework.
Meristem chlorosis:
Three plants per Col-0 × Bur-0 RIL (Simon et al. 2008) were scored for HopAM1-induced meristem chlorosis at 10 and 14 dpi, following vacuum infiltration with Pto DC3000D28E(hopAM1). CIM was performed again on the original genotyping data (89 markers) and the reannotated data (89 + 60 markers). QTL was detected as in Cell death above. Other parameters were set at default. The model for corrected means that was used before for HopAM1-induced cell death (see Table S2) was not suitable for the binary nature of phenotypes for HopAM1-induced meristem chlorosis and was not included in the model. Phenotypic scores for all 342 RILs were assigned as observed (with at least one responsive replicate resulting in a score of 1). Symptoms similar to HopAM1-induced cell death and meristem chlorosis have not been observed before in untreated Col-0 × Bur-0 RILs (Simon et al. 2008; Gery et al. 2011; Chavigneau et al. 2012).
Genome-wide association (GWA) analysis:
To obtain a species-wide view of HopAM1-induced cell death and meristem chlorosis, we assayed 98 A. thaliana accessions (ABRC). For most of our accessions (n = 64), whole-genome sequences were available (http://1001genomes.org; 1001 Genomes Consortium 2016). We used 4,004,754 SNPs with 0.98 genotyping rate and minor allele frequency (MAF) >5% to perform GWA analyses, using a linear mixed model to correct for population structure, implemented in the Efficient Mixed-Model Association eXpedited (EMMAX) package (Kang et al. 2010), as this has been proven to be efficient for Arabidopsis (Atwell et al. 2010). This model fits an additive genetic variance factor using a kinship matrix, which allows calculation of narrow sense heritability for the GWA population. To test the significance of the genetic variance factor, we used a likelihood ratio test comparing the fit of the full model against the null model without the kinship matrix. Allele effect sizes were reported as standardized regression coefficients or β. They are interpreted as the difference between genotypes carrying the MAF allele to those with the reference allele, and are expressed in standard deviations of the phenotype. Enrichment of top SNPs in QTL peaks was tested using a Fisher’s exact test with two-by-two tables of counts as follows:
Top GWA SNPs inside QTL range: total SNPs inside QTL range.
Top GWA SNPS in the chromosome of the QTL: total SNPs in the chromosome of the QTL.
Confocal microscopy:
Small leaves (5-mm diameter) were collected 14 days after vacuum infiltration with Pto DC3000(hopAM1). The abaxial side of Col-0 and Bur-0 leaves was observed using a C-Apochromat 40×/NA1.2 water immersion lens on a Carl Zeiss (Thornwood, NY) LSM710 confocal laser scanning microscope. Autofluorescence was observed using a 560-nm diode laser line for excitation and the photomultiplier tube detector collected emission bandwidth set at 568–740 nm. The confocal images were edited with ZEN 2009 software.
RNA sequencing:
We used RNA sequencing (RNA-seq) to investigate HopAM1-associated changes in the transcriptome of infected Arabidopsis leaves. Plants that were 5 weeks old and grown under short days were hand-infiltrated (OD600 = 0.1, ∼2.5 × 107 CFU/ml) with the control strain Pto DC3000D28E transformed with the empty vector pJC531; or with Pto DC3000D28E(hopAM1) (Goel et al. 2008). The accessions Col-0 and Bur-0 were used in the experiment. Samples were harvested immediately before (time = 0 hr) and 2, 4, 6, 8, 10, and 12 hr after bacteria infiltration. Three biological replicates were analyzed per condition, each consisting of three 7-mm disks collected from the inoculated leaf of one individual plant. Samples were frozen in liquid nitrogen and ground using glass beads and the Tissue Lyser II system (QIAGEN, Valencia, CA). Total RNA was extracted using TRIzol (Invitrogen, Purchase, NY), DNase (Ambion Turbo DNase) treated, and purified using the RNeasy Mini Kit (QIAGEN). A 1 µg amount was used to prepare Illumina-based mRNA-seq libraries. Quality control and quantification of the final libraries were performed using the 2100 Bioanalyzer (Agilent) and the Quant-iT PicoGreen dsDNA Reagent (Invitrogen). Barcoded libraries were combined into a single pool and sequenced in two lanes of an Illumina HiSeq2500, resulting in an average of 2.1 million 50-bp single-end reads per library.
RNA-seq reads were mapped using TopHat (Trapnell et al. 2009), allowing only one mismatch and discarding any read that mapped to multiple positions. The TAIR10 assembly was used as the reference genome for Col-0, whereas the assembly available at http://mus.well.ox.ac.uk/19genomes/ was used as the reference for Bur-0 (Schneeberger et al. 2011). Approximately 90% of reads could be mapped to the Arabidopsis genome. Reads mapped to nuclear protein-coding genes were counted using HTSeq (Anders et al. 2015) and the package edgeR (Robinson et al. 2010) was used to define differentially expressed genes between plants infiltrated with Pto DC3000D28E(hopAM1) and Pto DC3000D28E(EV) at each time point. Genes with a false discovery rate (FDR) <0.05 and a fold-change variation >2 were considered differentially expressed between conditions. All nuclear protein-coding genes were tested for differential expression without the adoption of thresholds to filter out weakly expressed genes. Gene ontology (GO) enrichment analyses were performed with the PlantGSEA platform (Yi et al. 2013) using the sets of differentially expressed genes. Transcriptional activation of chosen gene sets was represented as the median z-score transformed expression values [in reads per kilobase of transcript per million (RPKM) mapped reads].
Data availability
Bacterial strains are available upon request. All data necessary for reproducing QTL results and complete edgeR results for RNA-seq data can be found as supplemental materials. Raw RNA-seq data are available at the National Center for Biotechnology Information Sequence Read Archive under the accession number SRP075162.
Results
The quantitative nature of HopAM1-induced cell death
Previously, we cloned 93 type III secretion system effectors from P. syringae isolates (Baltrus et al. 2011) and transferred them individually into an engineered strain of the nonpathogenic P. fluorescens bacterium carrying a heterologous type III secretion system apparatus (Thomas et al. 2009). Disease resistance responses are often qualitative, and we hoped to identify several such host responses across this survey. HopAM1 induced obvious cell death in some accessions. To better define the distribution of phenotypes across the host species and to identify accessions that would be suitable for mapping in experimental crosses, we rescreened a collection of 98 accessions delivering HopAM1 by infection with the disarmed bacterial strain Pto DC3000D28E (Cunnac et al. 2011) bearing a plasmid with the hopAM1 gene with a C-terminal HA tag expressed from its own promoter (Goel et al. 2008). Although its virulence is severely reduced, Pto DC3000D28E stimulates MTI effectively and is at least as effective as P. fluorescens for delivery of type III effectors. Following hand inoculation of leaves with Pto DC3000D28E(hopAM1), we observed a range of cell death responses at the site of inoculations that varied between Arabidopsis accessions in the timing of onset of cell collapse (see Figure S1 and Table S3). Bur-0 showed strong cell death symptoms at 24 hpi; and Col-0 did not suffer from any symptoms, not even microscopic lesions visible with trypan blue staining (see Figure S2), until 6 dpi (∼144 hpi) (Figure 1, A and B). Analysis of an F2 population (468 plants) of Bur-0 × Col-0 demonstrated non-Mendelian segregation of HopAM1-induced cell death (see Table S4). Therefore, we followed the timing of appearance of cell death symptoms after inoculation to derive a quantitative cell death score (Figure 1C).
Screening and reannotation of Bur-0 × Col-0 RILs
We explored the genetic architecture driving HopAM1-induced cell death using an established Bur-0 × Col-0 F8 RIL population (Simon et al. 2008) screened with Pto DC3000D28E(hopAM1). Cell death symptoms developed continuously over time after inoculation (see Figure S2) in the RIL population and phenotyping was performed following the numerical scoring system illustrated in Figure 1C. Two individual plants from each RIL were infiltrated with Pto DC3000D28E(hopAM1) and tissue was harvested from both for genotyping (see Table S5). The distribution of phenotypic scores among the RILs was similar (see Figure S3) to the distribution of the 98 A. thaliana accessions screened previously, where most lines presented with intermediate cell death phenotypes (Kolmogorov–Smirnov test for similarity between distributions; D = 0.18, P = 0.014) (see Figure S1). The parental accessions Col-0 and Bur-0 had the most extreme phenotypes, and there was no transgressive segregation among RILs. To evaluate the heritability of the cell death response in the RILs, a second screen of the RIL population was performed, and the initial phenotypic scores were adjusted to control for environmental effects using a general linear model (see Table S2) (Chan et al. 2011; Brady et al. 2015). The broad sense heritability (H2) within the RILs for HopAM1-induced cell death was high (H2 = 95.4%, P < 2 × 10−16), with a very low amount of phenotypic variation being controlled by combined experimental factors (H2 = 0.06%) (see Table S2).
Five Arabidopsis loci additively control HopAM1-dependent cell death in the Bur-0 × Col-0 cross
CIM performed among the RILs identified five loci on chromosomes 1 (QTL1 = 0.0–8.4 Mb), 2 (QTL2 = 8.7–11.1 Mb), 4 (QTL4 = 0.0 – 5.79 Mb), and 5 (QTL5A = 16.3–21.3 Mb; QTL5B = 25.9–26.9 Mb) (Figure 1D; Table S6) (Wang et al. 2012). We screened putative heterogeneous inbred families (HIFs) derived from the RIL population for segregation at QTL4, QTL5A, and QTL5B to fine map the loci controlling HopAM1-induced cell death in the Bur-0 × Col-0 background. We progeny tested several RILs with a heterozygous haplotype for the QTL region of interest to identify segregating lines and then chose a few lines for subsequent fine mapping of each QTL (see Figure S4). QTL4 was delimited within the centromeric region of chromosome 4 (in Col-0 containing many genes of unknown function, several cysteine-rich receptor-like kinases, and one TIR-NLR, At4g04110). However, due to low recombination near the centromere, we could not reduce the size of the associated interval further. Thus, we focused our fine-mapping efforts on QTL5A and QTL5B. QTL5A was delimited within 296 kb (18.682–18.978 Mb) [containing genes of various function, including many TIR-NLR genes and the plasma-membrane localized receptor kinase protein FLS2 (At5g46330), which recognizes bacterial flagellin peptides that stimulate MTI responses (Zipfel et al. 2004)]. QTL5B was delimited to within 110 kb (26.639–26.749 Mb) (containing 30 genes, including MAPKK kinases and CC-NLR genes). The Col-0 gene annotations for the regions in QTL4, 5A and 5B, are listed in Table S7. We found only additive effects between the QTL using an ANOVA for each QTL pair from the initial CIM mapping to identify pairwise epistasis contributing to the HopAM1-induced cell death response (see Figure S5; Table S8) (Brady et al. 2015). However, our population sizes may be too small to effectively evaluate higher order epistasis (Joseph et al. 2013; Taylor and Ehrenreich 2014).
HopAM1-induced cell death is affected by EDS1, HSP90.2, and SGT1B
To assess if HopAM1-induced cell death is affected by genes known to be required for disease resistance, we made several crosses between the Bur-0 accession and Col-0 loss-of-function mutants in defense-related genes: sid2-1 (Wildermuth et al. 2001), sgt1b (Austin et al. 2002), rar1-21 (Tornero et al. 2002), ndr1-1 (Century et al. 1995), eds1-2 (Bartsch et al. 2006), hsp90.2-3 (Hubert et al. 2003), pad4-1 (Glazebrook et al. 1996), and adr1-1 (Grant et al. 2003). A total of 96 F2 progeny from each cross were genotyped and plants homozygous at markers flanking each Col-0-derived mutation in the respective crosses were selected for cell death phenotyping. As a comparative control, 96 plants from a Bur-0 × Col-0 F2 progeny were screened for cell death and subsequently genotyped with the same markers used for the specific mutation in each cross. In Figure 1E, the phenotypes of the F2 plants homozygous for a Col-0-derived mutant allele from the Col-0 mutant × Bur-0 cross are presented next to the phenotypes of plants from the subset of control Bur-0 × Col-0 F2s which were homozygous for the wild-type Col-0 allele for the relevant gene. Our results indicated that eds1-2, hsp90.2-3, and sgt1b affect HopAM1-induced cell death symptoms from Bur-0 (Figure 1E). Homozygosity for either the hsp90.2 (hsp90.2Col-0) or sgt1b mutant allele (sgt1bCol-0) in Bur-0 × Col-0 background attenuated HopAM1-induced cell death symptoms compared to their respective Bur-0 × Col-0 control populations. Because HSP90.2 and SGT1b are part of a steady-state NLR chaperone complex required for NLR ETI receptor activation (Hubert et al. 2003; Holt et al. 2005; Shirasu 2009), this is consistent with the hypothesis that at least one of the cell death QTL loci (Figure 1D) contains a functionally relevant NLR gene. The eds1-2 mutation in the Bur-0 × Col-0 background enhanced the onset of HopAM1-induced cell death symptoms (Figure 1E), which was unexpected because EDS1 is required for the function of TIR-NLR proteins (Feys et al. 2001). The other defense mutations we tested had no effect.
GWA reveals additional loci associated with HopAM1 cell death
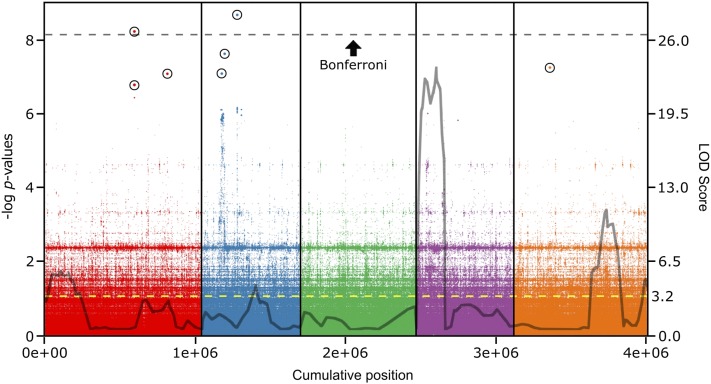
A disadvantage of QTL experiments is that the identified loci may represent rare, deleterious alleles compared to the entire population surveyed. GWA screens can complement QTL studies by identification of loci that do not have segregating alleles in the two RIL founder populations (Gibson 2012; Weigel and Nordborg 2015). We used 64 of the 98 accessions we had screened previously for a GWA study (see Table S3) because whole-genome resequencing data were available for them (http://1001genomes.org; 1001 Genomes Consortium 2016). The most significantly associated SNPs did not overlap with the intervals defined by the Bur-0 × Col-0 QTL analysis (Figure 2) but we found enrichment in the QTL regions when we considered the 50 (or more) most significantly associated SNPs in the same chromosome as the QTL (Table 1). The narrow sense heritability calculated for this study was h2 = 34.5% (likelihood-ratio test, P = 0.430) and the allele frequencies of the SNPs with the strongest allele effects were generally shifted toward low MAF (see Figure S6A), consistent with a polygenic architecture for the trait according to an infinitesimal model (Gibson 2012). The SNPs most significantly associated with variation in HopAM1-induced local cell death were two peaks with multiple significantly associated SNPs on chromosome 2, and single SNPs on chromosomes 1 and 2 (Figure 2). A total of 5 out of the 10 most significant SNPs fell within a 154-bp noncoding region between At1g50420, encoding the scarecrow-like 3 (SCL3) transcription factor, a positive regulator of gibberellic acid signaling (Zhang et al. 2011) and At1g50430 (STEROL Δ-7 REDUCTASE), which encodes a sterol reductase involved in brassinosteroid synthesis (see Table S9) (Lecain et al. 1996). The most significantly associated SNP was on chromosome 2 in the fourth exon of CYCLOPHILIN40 (At2g15790), which encodes a protein isomerase that functions in maturation of the AGO1-containing RNA-induced silencing complex important to epigenetic gene regulation (Romano et al. 2004; Smith et al. 2009; Iki et al. 2012). Aligning the haplotypes available from the 1001 Genomes Project that were used in our GWA over a 30-kb region spanning the CYCLOPHILIN40 gene showed a correlation between haplotype and the HopAM1-dependent cell death phenotype (see Figure S7 and File S1).
Figure 2.
The genetic architecture of HopAM1 cell death response defined by GWA depends on several loci with strong to moderate effects. HopAM1-induced cell death response was scored in a collection of 64 Arabidopsis accessions. EMMAX was used for GWA mapping based on whole-genome sequence data of >4 million SNPs. The Manhattan plot shows the association of each SNP and its P-value across the five Arabidopsis chromosomes (1 through 5 from left to right, separated by vertical lines). The dotted line designates the significance threshold for Bonferroni correction (P ≤ 1.2 × 10−8). All SNPs over the FDR threshold are highlighted with a circle. Gray outline reprises the QTL regions identified in Figure 1D in the Bur-0 × Col-0 background (cM converted to Mb only for each genetic marker’s coordinates on the genome and shows LOD score at that specific location; not identical, but very similar to peak LOD scores shown in previous QTL analysis in Figure 1D). Right y-axis, LOD score; yellow dotted line, significance threshold for CIM.
Table 1. P-values of enrichment tests of increasing numbers of most significant SNPs for each HopAM1-induced cell death QTL.
| Number of top SNPS | 10 | 50 | 100 | 500 | 1000 | 10000 |
|---|---|---|---|---|---|---|
| QTL1 | 0.489 | 0.995 | 0.579 | 0.643 | 0.909 | 1 |
| QTL2 | 0.818 | 1 | 1 | 1 | 1 | 1 |
| QTL4 | 0.0906 | 0.0278 | 3.10 × 10−13 | 2.88 × 10−46 | 2.14 × 10−66 | 1.52 × 10−08 |
| QTL5A | 0.537 | 0.0434 | 1.55 × 10−06 | 0.00942 | 0.0088 | 3.73 × 10−07 |
| QTL5B | 1 | 1 | 1 | 7.04 × 10−06 | 0.00021 | 0.00021 |
Calculated using a Fisher’s exact test with a two-by-two test table design.
Other biparental populations have HopAM1-induced cell death QTL that overlap with Bur-0 × Col-0 QTL5A
To explore the differential genetic architecture driving HopAM1-induced cell death among the accessions, we screened three different biparental mapping populations to investigate whether HopAM1-induced cell death is also a quantitative trait in other accessions. We scored plants from three other populations with parents divergent in their cell death phenotypes, but not as divergent as Bur-0 × Col-0: a Ler-0 × Col-4 RIL population (CS1899), a Sha × Col-0 F2 population, and a Ws-2 × Col-0 F2 population for HopAM1-induced cell death (see Table S10). Ler-0 and Ws-2 had cell death scores of 2 and the Sha score was 4 (see Table S3). All three populations exhibited a continuous phenotypic response similar to the Bur-0 × Col-0 background, but with weaker phenotypes. We again observed no transgressive segregation. We performed a CIM analysis to identify the loci controlling the trait in all three backgrounds and to search for overlap with the loci identified in the Bur-0 × Col-0 background (see Figure S8). In the Ler-0 × Col-4 background, two QTL were identified on chromosome 1 (16.1–22.2 Mb) and chromosome 5 (15.4–20.9 Mb), and in the Ws-2 × Col-0 and Sha × Col-0 backgrounds, one QTL was identified that was solely responsible for the HopAM1-induced cell death (chromosome 5; 14.7–23.8 Mb for Ws-2 × Col-0, and 8.4–20.1 Mb for Sha × Col-0). The QTL identified on chromosome 5 for all three mapping populations (Ler-0 × Col-4, Ws-2 × Col-0, and Sha × Col-0) overlapped with QTL5A from the Bur-0 × Col-0 RIL population (16.3–21.3 Mb), possibly suggesting that the common parent, Col-0/Col-4, carries an unusual allele at this locus.
HopAM1-induced cell death and chlorosis are not associated with bacterial growth restriction in both Col-0 and Bur-0
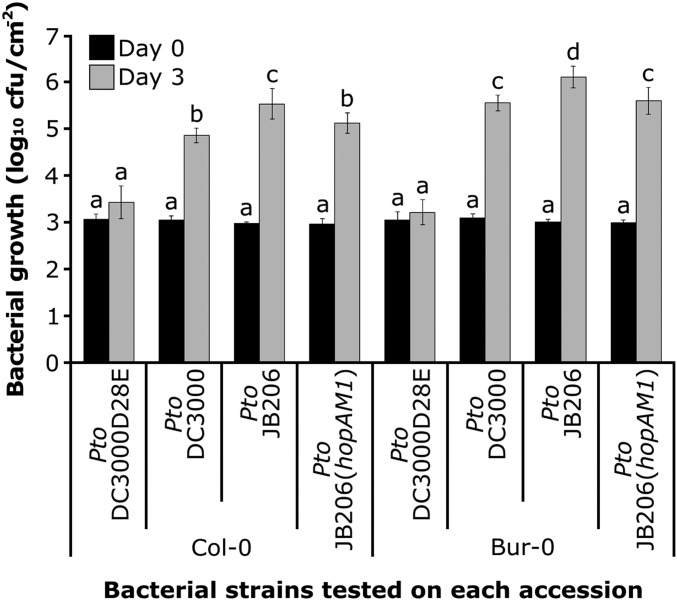
HopAM1 induces obvious phenotypes, but it is unclear how these might be related to the restriction of pathogen growth, the effect most relevant to the host. HopAM1 enhances the growth of a weakly virulent P. syringae strain Pma (M6CdE) in the Ws-2 accession, but not in several other accessions tested (Goel et al. 2008). Thus, HopAM1 is a weak virulence factor, at least on Ws-2. To determine if the different cell death responses of Bur-0 and Col-0 to HopAM1 were correlated with differences in the ability to restrict pathogen growth via recognition of HopAM1, we infected Bur-0 and Col-0 with Pto DC3000. This strain grew to levels associated with disease in both accessions (Figure 3), with slightly higher titers in Bur-0. To determine if the endogenous hopAM1 genes affect the growth of Pto DC3000 in these accessions, we infected Col-0 and Bur-0 with a Pto DC3000 derivative, Pto JB206, which lacks both the chromosomal (PSPTO_1022) (Boch et al. 2002) and plasmid-borne (PSPTO_A0005) (Landgraf et al. 2006) copies of hopAM1. We found a small, statistically significant, growth advantage of Pto JB206 over Pto DC3000 in both Bur-0 and Col-0 (Figure 3). This growth advantage disappeared from both accessions when Pto JB206 was complemented with the plasmid carrying hopAM1 (Figure 3). Thus, HopAM1 is recognized by both accessions, at least to an extent sufficient to trigger growth restriction and there is no genetic difference between Bur-0 and Col-0 for this pathogen growth restriction. We infer that both accessions recognize and respond to HopAM1 at levels sufficient to trigger weak ETI; however only in Bur-0 does the signal delivered by HopAM1 result in ETI-associated cell death.
Figure 3.
HopAM1 causes reduced growth of P. syringae in Col-0 and Bur-0 accessions, independent of cell death symptoms. Bacterial growth in Col-0 and Bur-0 accessions. The experiment is representative of five independent replicates; two times the SE between the means is noted by error bars. A weighted ANOVA test was applied to the difference in growth of the strains carrying hopAM1 (Pto DC3000 and Pto JB206+AM1) compared with the growth of the hopAM1 deleted strain (Pto JB206). Statistical significance is indicated by letters based on the weighted ANOVA test (P < 0.1).
Overlapping and distinct QTL for HopAM1-induced meristem chlorosis and cell death
HopAM1 induces a chlorotic phenotype in newly developed leaves when delivered by a bacterial strain (Pma M6CdE) via hand infiltration or following transgenic expression of hopAM1 in Arabidopsis (Goel et al. 2008). An improved vacuum infiltration assay was devised using Pto DC3000D28E(hopAM1) (see Materials and Methods). All 64 accessions used in our GWA for HopAM1-induced cell death response (see Table S3) were rescreened for HopAM1-induced meristem chlorosis. While most of the accessions exhibited meristem chlorosis at 7 dpi, 12 remained nonchlorotic (see Table S11). In fact, Col-0 exhibited strong meristem chlorosis and Bur-0 was nonresponsive, highlighting the contrasting underlying genetics of these two accessions in response to HopAM1 (Figure 4A). Of the three accessions that exhibited the highest cell death scores (Bur-0, Hov-4, and Wil-2; see Table S11), only Bur-0 did not exhibit HopAM1-induced meristem chlorosis. Hence, HopAM1 cell death and meristem chlorosis-inducing activities are not mutually exclusive. Confocal laser microscopy of Col-0 and Bur-0 newly emerged chlorotic leaves at 14 days after infection revealed that the chloroplasts in Col-0 were smaller, deformed, and exhibited a much weaker fluorescence signal; while the chloroplasts in the corresponding leaves in Bur-0 appeared normal (Figure 4B). Furthermore, Col-0 chlorotic leaf tissue had smaller, round-shaped mesophyll cells, while Bur-0 had wild-type cell morphology (Figure 4B). We used the Bur-0 × Col-0 RIL population (Simon et al. 2008) again to identify loci controlling the HopAM1-induced meristem chlorosis. To assess the heritability of the meristem chlorosis trait in the RILs, we performed a second screen of a subset of the RIL population (162 lines with the highest number of recombination events were chosen). We tested the influence of environmental effects on our meristem chlorosis phenotype scores using a general linear model as before (see Table S12) (Chan et al. 2011). The broad sense heritability of the HopAM1-induced meristem chlorosis trait was H2 = 60.7% (P < 2 × 10−16). Using the reannotated Bur-0 × Col-0 RILs, we performed a CIM that revealed three loci on chromosomes 4 (QTL4c1 = 0.9–5.2 Mb, QTL4c2 = 6.9–15.7 Mb) and 5 (QTL5c = 18.3–24.1 Mb) (Figure 4C; Table S13). The genomic regions of QTL4c1 and QTL5c overlap with the HopAM1-induced cell death QTL4 (0.0–5.79 Mb) and QTL5A (16.3–21.3 Mb), but the most significant chlorosis locus, QTL4c2, was distinct from all QTL identified for HopAM1-induced cell death.
Figure 4.
HopAM1 induces rosette chlorosis affecting chloroplast development and cell morphology controlled by three QTL in Col-0 × Bur-0. (A) Meristem chlorosis response following bacterial vacuum infiltration with Pto DC3000D28E(hopAM1). Col-0 exhibits strong chlorosis symptoms, while Bur-0 does not. F1 progeny exhibit mild symptoms. Chlorosis appears in newly emerging leaf and meristem in Col-0 at 10 dpi following vacuum infiltration. Chlorosis can be observed up to 21 dpi. (B) Confocal laser scanning microscopy of Arabidopsis accessions Col-0 and Bur-0 leaf tissue, harvested 14 dpi following vacuum infiltration with Pto DC3000D28E(hopAM1). Chlorophyll autofluorescence (enlarged and brightened insets with white dotted outline shown on the top left corner of respective panel) (red channel), plant cell shape (black dotted outline of cells) (green channel), and merged channels are shown for each accession. Upper and lower panels taken at equal light intensity. Bar, 10 μm. (C) CIM on 342 reannotated RILs from a Col-0 × Bur-0 collection. The x-axis displays chromosomes 1 through 5 with map distances in cM. Peak LOD scores are shown by ▾ along with their values. Effects on phenotype variation are shown as percentages for all three QTL. The global permutation level of significance was set on LOD 3.2. Bar, 50 cM.
HopAM1-induced cell death and meristem chlorosis traits are independent among wild accessions
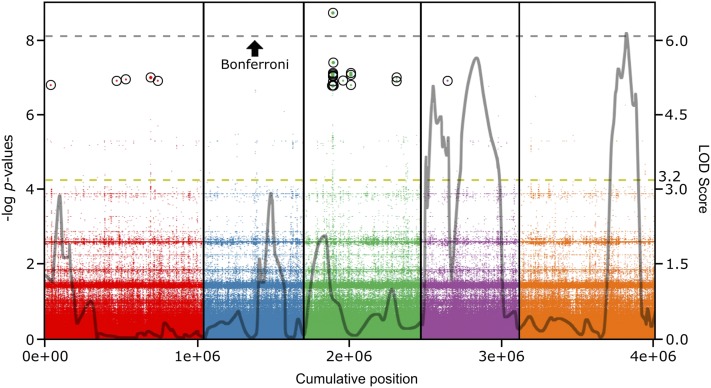
We also performed GWA mapping for HopAM1-induced meristem chlorosis among the 64 accessions that we had screened for HopAM1-induced cell death (see Table S11). The narrow sense heritability for the GWA study was h2 = 20.9% (likelihood-ratio test, P = 0.625) and the most significant SNPs tended to have low minor allele frequencies (see Figure S6B). There was an enrichment of the most significantly associated SNPs with QTL4C2 (Table 2). However, the only peak above the stringent Bonferroni significance level was outside of the QTL intervals (Figure 5). This SNP and 3 more of the 10 most significant meristem chlorosis SNPs (FDR ≤ 0.05; including the top hit) fell within a 235-bp region between the third and fourth exon of TOC64-III (At3g17970), which encodes a protein of the chloroplast outer envelope membrane transport complex (see Table S14) (Sommer et al. 2013). In total, 27 out of the 50 top hits fell within a 2-kb region of TOC64-III (third to ninth exon). Alignment of all the 1001 Genomes haplotypes that were used in the GWA study across a 30-kb region spanning the top SNP indicated a correlation between TOC64-III haplotype and HopAM1-induced meristem chlorosis response (see Figure S9 and File S2). Single toc64-III mutants, and a toc64 triple mutant (see Materials and Methods) were tested in the background of the meristem chlorosis-responsive accession Col-0. All toc64 mutants, including the triple mutant, were responsive for HopAM1-induced meristem chlorosis, indicating that lack of TOC64 is not sufficient to prevent the effect.
Table 2. P-values of enrichment tests of increasing numbers of most significant SNPs for each HopAM1-induced chlorosis QTL.
| Number of top SNPS | 10 | 50 | 100 | 500 | 1000 | 10000 |
|---|---|---|---|---|---|---|
| QTL4C1 | 1 | 0.982 | 0.959 | 0.93 | 0.502 | 0.993 |
| QTL4C2 | 0.114 | 0.0253 | 0.0145 | 0.0006 | 0.0002 | 0.0002 |
| QTL5C | 0.898 | 0.999 | 1 | 07.93 × 10−01 | 1 | 1 |
Calculated as in Table 1.
Figure 5.
The genetic architecture of the HopAM1 chlorosis response defined by GWA is associated with a single locus on chromosome 3. HopAM1-induced chlorosis was scored in a collection of 64 Arabidopsis accessions as for Figure 2. Nearly all top 10 hits were inside genes. Gray outline reprises the QTL regions identified in Figure 4C in the Bur-0 × Col-0 background (cM converted to Mb only for each genetic marker’s coordinates on the genome and shows LOD score at that specific location; not identical, but very similar to peak LOD scores shown in previous QTL analysis in Figure 4C). Right y-axis, LOD score; yellow-dotted line, significance threshold for CIM.
We also screened additional Arabidopsis mutants for lack of HopAM1-induced meristem chlorosis (see Table S15). The double mutant rbohd rbohf (CS68522) (Torres et al. 2002) was not responsive, suggesting that superoxide derived from this NADPH oxidase complex is required for the phenotype. Both eds1-2 and an ein2-1 pad4-1 sid2-2 (CS66006) triple mutant enhanced the meristem chlorosis by extending the duration of time in which plants produced chlorotic leaves compared to controls (see Figure S10). Neither the ein2-1 sid2-2 double mutant nor pad4-1 exhibited this enhanced chlorosis. EDS1 and PAD4 proteins interact and potentiate accumulation of salicylic acid (SA) in pathogen-infected tissues, potentially through regulation of TIR-NLR action (Feys et al. 2001; Rietz et al. 2011; Wagner et al. 2013). SID2 encodes isochorismate synthase 1 (ICS1) required for production of SA (Wildermuth et al. 2001). The extended chlorosis response of these two mutant lines suggests HopAM1 chlorosis is limited in timing through EDS1 function. All other Col-0 background mutant lines were similar to the parental Col-0 accession.
Host transcriptional signatures indicate that HopAM1 suppresses MTI but also induces ETI in Bur-0
To further investigate how HopAM1 interferes with host cellular functions, we performed genome-wide transcriptome analysis of infected plants. Col-0 and Bur-0 plants that were 5 weeks old were hand inoculated with Pto DC3000D28E(EV) or Pto DC3000D28E(hopAM1). RNA-seq libraries were prepared from plants harvested immediately before infiltration and at six time points postinfiltration spanning a 12 hr period (see Materials and Methods). The number of differentially expressed genes between plants inoculated with Pto DC3000D28E(EV) and Pto DC3000D28E(hopAM1) was relatively low at 2 hpi, a time point just preceding delivery of type III effectors (de Torres Zabala et al. 2009), for both accessions (306 in Col-0 and only 4 in Bur-0; Figure 6A). The number of differentially expressed genes greatly increased over the remainder of the infection time course, ranging from 1432 to 2981 in Col-0 and from 522 to 5762 in Bur-0 (Figure 6A). The complete edgeR results are presented in Table S16. HopAM1-dependent differential expression was minimal for the 10 most highly associated loci identified in each GWA study. The HopAM1 chlorosis QTL regions still include hundreds of genes and many of them showed HopAM1-dependent transcriptional responses at some time points. The more narrowly defined cell death related QTL, QTL4, QTL5A, and QTL5B, also included multiple genes with differential expression in response to HopAM1 (see Table S7).
Figure 6.
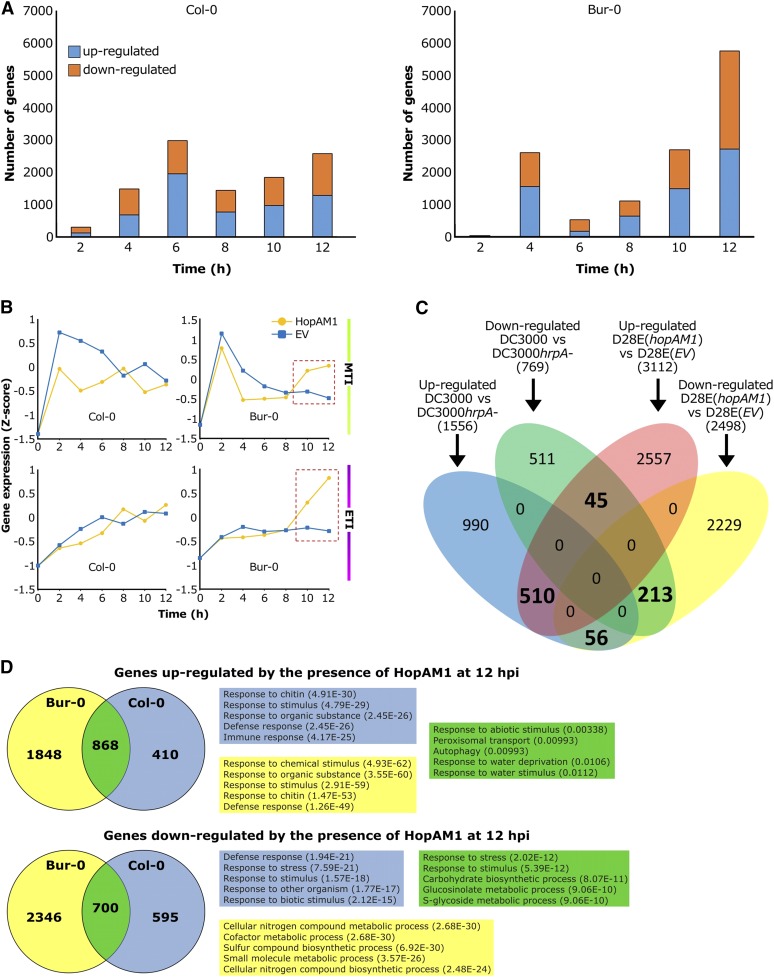
HopAM1 induces intense transcriptional reprogramming for thousands of genes in Col-0 and Bur-0, results in initial suppression of MTI in both ecotypes and later induction of ETI in Bur-0 only. (A) Number of genes (y-axis) differentially expressed as a result of HopAM1 delivery in Col-0 and Bur-0 every 2 hr (x-axis) in the first 12 hr postinfiltration. Orange bar, downregulated; blue bar, upregulated. (B) Upper panel: average expression profile of flg22-induced genes within the same time period in Col-0 and Bur-0. Lower panel: average expression profile of genes upregulated by ETI induced upon recognition of the avrRps4 avirulence factor by the NLR protein Rps4 in Col-0. Gene expression is shown as the median z-scored transformed RPKM values of the 1418 and 718 marker genes for MTI and ETI, respectively. (C) Overlap between genes differentially expressed by Pto DC3000D28E(hopAM1) infection and those specifically misregulated by Pto DC3000 effectors. A total of 5610 genes that were differentially expressed in Col-0 in at least one time point of our experiment were included in the analysis. A set of 517 genes showing ambiguous regulation (i.e., both up- and downregulation at different time points) was discarded. (D) Venn diagrams showing number of genes upregulated and downregulated at 12 hpi for both Bur-0 and Col-0. GOs for the top five sets are shown for each group.
GO-enrichment analyses showed that HopAM1 downregulated defense-related genes at most time points in Col-0, and at the early time points in Bur-0 (see Figure S11; Table S17, Table S18, Table S19, Table S20, and Table S21). We determined if the changes in expression of defense-responsive genes were related to MTI or ETI by focusing on the expression of 1418 genes induced by the well-characterized MAMP flg22 (Rallapalli et al. 2014) for MTI and for ETI, on expression profiles of a set of 747 markers corresponding to genes upregulated in a comparison of Col-0 plants infected with ETI-inducing Pto DC3000(avrRps4) (triggering the RPS4 TIR-NLR) vs. plants infected with virulent Pto DC3000 (Howard et al. 2013). The average expression profile of the flg22-induced genes after infection with Pto DC3000(hopAM1) compared to Pto DC3000(EV) is consistent with HopAM1 suppressing MTI in both accessions at early time points. At 10–12 hpi, ETI-induced genes were upregulated by HopAM1 in Bur-0 but not in Col-0 (Figure 6B). Specific genes associated with typical immune responses such as GSL5 (At4g03550), RBOHD (At5g47910), SAG101 (At5g14930), NDR1 (At3g20600), and SID2 (At1g74710) were upregulated in Bur-0 from 10 hpi onwards, but not in Col-0 (see Figure S12). Our gene expression profiling indicates that HopAM1 induces an ETI response in the Bur-0 accession, which supports rapid HopAM1-induced cell death; but not in Col-0, which does not. Notably, this difference in ETI response is not correlated with restriction of bacterial growth in Bur-0 compared to Col-0 (Figure 3).
P. syringae type III effectors suppress MTI transcriptional responses in the host plant (Truman et al. 2007; Lewis et al. 2015). Since HopAM1 is delivered to host plants by Pto DC3000 along with the rest of its suite of type III effectors, we expected that the transcriptional response to infection with Pto DC3000 should overlap with the response to Pto DC3000D28E(hopAM1). Lewis et al. (2015) identified a set of 2325 Arabidopsis genes that were differentially expressed (769 downregulated, 1556 upregulated) during the infection with Pto DC3000, but not with the mutant Pto DC3000hrpA. Since the hrpA mutant does not deliver effectors into the host cells, the differential gene expression was dependent on the bacterial effectors of Pto DC3000. A total of 213 of the 769 Pto DC3000 effector downregulated genes (28%) were also downregulated by HopAM1. These genes were enriched in biological processes related to nucleosome organization and chromatin assembly (FDR < 0.001; see Table S22). Furthermore, 510 of the 1556 genes (33%) activated by Pto DC3000 effectors were also upregulated by HopAM1 in our experiment. These genes were enriched in biological processes related to autophagy and photoperiodism (FDR < 0.001; see Table S22). MAMP perception has been shown to trigger large-scale suppression of genes for chloroplast-localized proteins in Arabidopsis, but that suppression was attenuated in plants infected with Pto DC3000, due to the combined functions of its collection of type III effectors (de Torres Zabala et al. 2015). When compared to Pto DC3000D28E(EV), infection with Pto DC3000D28E(hopAM1) resulted in higher expression of genes encoding chloroplast-localized proteins in both Col-0 and Bur-0 at 4 and 6 hpi, followed by reduced expression in both accessions by 12 hpi (see Figure S13); indicating that HopAM1 may affect chloroplast responses to MTI (Figure 4B) in both accessions even though Bur-0 does not suffer from meristem chlorosis.
Discussion
HopAM1-induced local cell death is associated with HopAM1-induction of ETI response
Using RNA-seq, we compared gene expression changes induced by Pto DC3000D28E(hopAM1) to changes induced by Pto DC3000D28E(EV). We found that HopAM1 repressed gene sets related to MTI immune responses in both Col-0 and Bur-0 at early time points (Figure 6B). Our results are consistent with our previous report that HopAM1 suppresses basal defense responses (Goel et al. 2008). Genes suppressed by HopAM1 overlapped with genes suppressed by Pto DC3000 delivering its full suite of type III effectors that includes HopAM1 (Truman et al. 2007; de Torres Zabala et al. 2015; Lewis et al. 2015). However, at 10–12 hpi, we defined a Bur-0-specific induction of genes previously associated with RPS4-dependent ETI (Howard et al. 2013) (Figure 6C; see Figure S11). As expected, if the early onset cell death seen in Bur-0 is the result of induced ETI, it is attenuated in F2 plants from crosses of Bur-0 with Col-0-derived mutants that were homozygous for loss-of-function mutations in HSP90.2 and SGT1b (Figure 1E) required for NLR receptor activation (Hubert et al. 2003; Holt et al. 2005; Shirasu 2009).
Our QTL analysis for HopAM1-induced cell death on the Bur-0 × Col-0 RIL population revealed multiple loci contributing additively to the timing of onset of cell death (Figure 1D). The three loci that account for most of the variation (QTL4, QTL5A, and QTL5B) each contain genes encoding NLR proteins, ETI receptors that are often highly variable in Arabidopsis (Shen et al. 2006; Cui et al. 2015) (see Table S3 and Table S7). Additional fine mapping and mutation analysis of individual candidates will be necessary to confirm candidate genes as responsible for the QTL traits. Specific alleles at each of the cell death QTL loci contribute additively to induce cell death earlier in Bur-0 than in other accessions as if each locus has an independent ETI function and the timing of cell death depends on the accumulation of effects from each locus. This is unlike the well-characterized ETI responses to pathogen effector proteins that present as gene-for-gene resistance scenarios, where one NLR gene is sufficient to mount an effective ETI response when its product recognizes the action of one type III effector (Dangl and Jones 2001).
GWA studies complement QTL mapping in biparental populations
Since they work at the population level, GWA studies have maximal power to detect associations with common allele variants and might give additional clues to understand species-wide adaptation patterns. GWA studies can identify loci even when they have relatively low power (Atwell et al. 2010) or when heritability is apparently low (Fournier-Level et al. 2011), and provide much higher mapping resolution with fewer individuals than QTL analysis (Balasubramanian et al. 2009). Successful examples are GWA studies of plant fitness in field experiments in which a myriad of causal loci with environmental-dependent effects lowered heritability estimates below 50%, usually closer to zero, but loci underlying relevant ecological traits were still found (Fournier-Level et al. 2011; Fournier-Level et al. 2013). In humans, GWA with polygenic and complex disease traits such as human height or schizophrenia keep providing new gene candidates and promote advances in quantitative genetic theory (Eichler et al. 2010; Visscher et al. 2010; Yang et al. 2010; Lee et al. 2014). The most significant candidate genes from our HopAM1-induced cell death GWA have functions consistent with affecting an ETI response. The most significantly associated SNPs were in the gene for cyclophilin 40, involved in epigenetic gene regulation, which is important to most cellular functions and certainly relevant to mounting effective ETI responses (Navarro et al. 2006; Zhai et al. 2011; Shivaprasad et al. 2012). Other significant cell death SNPs were located between a gene for brassinosteroid synthesis (STEROL Δ-7 REDUCTASE) and a gene for regulation of gibberellic acid signaling (SCL3). Both hormones contribute to the interplay of plant hormones that balances plant growth and defense (Bari and Jones 2009; Lozano-Duran and Zipfel 2015).
Local cell death is associated with activation of ETI responses even though it is not linked to changes in pathogen growth
The HopAM1-induced ETI response does not result in less growth of Pto DC3000 in Bur-0 than in Col-0 (Figure 3). Thus, while there is a clear ETI transcriptional signature from HopAM1 in Bur-0 that is cell death related, this ETI signature is decoupled from growth restriction itself. There are several examples of NLR-mediated ETI in which cell death and pathogen growth are uncoupled (reviewed in Coll et al. 2011), such as the extreme resistance to potato virus X by the Rx NLR protein (Bendahmane et al. 1999). Some ecotypes of Arabidopsis are resistant to P. syringae encoding the type III effectors AvrRps4 or HopA1 without developing obvious local lesions (Gassmann 2005; Kim et al. 2009). Uncoupling of effector-triggered cell death and pathogen growth restriction has also been observed for a number of Arabidopsis mutants such as cyclic nucleotide gated ion channel mutants dnd1 (Clough et al. 2000), hlm1 (Balague et al. 2003), dnd2 (Jurkowski et al. 2004), and metacaspase 1 (Coll et al. 2010). The extent of cell death can also vary depending on physiological conditions, such as light (Zeier et al. 2004; Bruggeman et al. 2015) and temperature (Menna et al. 2015) without loss of bacterial growth restriction.
A further hint that HopAM1 targets NLR-dependent downstream pathways comes from results showing that EDS1 plays a role both in HopAM1-induced cell death (Figure 1E) and meristem chlorosis (see Figure S10). In most examples, eds1 loss-of-function mutants cause reduction in pathogen- or stress-induced TIR-NLR-mediated ETI and cell death responses (Bartsch et al. 2006; Ochsenbein et al. 2006; Bhattacharjee et al. 2011). Interestingly, HopAM1 induced a constant transcriptional upregulation in Col-0 and Bur-0 from 6 hpi of both EDS1 (At3g48090) and PAD4 (At3g52430) (see Table S16). However, if HopAM1-mediated cell death is the consequence of ETI activation of a TIR-NLR, a loss-of-function mutant like eds1-2 is expected to be unable to mount the ETI response. Instead, homozygosity for the eds1-2 allele enhanced cell death in our F2 plants (Figure 1E). Such a counterintuitive observation of a loss-of-function eds1 mutation resulting in stronger NLR-mediated cell death has been observed before: the eds1-2 mutation has been shown to enhance the immune response of an autoactive mutant derived from the CC-NLR gene ADR1 (Roberts et al. 2013). In this regard, our finding of a cluster of CC-NLR genes in QTL5B, which we reduced to an interval of 100 kb containing only 30 genes, may suggest that one or more of these genes is causally related to HopAM1-induced cell death in Bur-0. Further fine mapping will address this possibility.
HopAM1 may reduce MTI responses through changes in chloroplast metabolism
Reactive oxygen, specifically superoxide, is required for HopAM1-induced meristem chlorosis since the double mutant of RBOHD and RBOHF did not support it (see Figure S10). Reactive oxygen species (ROS) accumulation is an integral part of MTI and ETI responses and high levels of ROS damage chloroplasts (Wagner et al. 2004; Kim et al. 2012). Chloroplasts play a key role in defense by producing ROS (Shapiguzov et al. 2012) and balancing hormone levels (Robert-Seilaniantz et al. 2011), thus constituting a target of preference for pathogens. Pto DC3000 effectors can suppress the expression of nuclear-encoded chloroplast genes in Arabidopsis and target the chloroplast (de Torres Zabala et al. 2015). Our expression analyses demonstrate that HopAM1 modifies the transcription of genes for chloroplast-localized proteins (see Figure S13). Our GWA study identified TOC64III, as significantly associated with variation in chlorosis phenotype (Figure 4; Table S14). TOC64III encodes a chloroplast TOC transporter accessory protein (Sohrt and Soll 2000; Aronsson et al. 2007) which modulates the translocation efficiency of proteins into the chloroplast (Sommer et al. 2013). TOC64 protein is not essential for chloroplast function; no detrimental phenotype is observed under standard growth conditions for Arabidopsis insertion mutants defective in TOC64III. (Aronsson et al. 2007). Likewise, TOC64 protein is not necessary for response to HopAM1 although allelic variation is associated with different chlorotic responses to HopAM1. We suggest that allelic variation in TOC64 could render chloroplasts more or less resistant to damage caused by ROS induced during MTI defense responses.
Do meristem chlorosis and HopAM1 local cell death share a common mechanism?
HopAM1-induced local cell death and meristem chlorosis are not mutually exclusive. Both early cell death and chlorosis were observed in the Hov-4 and Wil-2 accessions (see Figure S7 File S1). Nevertheless, two of the three QTL loci related to meristem chlorosis from the Bur-0 × Col-0 RIL population overlap. Until the genes responsible for each phenotype have been identified, it is possible that independent genes and independent processes are responsible for chlorosis or cell death. However, the fact that eds1-2 mutants both enhance cell death and meristem chlorosis suggests that EDS1 is a negative regulator of both HopAM1 responses. EDS1 regulates cell death by balancing phytohormone levels that lead to enhanced ROS production, likely involving modulation of TIR-NLR activation (Wiermer et al. 2005; Ochsenbein et al. 2006; Muhlenbock et al. 2008; Kim et al. 2012). Although our QTL and GWA analysis showed that HopAM1-induced chlorosis and cell death require distinct loci, EDS1 appears to play an integral role in a critical biochemical junction where these two responses converge.
Conclusions
Pathogen effectors interact with and alter components of protein complexes important to plant immunity (Mukhtar et al. 2011; Weßling et al. 2014; Macho and Zipfel 2015). Therefore, individual pathogen effector proteins can serve as probes to identify interacting partners in immune complexes. To our knowledge, this is the first time that responses to a single pathogen effector have generated quantitative trait responses in the host. Previously defined ETI-related gene transcriptional alterations were also not associated with HopAM1-dependent bacterial growth restriction, which challenges the concept of what constitutes an ETI response, and more importantly, what is required to halt pathogen proliferation. Our study demonstrates that dissection of natural variation of host plants for quantitative responses to individual effectors, such as HopAM1, can reveal multiple genes involved in plant immune responses and help to dissect the quantitative features of what is often assumed to be a qualitative response.
Acknowledgments
We thank Marc Nishimura for immeasurable assistance with materials and advice. We also thank Farid El Kasmi, Eui-Hwan Chung, Freddy Monteiro, Scott Yourstone, and Sur Herrera-Paredes for general assistance and advice; and Joseph Kieber, Jason Reed, Li Yang, Christine Camilleri, Salomé Prat, and Úrsula Flores Pérez for providing various mutant lines used in this study. M.I., M.G.C., C.-M.V., T.D., and S.R.G. were supported by National Science Foundation grant IOS-1022286. Work at the Max Planck Institute was supported by Energy Regulatory Commission Advanced Grant IMMUNEMESIS. J.L.D. is an investigator of the Howard Hughes Medical Institute (HHMI), supported by the HHMI and the Gordon and Betty Moore Foundation (GBMF3030) and was also supported by National Science Foundation grant IOS-1257373. P.J.P.L.T. was supported by a fellowship from the Pew Latin American Fellows Program in the Biomedical Sciences.
Footnotes
Communicating editor: T. Juenger
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.190678/-/DC1.
Literature Cited
- 1001 Genomes Project Consortium , 2016. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W., 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold D. L., Jackson R. W., Fillingham A. J., Goss S. C., Taylor J. D., et al. , 2001. Highly conserved sequences flank avirulence genes: isolation of novel avirulence genes from Pseudomonas syringae pv. pisi. Microbiology 147: 1171–1182. [DOI] [PubMed] [Google Scholar]
- Aronsson H., Boij P., Patel R., Wardle A., Töpel M., et al. , 2007. Toc64/OEP64 is not essential for the efficient import of proteins into chloroplasts in Arabidopsis thaliana. Plant J. 52: 53–68. [DOI] [PubMed] [Google Scholar]
- Atwell S., Huang Y. S., Vilhjálmsson B. J., Willems G., Horton M., et al. , 2010. Genome-wide association study of 107 phenotypes in a common set of Arabidopsis thaliana inbred lines. Nature 465: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin M. J., Muskett P., Kahn K., Feys B. J., Jones J. D. G., et al. , 2002. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295: 2077–2080. [DOI] [PubMed] [Google Scholar]
- Balague C., Lin B., Alcon C., Flottes G., Malmstrom S., et al. , 2003. HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15: 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S., Schwartz C., Singh A., Warthmann N., Kim M. C., et al. , 2009. QTL mapping in new Arabidopsis thaliana advanced intercross-recombinant inbred lines. PLoS One 4: e4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus D. A., Nishimura M. T., Romanchuk A., Chang J. H., Mukhtar M. S., et al. , 2011. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 7: e1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R., Jones J. D., 2009. Role of plant hormones in plant defence responses. Plant Mol. Biol. 69: 473–488. [DOI] [PubMed] [Google Scholar]
- Bartsch M., Gobbato E., Bednarek P., Debey S., Schultze J. L., et al. , 2006. Salicylic acid–independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A., Kanyuka K., Baulcombe D. C., 1999. The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11: 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S., Halane M. K., Kim S. H., Gassmann W., 2011. Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334: 1405–1408. [DOI] [PubMed] [Google Scholar]
- Boch J., Joardar V., Gao L., Robertson T. L., Lim M., et al. , 2002. Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol. Microbiol. 44: 73–88. [DOI] [PubMed] [Google Scholar]
- Bonardi V., Dangl J. L., 2012. How complex are intracellular immune receptor signaling complexes? Front. Plant Sci. 3: 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes D. C., Nam J., Dangl J. L., 1998. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl. Acad. Sci. USA 95: 15849–15854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady S. M., Burow M., Busch W., Carlborg Ö., Denby K. J., et al. , 2015. Reassess the t test: interact with all your data via ANOVA. Plant Cell 27: 2088–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman Q., Raynaud C., Benhamed M., Delarue M., 2015. To die or not to die? Lessons from lesion mimic mutants. Front. Plant Sci. 6: 24–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell C. R., Joardar V., Lindeberg M., Selengut J., Paulsen I. T., et al. , 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100: 10181–10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century K. S., Holub E. B., Staskawicz B. J., 1995. NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA 92: 6597–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E. K. F., Rowe H. C., Corwin J. A., Joseph B., Kliebenstein D. J., 2011. Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol. 9: e1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. H., Urbach J. M., Law T. F., Arnold L. W., Hu A., et al. , 2005. A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc. Natl. Acad. Sci. USA 102: 2549–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavigneau H., Goue N., Delaunay S., Courtial A., Jouanin L., et al. , 2012. QTL for floral stem lignin content and degradability in three recombinant inbred line (RIL) progenies of Arabidopsis thaliana and search for candidate genes involved in cell wall biosynthesis and degradability. Open J. Genetics 2: 7–30. [Google Scholar]
- Cheng H., Qin L., Lee S., Fu X., Richards D. E., et al. , 2004. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131: 1055–1064. [DOI] [PubMed] [Google Scholar]
- Cheong M. S., Kirik A., Kim J.-G., Frame K., Kirik V., et al. , 2014. AvrBsT acetylates Arabidopsis ACIP1, a protein that associates with microtubules and is required for immunity. PLoS Pathog. 10: e1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E.-H., da Cunha L., Wu A.-J., Gao Z., Cherkis K., et al. , 2011. Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 9: 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Fengler K. A., Yu I. C., Lippok B., Smith R. K., Jr, et al. , 2000. The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. USA 97: 9323–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll N. S., Vercammen D., Smidler A., Clover C., Van Breusegem F., et al. , 2010. Arabidopsis type I metacaspases control cell death. Science 330: 1393–1397. [DOI] [PubMed] [Google Scholar]
- Coll N. S., Epple P., Dangl J. L., 2011. Programmed cell death in the plant immune system. Cell Death Differ. 18: 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cournoyer B., Sharp J. D., Astuto A., Gibbon M. J., Taylor J. D., et al. , 1995. Molecular characterization of the Pseudomonas syringae pv. pisi plasmid-borne avirulence gene avrPpiB which matches the R3 resistance locus in pea. Mol. Plant Microbe Interact. 8: 700–708. [DOI] [PubMed] [Google Scholar]
- Cui H., Tsuda K., Parker J. E., 2015. Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66: 487–511. [DOI] [PubMed] [Google Scholar]
- Cunnac S., Chakravarthy S., Kvitko B. H., Russell A. B., Martin G. B., et al. , 2011. Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc. Natl. Acad. Sci. USA 108: 2975–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J. L., Jones J. D. G., 2001. Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Deslandes L., Rivas S., 2012. Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci. 17: 644–655. [DOI] [PubMed] [Google Scholar]
- de Torres Zabala M., Bennett M. H., Truman W. H., Grant M. R., 2009. Antagonism between salicylic and abscisic acid reflects early host–pathogen conflict and moulds plant defence responses. Plant J. 59: 375–386. [DOI] [PubMed] [Google Scholar]
- de Torres Zabala M., Littlejohn G., Jayaraman S., Studholme D., Bailey T., et al. , 2015. Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nature Plants 1: 15074. [DOI] [PubMed] [Google Scholar]
- Dodds P. N., Rathjen J. P., 2010. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Eichler E. E., Flint J., Gibson G., Kong A., Leal S. M., et al. , 2010. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 11: 446–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B. J., Moisan L. J., Newman M.-A., Parker J. E., 2001. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20: 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier-Level A., Korte A., Cooper M. D., Nordborg M., Schmitt J., et al. , 2011. A map of local adaptation in Arabidopsis thaliana. Science 334: 86–89. [DOI] [PubMed] [Google Scholar]
- Fournier-Level A., Wilczek A. M., Cooper M. D., Roe J. L., Anderson J., et al. , 2013. Paths to selection on life history loci in different natural environments across the native range of Arabidopsis thaliana. Mol. Ecol. 22: 3552–3566. [DOI] [PubMed] [Google Scholar]
- Fu Z. Q., Guo M., Jeong B.-r., Tian F., Elthon T. E., et al. , 2007. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 447: 284–288. [DOI] [PubMed] [Google Scholar]
- Gassmann W., 2005. Natural variation in the Arabidopsis response to the avirulence gene hopPsyA uncouples the hypersensitive response from disease resistance. Mol. Plant Microbe Interact. 18: 1054–1060. [DOI] [PubMed] [Google Scholar]
- Gery C., Zuther E., Schulz E., Legoupi J., Chauveau A., et al. , 2011. Natural variation in the freezing tolerance of Arabidopsis thaliana: effects of RNAi-induced CBF depletion and QTL localisation vary among accessions. Plant Sci. 180: 12–23. [DOI] [PubMed] [Google Scholar]
- Gibson G., 2012. Rare and common variants: twenty arguments. Nat. Rev. Genet. 13: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Rogers E. E., Ausubel F. M., 1996. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143: 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A. K., Lundberg D., Torres M. A., Matthews R., Akimoto-Tomiyama C., et al. , 2008. The Pseudomonas syringae type III effector HopAM1 enhances virulence on water-stressed plants. Mol. Plant Microbe Interact. 21: 361–370. [DOI] [PubMed] [Google Scholar]
- Grant J. J., Chini A., Basu D., Loake G. J., 2003. Targeted activation tagging of the Arabidopsis NBS-LRR gene, ADR1, conveys resistance to virulent pathogens. Mol. Plant Microbe Interact. 16: 669–680. [DOI] [PubMed] [Google Scholar]
- Holt B. F., 3rd, Boyes D. C., Ellerström M., Siefers N., Wiig A., et al. , 2002. An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Dev. Cell 2: 807–817. [DOI] [PubMed] [Google Scholar]
- Holt B. F., 3rd, Belkhadir Y., Dangl J. L., 2005. Antagonistic control of disease resistance protein stability in the plant immune system. Science 309: 929–932. [DOI] [PubMed] [Google Scholar]
- Howard B. E., Hu Q., Babaoglu A. C., Chandra M., Borghi M., et al. , 2013. High-throughput RNA sequencing of Pseudomonas-infected Arabidopsis reveals hidden transcriptome complexity and novel splice variants. PLoS One 8: e74183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert D. A., Tornero P., Belkhadir Y., Krishna P., Takahashi A., et al. , 2003. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 22: 5679–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iki T., Yoshikawa M., Meshi T., Ishikawa M., 2012. Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J. 31: 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D. G., Dangl J. L., 2006. The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Joseph B., Corwin J. A., Li B., Atwell S., Kliebenstein D. J., 2013. Cytoplasmic genetic variation and extensive cytonuclear interactions influence natural variation in the metabolome. eLife 2: e00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkowski G. I., Smith R. K., Jr, Yu I. C., Ham J. H., Sharma S. B., et al. , 2004. Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. Mol. Plant Microbe Interact. 17: 511–520. [DOI] [PubMed] [Google Scholar]
- Kang H. M., Sul J. H., Service S. K., Zaitlen N. A., Kong S.-y., et al. , 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Meskauskiene R., Zhang S., Lee K. P., Lakshmanan Ashok M., et al. , 2012. Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 24: 3026–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Kwon S. I., Saha D., Anyanwu N. C., Gassmann W., 2009. Resistance to the Pseudomonas syringae effector HopA1 is governed by the TIR-NBS-LRR protein RPS6 and is enhanced by mutations in SRFR1. Plant Physiol. 150: 1723–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimyuk V. I., Carroll B. J., Thomas C. M., Jones J. D. G., 1993. Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J. 3: 493–494. [DOI] [PubMed] [Google Scholar]
- Kroj T., Chanclud E., Michel-Romiti C., Grand X., Morel J. B., 2016. Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 210: 618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf A., Weingart H., Tsiamis G., Boch J., 2006. Different versions of Pseudomonas syringae pv. tomato DC3000 exist due to the activity of an effector transposon. Mol. Plant Pathol. 7: 355–364. [DOI] [PubMed] [Google Scholar]
- Lecain E., Chenivesse X., Spagnoli R., Pompon D., 1996. Cloning by metabolic interference in yeast and enzymatic characterization of Arabidopsis thaliana sterol Δ7-reductase. J. Biol. Chem. 271: 10866–10873. [DOI] [PubMed] [Google Scholar]
- Lee S., Abecasis G. R., Boehnke M., Lin X., 2014. Rare-variant association analysis: study designs and statistical tests. Am. J. Hum. Genet. 95: 5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L. A., Polanski K., de Torres-Zabala M., Jayaraman S., Bowden L., et al. , 2015. Transcriptional dynamics driving MAMP-triggered immunity and pathogen effector-mediated immunosuppression in Arabidopsis leaves following infection with Pseudomonas syringae pv tomato DC3000. Plant Cell 27: 3038–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, S., M. Daly, and E. Lander, 1992 Constructing genetic maps with MAPMAKER/EXP 3.0. Whitehead Institute Technical Report. 3rd Edition. Whitehead Institute, Cambridge, MA.
- Lozano-Duran R., Zipfel C., 2015. Trade-off between growth and immunity: role of brassinosteroids. Trends Plant Sci. 20: 12–19. [DOI] [PubMed] [Google Scholar]
- Macho A. P., Zipfel C., 2015. Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr. Opin. Microbiol. 23: 14–22. [DOI] [PubMed] [Google Scholar]
- Menna A., Nguyen D., Guttman D. S., Desveaux D., 2015. Elevated temperature differentially influences effector-triggered immunity outputs in Arabidopsis. Front. Plant Sci. 6: 995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenbock P., Szechynska-Hebda M., Plaszczyca M., Baudo M., Mateo A., et al. , 2008. Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20: 2339–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar M. S., Carvunis A. R., Dreze M., Epple P., Steinbrenner J., et al. , 2011. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333: 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkvold K. R., Martin M. E., Bronstein P. A., Collmer A., 2008. A survey of the Pseudomonas syringae pv. tomato DC3000 type III secretion system effector repertoire reveals several effectors that are deleterious when expressed in Saccharomyces cerevisiae. Mol. Plant Microbe Interact. 21: 490–502. [DOI] [PubMed] [Google Scholar]
- Nagpal P., Walker L. M., Young J. C., Sonawala A., Timpte C., et al. , 2000. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., et al. , 2006. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439. [DOI] [PubMed] [Google Scholar]
- Nordberg H., Cantor M., Dusheyko S., Hua S., Poliakov A., et al. , 2014. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 42: D26–D31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenbein C., Przybyla D., Danon A., Landgraf F., Göbel C., et al. , 2006. The role of EDS1 (enhanced disease susceptibility) during singlet oxygen-mediated stress responses of Arabidopsis. Plant J. 47: 445–456. [DOI] [PubMed] [Google Scholar]
- Rallapalli G., Kemen E. M., Robert-Seilaniantz A., Segonzac C., Etherington G. J., et al. , 2014. EXPRSS: an Illumina based high-throughput expression-profiling method to reveal transcriptional dynamics. BMC Genomics 15: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietz S., Stamm A., Malonek S., Wagner S., Becker D., et al. , 2011. Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 191: 107–119. [DOI] [PubMed] [Google Scholar]
- Roberts M., Tang S., Stallmann A., Dangl J. L., Bonardi V., 2013. Genetic requirements for signaling from an autoactive plant NB-LRR intracellular innate Immune receptor. PLoS Genet. 9: e1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J. D., 2011. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49: 317–343. [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano P., He Z., Luan S., 2004. Introducing immunophilins. From organ transplantation to plant biology. Plant Physiol. 134: 1241–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter L. M., Vencato M., Jordan K. L., Schneider S. E., Schneider D. J., et al. , 2006. Multiple approaches to a complete inventory of Pseudomonas syringae pv. Tomato DC3000 type III secretion system effector proteins. Mol. Plant Microbe Interact. 19: 1180–1192. [DOI] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Ott F., Klein J. D., Wang X., et al. , 2011. Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc. Natl. Acad. Sci. USA 108: 10249–10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiguzov A., Vainonen J. P., Wrzaczek M., Kangasjarvi J., 2012. ROS-talk – how the apoplast, the chloroplast, and the nucleus get the message through. Front. Plant Sci. 3: 292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Araki H., Chen L., Chen J.-Q., Tian D., 2006. Unique evolutionary mechanism in R-genes under the presence/absence polymorphism in Arabidopsis thaliana. Genetics 172: 1243–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K., 2009. The HSP90–SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 60: 139–164. [DOI] [PubMed] [Google Scholar]
- Shivaprasad P. V., Chen H. M., Patel K., Bond D. M., Santos B. A., et al. , 2012. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24: 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Loudet O., Durand S., Bérard A., Brunel D., et al. , 2008. Quantitative trait loci mapping in five new large recombinant inbred line populations of Arabidopsis thaliana genotyped with consensus single-nucleotide polymorphism markers. Genetics 178: 2253–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. R., Willmann M. R., Wu G., Berardini T. Z., Möller B., et al. , 2009. Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 5424–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrt K., Soll J., 2000. Toc64, a new component of the protein translocon of chloroplasts. J. Cell Biol. 148: 1213–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer M., Rudolf M., Tillmann B., Tripp J., Sommer M. S., et al. , 2013. Toc33 and Toc64-III cooperate in precursor protein import into the chloroplasts of Arabidopsis thaliana. Plant Cell Environ. 36: 970–983. [DOI] [PubMed] [Google Scholar]
- Tao Y., Xie Z., Chen W., Glazebrook J., Chang H. S., et al. , 2003. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. B., Ehrenreich I. M., 2014. Genetic interactions involving five or more genes contribute to a complex trait in yeast. PLoS Genet. 10: e1004324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas W. J., Thireault C. A., Kimbrel J. A., Chang J. H., 2009. Recombineering and stable integration of the Pseudomonas syringae pv. syringae 61 hrp/hrc cluster into the genome of the soil bacterium Pseudomonas fluorescens Pf0–1. Plant J. 60: 919–928. [DOI] [PubMed] [Google Scholar]
- Tornero P., Merritt P., Sadanandom A., Shirasu K., Innes R. W., et al. , 2002. RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell 14: 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M. A., Dangl J. L., Jones J. D. G., 2002. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99: 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W., Bennett M. H., Kubigsteltig I., Turnbull C., Grant M., 2007. Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc. Natl. Acad. Sci. USA 104: 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn R. A. L., Kamoun S., 2008. From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher P. M., McEvoy B., Yang J., 2010. From Galton to GWAS: quantitative genetics of human height. Genet. Res. 92: 371–379. [DOI] [PubMed] [Google Scholar]
- Wagner D., Przybyla D., Op den Camp R., Kim C., Landgraf F., et al. , 2004. The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306: 1183–1185. [DOI] [PubMed] [Google Scholar]
- Wagner S., Stuttmann J., Rietz S., Guerois R., Brunstein E., et al. , 2013. Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe 14: 619–630. [DOI] [PubMed] [Google Scholar]
- Wang, S., C. J. Basten, and Z.-B. Zeng, 2012 Windows QTL Cartographer 2.5 Department of Statistics, North Carolina State University, Raleigh, NC. Available at: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm. [Google Scholar]
- Wang Y., Liu C., Li K., Sun F., Hu H., et al. , 2007. Arabidopsis EIN2 modulates stress response through abscisic acid response pathway. Plant Mol. Biol. 64: 633–644. [DOI] [PubMed] [Google Scholar]
- Weigel D., Nordborg M., 2015. Population genomics for understanding adaptation in wild plant species. Annu. Rev. Genet. 49: 315–338. [DOI] [PubMed] [Google Scholar]
- Weßling R., Epple P., Altmann S., He Y., Yang L. et al, 2014. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 16: 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M., Feys B. J., Parker J. E., 2005. Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8: 383–389. [DOI] [PubMed] [Google Scholar]
- Wildermuth M. C., Dewdney J., Wu G., Ausubel F. M., 2001. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565. [DOI] [PubMed] [Google Scholar]
- Wright C. A., Beattie G. A., 2004. Pseudomonas syringae pv. tomato cells encounter inhibitory levels of water stress during the hypersensitive response of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 101: 3269–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X.-F., He S. Y., 2013. Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51: 473–498. [DOI] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X., Du Z., Su Z., 2013. PlantGSEA: a gene set enrichment analysis toolkit for plant community. Nucleic Acids Res. 41: W98–W103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeier J., Pink B., Mueller M. J., Berger S., 2004. Light conditions influence specific defence responses in incompatible plant-pathogen interactions: uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 219: 673–683. [DOI] [PubMed] [Google Scholar]
- Zhai J., Jeong D.-H., De Paoli E., Park S., Rosen B. D., et al. , 2011. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25: 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Shao F., Li Y., Cui H., Chen L., et al. , 2007. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185. [DOI] [PubMed] [Google Scholar]
- Zhang Z.-L., Ogawa M., Fleet C. M., Zentella R., Hu J., et al. , 2011. SCARECROW-LIKE 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 2160–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E. J., Jones J. D. G., et al. , 2004. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767. [DOI] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J. D., et al. , 2006. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Bacterial strains are available upon request. All data necessary for reproducing QTL results and complete edgeR results for RNA-seq data can be found as supplemental materials. Raw RNA-seq data are available at the National Center for Biotechnology Information Sequence Read Archive under the accession number SRP075162.