Abstract
Reduced endothelial-pericyte interactions are linked to progressive small vessel loss in pulmonary arterial hypertension (PAH), but the molecular mechanisms underlying this disease remain poorly understood. To identify relevant gene candidates associated with aberrant pericyte behavior, we performed a transcriptome analysis of patient-derived donor control and PAH lung pericytes followed by functional genomics analysis. Compared with donor control cells, PAH pericytes had significant enrichment of genes involved in various metabolic processes, the top hit being PDK4, a gene coding for an enzyme that suppresses mitochondrial activity in favor of glycolysis. Given reports that link reduced mitochondrial activity with increased PAH cell proliferation, we hypothesized that increased PDK4 is associated with PAH pericyte hyperproliferation and reduced endothelial-pericyte interactions. We found that PDK4 gene and protein expression was significantly elevated in PAH pericytes and correlated with reduced mitochondrial metabolism, higher rates of glycolysis, and hyperproliferation. Importantly, reducing PDK4 levels restored mitochondrial metabolism, reduced cell proliferation, and improved endothelial-pericyte interactions. To our knowledge, this is the first study that documents significant differences in gene expression between human donor control and PAH lung pericytes and the link between mitochondrial dysfunction and aberrant endothelial-pericyte interactions in PAH. Comprehensive characterization of these candidate genes could provide novel therapeutic targets to improve endothelial-pericyte interactions and prevent small vessel loss in PAH.
Pulmonary arterial hypertension (PAH) is a rare disorder associated with progressive elevation of pulmonary pressures, which, if untreated, leads to right-sided heart failure and death.1, 2 The major pathologic features of PAH are obliterative vasculopathy and progressive loss of distal pulmonary microvessels that is irreversible and unresponsive to available therapies. It has been postulated that progressive loss of distal pulmonary microvessels is the result of inappropriate angiogenesis, but the mechanisms that regulate pulmonary angiogenesis remain poorly understood.3, 4, 5 Angiogenesis is the process by which new vessels are formed from existing vessels by endothelial cell proliferation and formation of vascular tubes and recruitment of pericytes, highly specialized cells that associate with newly formed vessels to provide mural support and preserve endothelial viability.6, 7, 8 Despite inappropriate rates of proliferation and survival, pulmonary microvascular endothelial cells (PMVECs) from patients with PAH form smaller vascular tube networks compared with donor control cells, suggesting an intrinsic impairment in their angiogenic capacity.9, 10, 11, 12, 13 In addition, although recruitment of pericytes by PMVECs to developing vascular tubes is necessary for their maturation into functional blood vessels, the extent of involvement of pericytes in PAH pathogenesis remains poorly characterized. Disruption of pericyte-endothelial cell communication has been linked to various vascular disorders, such as diabetic retinopathy and stroke, in which loss of pericyte coverage leads to vascular dysfunction and loss of small vessels.14, 15, 16, 17 To date, most studies have centered on elucidating the contribution of pericytes to systemic microvessels, but little is known about their role in the pulmonary circulation.
We recently published a report describing for the first time that pericytes purified from the lungs of patients with PAH to properly associate with donor control PMVECs when co-cultured in Matrigel, a well-established assay to study angiogenesis in vitro.18 Using live imaging video microscopy, we identified a defect in PAH pericyte motility and polarity that led us to investigate signaling pathways, such as the Wnt/planar cell polarity, which is responsible for orchestrating PMVEC motility during angiogenesis.9 Genetic and molecular studies confirmed that reduced expression of the Wnt/planar cell polarity genes Fzd7 and cdc42 was present in PAH pericytes, and restoring their expression could significantly improve endothelial-pericyte interactions and promote angiogenesis. On the basis of our findings, we speculate that differential patterns of gene expression could account for the abnormal phenotype of PAH pericytes during pulmonary angiogenesis.
In this study, we present for the first time a bioinformatic analysis of the transcriptomes of patient-derived donor control and PAH pericytes. Our analysis provides evidence of different gene expression profiles between donor control and PAH pericytes in favor of genes involved in cell metabolism. Among the genes with the highest differential expression, we chose to focus on PDK4, a gene coding for a mitochondrial enzyme responsible for suppressing glucose oxidation.19, 20, 21, 22 We found that increased PDK4 expression in PAH pericytes results in higher rates of glycolysis, which correlate with increased PAH pericyte proliferation and survival. This observation is particularly relevant to PAH because increased expression of PDK family proteins and high rates of glycolysis have been linked to abnormal vascular remodeling in rat models of PAH.23, 24 We found that reducing PDK4 expression with siRNAs in PAH pericytes can restore mitochondrial metabolism and normalize rates of proliferation and survival. A major benefit of restoring mitochondrial metabolism in PAH pericytes is that their capacity to establish adequate endothelial-pericyte interaction is enhanced, resulting in vascular tube networks with morphologic features similar to that of donor control cells. We conclude that PAH pericytes have a unique gene expression profile associated with their abnormal phenotype and their study can provide opportunities for the development of therapeutic strategies capable of treating small vessel loss in PAH.
Materials and Methods
Cell Culture
Donor control PMVECs (Promokine catalog number C-12282; PromoCell GmbH, Heidelberg, Germany) were grown in EC media (Promokine catalog number C-22120; PromoCell GmbH) and used between passages 4 and 8.
Pericyte Isolation and Culture
For primary pericyte culture, the day before isolation, 40 μL of M280 sheep anti-mouse IgG dynabeads (11201D; Life Technologies Corporation, Carlsbad, CA) were incubated with 10 μg of 3G5 IgG (LakePharma, Belmont, CA) overnight. The next day, fresh donor control or PAH lung tissue was washed with 1× Hanks' balanced salt solution, minced into small pieces, and digested in a solution containing collagenase I, dispase, and DNase I (all from Sigma-Aldrich, St. Louis, MO) for 15 to 30 minutes at 37°C on an orbital shaker. An equivalent amount of ice-cold Pericyte Medium (ScienCell catalog number 1201; ScienCell, Carlsbad, CA) containing 2% fetal bovine serum (FBS) was added immediately after digestion. The cell suspension was passed through a 100-μm mesh filter (BD Biosciences, San Jose, CA) to remove undigested fibrous tissue. The cell pellet was resuspended in 1 mL of 1× phosphate-buffered saline (PBS) containing 0.2% FBS and 40 μL of 3G5 IgG-coated magnetic beads and gently rotated at 4°C for 45 minutes. Beads were then washed five times with 1× PBS and seeded in gelatin coated cell culture plates with pericyte medium. Fresh pericyte medium was replaced every other day to promote pericyte attachment and propagation. Clinical and hemodynamic data from control donors and patients with PAH who served as source of pericytes can be found in Supplemental Table S1. All lung tissue was obtained through the Pulmonary Hypertension Breakthrough Initiative. Validation of pericyte identity was performed using i) cell morphologic features, ii) location in lung tissue, and iii) positivity for various cell markers (calponin, CD90, NG2, platelet-derived growth factor-β, CD146, α-smooth muscle actin, and SM22α) as previously described.18
Microarray and Bioinformatics Analysis
The Illumina HumanHT-12 version 3.0 Gene Expression BioChip (catalog number BD-103-0204; Illumina, San Diego, CA) was used to capture transcriptome data.
The array data was deposited in Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession number GSE79786). The differential gene expression analysis was implemented by limma version 3.26.9 (Bioconductor, Seattle, WA; http://bioconductor.org/packages/release/bioc/html/limma.html).25 The P value cutoff was 0.05, and the fold-change cutoff was 2. The normalized expression levels of differentially expressed genes were extracted, and hierarchical clustering was implemented. The function enrichment analyses and concept-gene network analysis were implemented using the Bioconductor package GeneAnswers version 2.14.0 (Bioconductor; http://bioconductor.org/packages/release/bioc/html/GeneAnswers.html).26 All heat maps were generated by GENE-E version 3.0 (Broad Institute, Cambridge, MA; http://www.broadinstitute.org/cancer/software/GENE-E).
Immunofluorescence
The day before the studies, approximately 1.0 × 104 cells were seeded per well of a four-well EZ slides (Millipore Corporation, Billerica, MA). The next day, cells were fixed for 15 minutes in 4% paraformaldehyde, followed by three washes with 1× PBS. Cells were then permeabilized with ice-cold 1× PBS containing 0.1% Triton X-100 and goat serum for 1 hour, followed by overnight incubation with the primary antibodies at 4°C. The next day, cells were washed again in 1× PBS and incubated with Alexa Fluor 594 goat anti-mouse antibody and/or Alexa Fluor 488 goat anti-rabbit (Life Technologies Corporation) accordingly for 1 hour at room temperature. Slides were mounted with Prolong Gold antifade solution containing DAPI (Life Technologies Corporation). For lung tissue, OCT-embedded lung sections were labeled using CD31 (LS-B4737; LifeSpan Biosciences, Inc., Seattle, WA) and PDK4 (catalog number 63157; Abcam, Boston, MA). All staining was performed as previously indicated.18
Taqman Quantitative PCR
Total cellular RNA was extracted using the RNeasy mini Kit (Qiagen, Redwood City, CA) and converted to cDNA using the Fermentas Reverse Transcription Kit (Vilnius, Lithuania) following the manufacturer's protocol. Primers for PDK isoforms were purchased from Applied Biosciences (Foster City, CA). The PCR reaction mixture was denatured at 95°C for 15 minutes and then run for 40 cycles (94°C for 15 seconds, 55°C for 30 seconds, and 70°C for 30 seconds). Melting curve analysis was run at the same time to rule out nonspecific reactions or contamination. Glyceraldehyde-3-phosphate dehydrogenase was used for normalization. Agarose gel electrophoresis was used for verification of uncertain results. All real-time quantitative PCR studies were run in triplicates. The difference in mRNA expression was determined by ΔCT against glyceraldehyde-3-phosphate dehydrogenase.
Mitochondria Extraction
Mitochondria were purified from whole cell extracts (approximately 1.0 × 106 cells) centrifuged at 400 × g for 2 minutes using the Mitochondria Isolation Kit according to the manufacturer's instructions (catalog number 89874; Thermo Fisher, Lakeside Drive, CA). Total mitochondria were lyzed using radioimmunoprecipitation assay buffer, and PDK4 protein expression was analyzed by Western immunoblotting.
Western Immunoblot
Cells were washed three times with ice-cold 1× PBS and lysates prepared by adding lysis buffer (1× radioimmunoprecipitation assay buffer and 1 mmol/L phenylmethylsulfonyl fluoride) and boiling for 10 minutes before centrifugation. Supernatants were transferred to fresh microcentrifuge tubes and stored at −80°C. The protein concentration was determined by the Lowry assay (Bio-Rad Laboratories, Segrate, Italy). Equal amounts of protein were loaded onto each lane of a 4% to 12% Bis-Tris gel (Life Technologies) and subjected to SDS-PAGE electrophoresis under reducing conditions. After blotting, polyvinylidene difluoridemembranes were blocked for 1 hour in blocking buffer (5% milk powder in 0.1% PBS/Tween 20) and incubated with primary antibodies for PDK4 (catalog number ab63157; Abcam, Boston, MA) overnight at 4°C followed the next day by incubation with horseradish peroxidase–conjugated secondary antibodies. Bands were visualized using the ECL kit (Thermo Fisher). Signal normalization was performed against α-tubulin (catalog number ab7291; Abcam) or VDAC-1 (catalog number ab15895; Abcam).
Cell Proliferation
Cells were seeded in triplicate on collagen coated 96-well plates (5.0 × 103 cells per well) (Nunc, Rochester, NY) and cultured in complete medium overnight at 37°C and 5% CO2. The next day, cells were starved in 0.1% FBS reduced medium for 48 hours. Day 0 was the baseline right before changing back to complete medium. Cell proliferation was determined on days 1, 2, and 3 in the CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega, Sunnyvale, CA). BrdU cell proliferation assay (catalog number ab126556; Abcam) was performed following the manufacturer's instructions.
Cell Survival
Apoptosis was determined by measuring caspase 3/7 activation using Caspase-GLO 3/7 assay (Promega). Pericytes were plated into 96-well, white, clear bottom plates (5.0 × 103 cells per well) in complete medium overnight followed by exposure to 0.1% FBS reduced serum media for 48 hours. Caspase 3/7 levels were obtained by adding 100 μL of caspase 3/7 luciferase reagent mix (Caspase-Glo Reagent; Promega) for 1 hour followed by measurement of total luminescence using the GloMax luminometer (Promega). Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) apoptosis assay was performed using the TUNEL Detection Kit (catalog number S7101; Millipore, Billerica, CA) following the manufacturer's protocol. Briefly, 2.0 × 105 cells were seeded in 4-well EZ chamber slides. After 48 hours of 0.1% FBS reduced serum media, cells were fixed and treated with 150 μL of equilibration buffer. After washing, 100 μL of working-strength TdT enzyme was added and incubated for 1 hour at 37°C. The reaction was stopped by adding the stop/wash buffer followed by application of 150 μL of anti–digoxigenin peroxidase conjugate.
RNA Interference
To achieve gene knockdown, 2 μmol/L siRNA of PDK4 (catalog number L-019425-00; Dharmacon, Lafayette, CO) or nontargeting siRNA control were transfected into donor control pericytes. Knockdown efficiency was evaluated 72 hours after electroporation by measuring PDK4 protein levels in cell lysates via Western immunoblotting. Transfection was performed using a Nucleofector II (program U-25; Lonza Inc., Basel, Switzerland) with the Basic Smooth Muscle Cell Nucleofection kit (Lonza Inc.). All experiments were performed 72 hours after electroporation.
Fluorescence Activated Cell Sorting
A total of approximately 1.0 × 106 cells were washed three times with 1× PBS in a 15-mL tube and stained with 300 μL of propidium iodide (Life Technologies) containing 50 μg/mL of RNase A and 1% BSA. Samples were run in a flow cytometer (FACS Aria III; BD Biosciences) and analyzed with Flowjo software version 7.6.3 (Flowjo LLC, Ashland, OR).
MitoProbe 3,3-Diethyloxacarbocyanine Iodide Assay for Mitochondria Membrane Potential
Cells were stained with 3,3-diethyloxacarbocyanine iodide (Mitoprobe, M34150; Thermo Fisher) followed by flow cytometry (FACS Aria III; BD Biosciences). 3,3-Diethyloxacarbocyanine iodide is a cationic cyanide dye that accumulates in mitochondria with high mitochondria membrane potential (MMP).27 Carbonyl cyanide m-chlorophenyl hydrazone is a compound that reduces MMP and was used in comparison studies. Data points captured in the upper right quadrant represent cells with high MMP.
Mitochondrial CellROX Assay
Pericytes were seeded onto four-well EZ slides (Millipore) and treated with 100 nmol/L MitoTracker Red (Thermo Fisher) and 5 μmol/L CellROX Green (C10444; Thermo Fisher) to label mitochondria and superoxide production, respectively. Reactive oxygen species (ROS) production was normalized to cell number, and mitochondrial content was calculated as the ratio of CellROX to MitoTracker fluorescence (FCellRox/MitoTracker) using ImageJ software version 1.51d (NIH, Bethesda, MD; http://imagej.nih.gov/ij).
Matrigel Coculture Tube Formation Assay
Matrigel (catalog number 3432; Trevigen, Gaithersburg, MD) was thawed on ice overnight, dispersed on 96-well plates (55 μL per plate), and allowed to polymerize for 30 minutes at 37°C. PMVECs and pericytes were stained with the cell membrane dyes PKH67 green (MINI67; Sigma-Aldrich) and PKH26 red (MINI26; Sigma-Aldrich), respectively, following the manufacturer's protocol. Approximately 5.0 × 103 PMVECs were mixed with approximately 1.0 × 103 pericytes in 100 μL of pericyte medium and loaded per well. After 6 hours, images were taken and accessed by using a Leica DMRBII fluorescent microscope (Leico Microsystems, Wetzlar, Germany). Total tube number, branching points, and number of loops were quantified using Wimasis online image analysis (Wimasis, Munchen, Germany). Mean pericyte coverage of endothelial tubes was obtained from three independent experiments.
Mitochondrial Function Assays
Glycolytic capacity was measured using the XF Glycolysis Stress Test Kit (catalog number 1021194-100; Seahorse Bioscience, Billerica, MA) on the XF24 analyzer as described previously.28 For extracellular acidification rate (ECAR) measurements, approximately 3.0 ×104 cells were seeded and washed in glucose-free XF base medium (Seahorse Bioscience) adjusted to pH 7.35 at 37°C and containing 2 mmol/L l-glutamine. ECAR was measured after serial injections with 10 mmol/L d-glucose, 1 μmol/L oligomycin, and 100 mmol/L 2-deoxyglucose. Glycolysis was defined as ECAR response to oligomycin after glucose injection, and glycolytic reserve was calculated as ΔECAROligomycin-Glucose. Data were analyzed by the XF Glycolysis Stress Test Report Generator macro provided by Seahorse Biosciences.
Statistical Analysis
Values from multiple experiments are expressed as means ± SEM. Statistical significance was determined using an unpaired t-test or one-way analysis of variance followed by Bonferroni's multiple comparison tests unless stated otherwise. P < 0.05 was considered significant. The number of experiments and the sample number in each group are indicated in the figure legends.
Results
Microarray Analysis of Donor Control and PAH Pericytes Reveals Differences in Genes Associated with Cell Metabolism
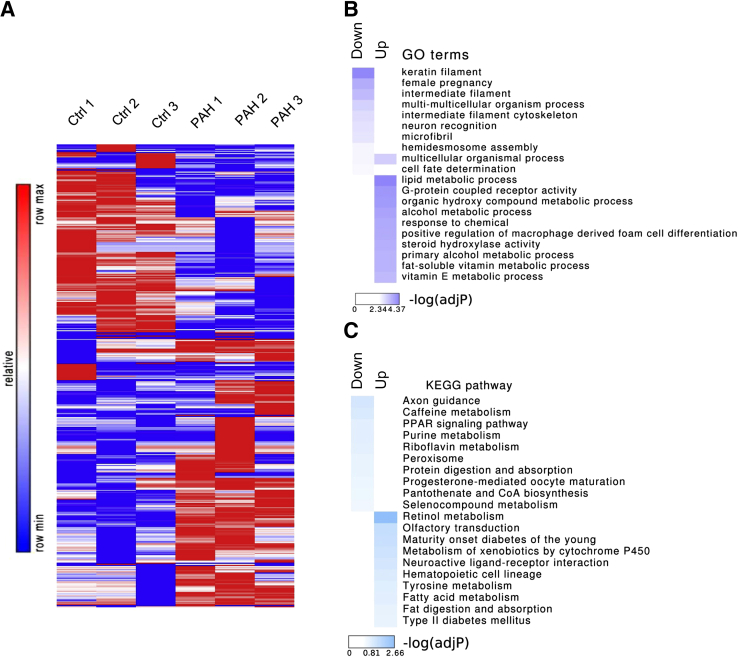
We used the Illumina HumanHT-12 version 3.0 Gene Expression BioChip to probe expression levels in human donor control (n = 3) versus PAH pericytes (n = 3). After rank invariant normalization, multidimensional scaling plot was generated to explore the relationships between the samples followed by differential gene expression analysis (Supplemental Figure S1A). Compared with donor control pericytes, 232 genes were up-regulated in PAH pericytes, whereas another 194 genes were down-regulated (P < 0.05) (Figure 1A). Gene Ontology (GO) enrichment analysis implied that functions related to metabolic processes, cell differentiation, and signaling were highly enriched in PAH pericytes (Figure 1B), which was further confirmed by KEGG pathway enrichment analysis (Figure 1C).
Figure 1.
Comparison of gene expression patterns of healthy donor control (Ctrl) and pulmonary arterial hypertension (PAH) pericytes by microarray analysis. A: The heat map of differently expressed genes (P < 0.05, fold change >2.0) between three PAH versus three donor control pericytes. B: Gene Ontology (GO) enrichment analyses of significantly up-regulated/down-regulated genes in PAH. C: Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of significantly up-regulated/down-regulated genes in PAH.
Analysis of the top 10 up-regulated genes in PAH pericytes revealed candidate genes involved in processes relevant to cell signaling (GPR64, RORB), angiogenesis (PF4V1, SPON1), and metabolism (PDK4, ADH1A, CHST15), whereas the top 10 down-regulated genes included those involved in cell motility and tissue morphogenesis (FBN2, HOXC8, ACTG2), oxidative stress (CYBG, NXN), and signaling (IGFBP2, RARRES3) (Table 1). Of the genes prioritized by our bioinformatic analysis, PDK4 stood out as having the highest up-regulation (eightfold) in PAH pericytes (Supplemental Figure S1B). PDK4 codes for one of the four PDK family proteins located in the mitochondrial matrix, where it regulates the rate of glucose oxidation and fatty acid metabolism in mammalian cells by direct inhibition of the pyruvate dehydrogenase (PDH) complex.21, 29, 30 Of the four family proteins, PDK1 and 2 are expressed in most cells, whereas PDK3 is mainly expressed in the testes, and PDK4 is mostly expressed in muscle under metabolic stress. Phosphorylation of PDH by PDKs reduces conversion of pyruvate into acetyl-CoA (the main substrate for the Krebs cycle), resulting in down-regulation of aerobic metabolism and a switch toward glycolysis driven (ie, anaerobic) energy production.21 This metabolic switch toward glycolysis (ie, Warburg effect) is a feature of malignant cells and is associated with their aggressive growth, survival, and metastasis.31, 32, 33 Most importantly, this phenomenon has also been described in PAH, where mitochondrial dysfunction, reduced glucose oxidation, and increased glycolysis have been linked to severity of pulmonary vascular remodeling, resulting from excessive pulmonary artery smooth muscle cell (PASMC) proliferation and survival and right ventricular dysfunction.21, 23, 24 Increased PDK expression contributes to this metabolic shift and use of dichloroacetate (DCA), a pharmacologic PDK inhibitor, can restore mitochondrial function and effectively reverse obliterative vascular remodeling in rat models of PAH by suppressing PASMC proliferation and survival.23, 34, 35 Of note, studies have found that PDK4 specifically is associated with impaired mitochondrial activity and severe right ventricular dysfunction in fawn-hooded rats that could be partially reversed with DCA.24 In light of these studies and based on the results of our microarray analysis, we validated PDK4 as a gene candidate in PAH pericytes.
Table 1.
Altered Top 10 Highly Expressed Genes with Significant Fold Change (P < 0.05) in Pulmonary Arterial Hypertension Pericytes Versus Donor Control
| Up-regulated | Down-regulated | Gene symbol | Gene name | Normalized fold change | P value |
|---|---|---|---|---|---|
| + | PDK4 | Pyruvate dehydrogenase kinase 4 | 7.8 | 0.02929 | |
| + | G0S2 | G0/G1 switch gene 2 | 6.6 | 0.02626 | |
| + | ADH1A | Alcohol dehydrogenase 1A | 12.7 | 0.02771 | |
| + | RORB | RAR-related orphan receptor B | 4.7 | 0.00037 | |
| + | SPON1 | Spondin 1 | 4.6 | 0.01427 | |
| + | SIPA1L2 | Signal-induced proliferation-associated 1 like 2 | 4.6 | 0.00038 | |
| + | GPR64 | G protein–coupled receptor 64 | 13.1 | 0.00499 | |
| + | C4ORF31 | Fibronectin type III domain-containing protein | 5.8 | 0.02348 | |
| + | PF4V1 | Platelet factor 4 variant 1 | 7.4 | 0.04415 | |
| + | CHST15 | Carbohydrate sulfotransferase 15 | 11.3 | 0.00399 | |
| + | IGFBP2 | Insulin-like growth factor binding protein 2 | 5.6 | 0.0038 | |
| + | FBN2 | Fibrillin 2 | 8.8 | 0.00047 | |
| + | CYGB | Cytoglobin | 5.9 | 0.01236 | |
| + | ACTG2 | Actin, gamma 2 | 8.4 | 0.02231 | |
| + | MARCH4 | MARCH of membrane-bound E3 ubiquitin ligases | 4.8 | 0.01468 | |
| + | TMEM35 | Transmembrane protein 35 | 6.0 | 0.00413 | |
| + | NXN | Nucleoredoxin | 4.5 | 0.00288 | |
| + | RARRES3 | Retinoic acid receptor 3 | 4.3 | 0.02779 | |
| + | CPA4 | Carboxypeptidase A4 | 7.4 | 0.00969 | |
| + | HOXC8 | Homeobox C8 | 7.1 | 0.01314 |
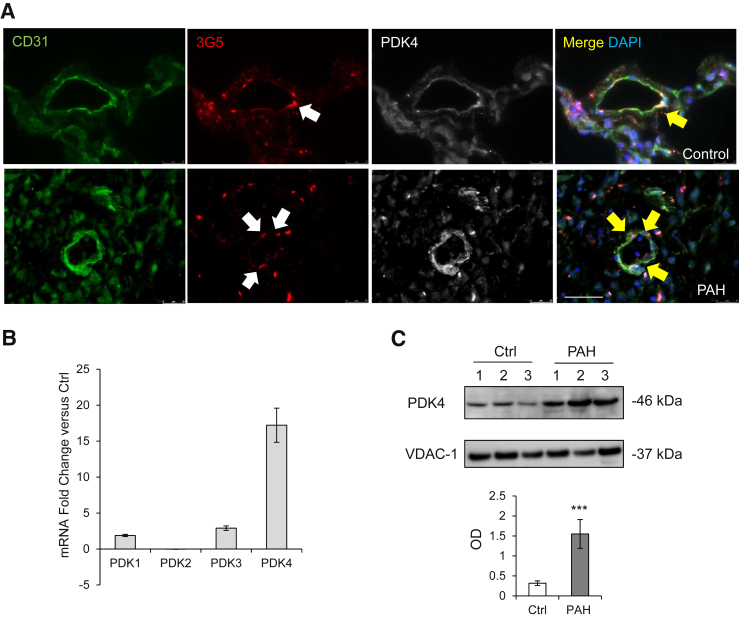
PAH Pericytes Have Increased PDK4 Gene and Protein Expression
We validated our microarray analysis findings by measuring PDK4 expression in lung tissue and lung derived pericytes from donor controls and patients with PAH. Immunofluorescence of healthy donor and PAH lung sections revealed strong expression of PDK4 in pericytes (Figure 2A). Next, we measured PDK4 mRNA and protein in pericytes purified from explanted lung using our previously described method.18 Compared with donor control, Taqman PCR analysis confirmed that PDK4 mRNA expression was significantly elevated (approximately 16-fold) in PAH pericytes, and its expression was the highest among all other PDK isoforms (Figure 2B). Although similar PDK4 protein expression was seen in whole cell lysates of donor control and PAH pericytes (data not shown), mitochondrial PDK4 protein levels were significantly increased in the latter (Figure 2C).
Figure 2.
PDK4 increases in pulmonary arterial hypertension (PAH) pericytes. A: Confocal images of PDK4 lung sections. Endothelium was stained with CD31 (green), pericytes with 3G5 (red), and lung tissue with PDK4 (white). White arrows indicate pericyte-positive staining, and yellow arrows indicate co-staining of 3G5 and PDK4. DAPI labels cell nuclei (blue). B: TaqMan quantitative PCR for the four PDK family proteins in donor control and PAH pericytes. The expression of each gene is shown relative to that in donor control pericytes. C: Representative Western immunoblot images of PDK4 in enriched mitochondria of pericytes from three donor control (Ctrl) versus three patients with PAH. ∗∗∗P < 0.001 versus unstimulated controls using the unpaired t-test. Scale bar = 50 μm. OD, optical density.
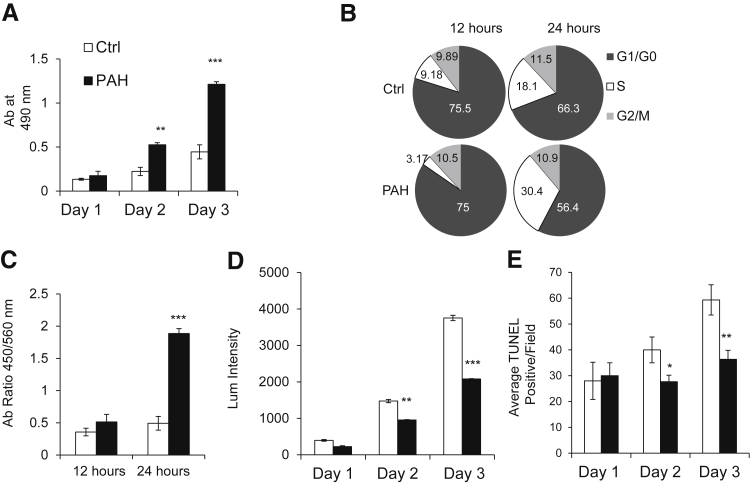
Increased PDK4 Expression Correlates with Increased PAH Pericyte Proliferation and Survival
Having established that PDK4 expression was higher in PAH pericytes, we wanted to determine whether this gene signature correlated with a hyperproliferative and apoptotic resistant phenotype similar to that documented in PAH PASMCs.23 Compared with donor control cells, PAH pericytes exposed to full media had increased rates of proliferation during 72 hours by MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) (Figure 3A). Cell cycle analysis revealed that 18.1% of donor control pericytes were in S-phase at 24 hours, which was a twofold larger proportion than at 12 hours. However, 30.4% of the PAH pericytes were in S-phase at 24 hours versus 3.17% at 12 hours, indicating faster cell cycle progression for PAH pericytes than donor controls (Figure 3B). This finding was further confirmed using the BrdU assay, which revealed significantly increased DNA synthesis at 12 and 24 hours in PAH pericytes (Figure 3C). Regarding survival, serum-starved PAH pericytes also had significantly lower rates of apoptosis during 72 hours compared with donor control cells as measured by both caspase 3/7 activity (Figure 3D) and TUNEL staining (Figure 3E).
Figure 3.
Pulmonary arterial hypertension (PAH) pericytes have greater proliferation and survival compared with healthy donor cells. A: Cell growth was evaluated by the MTS assay on days 1, 2, and 3 of culture. All values were normalized to the cells after 48 hours of starvation medium. B: Serum-starved cells were analyzed by flow cytometry on day 3 after the addition of serum of 12 and 24 hours. The percentage of cells in the G1, S, and G2/M phases is indicated. C: DNA analysis of control and PAH pericytes in 12 hours and 24 hours by BrdU. D: Cell apoptosis was evaluated by caspase 3/7 assay on days 1, 2, and 3 of culture. All values were normalized to the cells after 48 hours of starvation medium. E: Cell apoptosis level was further evaluated by the terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay on days 1, 2, and 3 of culture. All values were normalized after 48 hours of starvation. Average TUNEL-positive cells per field obtained from six independent microscopic fields. All experiments performed were repeated three times ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus control using one-way analysis of variance with Dunnett's posttest. Ctrl, control.
Taken together, our findings firmly establish that PAH pericytes have a proproliferative and prosurvival phenotype similar to that described in PAH PASMCs. Because this abnormal cellular phenotype has been associated with PDK-mediated suppression of mitochondrial activity,21 we hypothesized that PAH pericytes also have a similar metabolic signature directly linked to increased PDK4 expression.
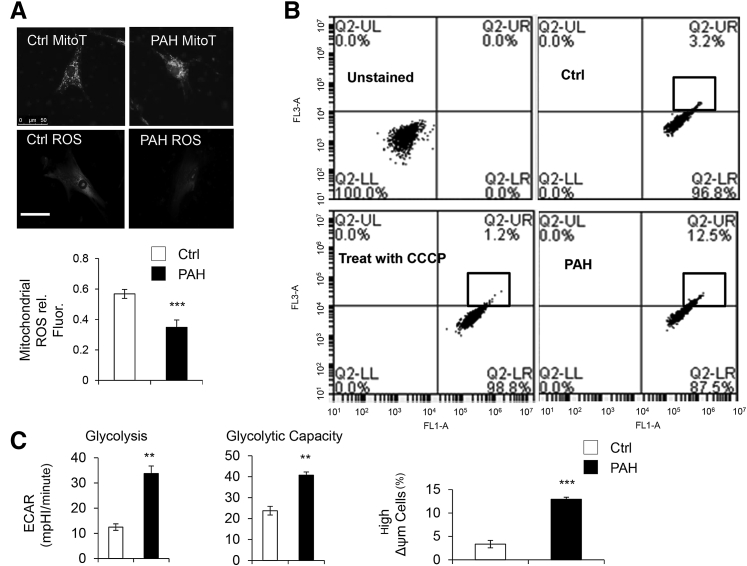
PAH Pericytes Demonstrate Reduced Mitochondrial Activity and Increased Glycolysis
Aerobic metabolism is the major source of ATP production in mammalian cells, and it is carried within the mitochondria.36, 37, 38 In PAH, cells have dysmorphic mitochondria with reduced respiratory chain coupling, inefficient use of oxygen, and increased glycolysis.28, 39, 40 To test whether PAH pericytes have the same alterations, we used MitoTracker to capture mitochondrial morphologic features and measure production of mitochondrial-derived ROS. Compared with donor controls, PAH pericytes have dense clusters of perinuclear mitochondria along with significantly reduced ROS production (Figure 4A) and higher MMP (a marker of reduced mitochondrial activity) (Figure 4B). To document differences in glycolysis between donor control and PAH pericytes, we measured the ECAR (a marker of lactate production) using a Seahorse analyzer. As expected, we found significantly greater aerobic glycolysis and glycolytic capacity in PAH pericytes compared with donor control cells (Figure 4C).
Figure 4.
Increased PDK4 expression correlates with reduced mitochondrial activity in pulmonary arterial hypertension (PAH) pericytes. A: Reactive oxygen species (ROS) production in PAH versus donor control (Ctrl) cells. Fluorescence intensity (rel. Fluor.) was normalized against MitoTracker (MitoT). B: Fluorescence activated cell sorting of healthy donor and PAH pericytes using mitochondrial membrane potential (MMP). FL1-A: fluorescein isothiocyanate; FL3-A: Texas Red. The percentage of high MMP is found in the block of Q2-UR. C: Aerobic glycolysis and glycolytic capacity in cells as measured by Seahorse analysis. ∗∗P < 0.01, ∗∗∗P < 0.001 compared with controls using unpaired t-test. Scale bar = 50 μm. CCCP, Carbonyl cyanide 3-chlorophenylhydrazone; ECAR, extracellular acidification rate; LL, lower left; LR, lower right; mpH, mili pH per minute; UL, up-left; UR, up-right.
In conclusion, we found that PAH pericytes have reduced mitochondrial activity and a glycolytic switch. As a next step, we sought to link these abnormalities to PDK4 using siRNA-mediated gene knockdown.
PDK4 Knockdown in PAH Pericytes Results in Reduced Proliferation and Increased Apoptosis
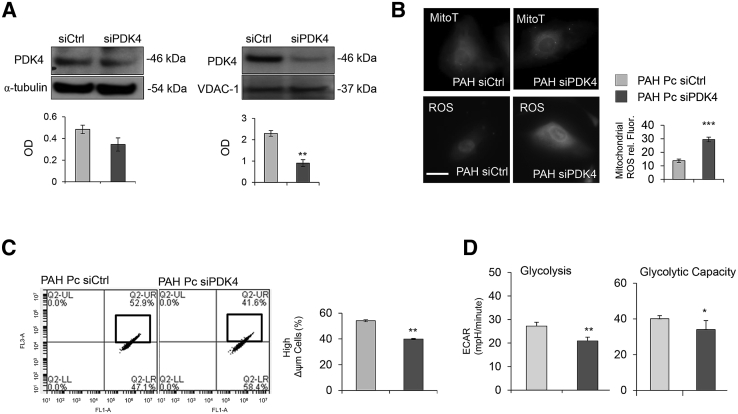
To reduce PDK4 expression in PAH pericytes, we transfected cells with either nontargeting (siCtrl) or PDK4-specific (siPDK4) siRNAs and measured protein expression in whole cell and mitochondrial lysates. Although we saw a mild but significant reduction in whole cell lysates, transfection of siPDK4 resulted in substantial PDK4 knockdown in PAH pericyte mitochondria (Figure 5A). Transfection of siPDK4 in PAH pericytes resulted in a significant increase in mitochondrial ROS (Figure 5B) and mild but significant reductions in MMP (Figure 5C) and EAR (Figure 5D). Of note, we did not see any change in ROS production and EAR in donor control pericytes transfected with either siCtrl or PDK4 siRNA (Supplemental Figure S2).
Figure 5.
Knockdown of PDK4 in pulmonary arterial hypertension (PAH) pericytes restores mitochondrial activity. A: Western immunoblot for PDK4 in total cell lysate (α-tubulin as an internal control) and mitochondria lysate (VDAC-1 as an internal control) from PAH pericytes transfected with either nontargeting (siCtrl) or PDK4-specific (siPDK4) siRNAs. B: Reactive oxygen species (ROS) production in PAH pericytes treated with either siCtrl or siPDK4. Fluorescence intensity (rel. Fluor.) was normalized against MitoTracker (MitoT). Mitochondrial membrane potential (MMP) (C) and aerobic glycolysis and glycolytic capacity (D) in donor control versus PAH with PDK4 siRNA treatment. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 compared with controls using the unpaired t-test. Scale bar = 25 μm. ECAR, extracellular acidification rate; LL, lower left; LR, lower right; mpH, mili pH per minute; OD, optical density; Pc, pericyte; UL, up-left; UR, up right.
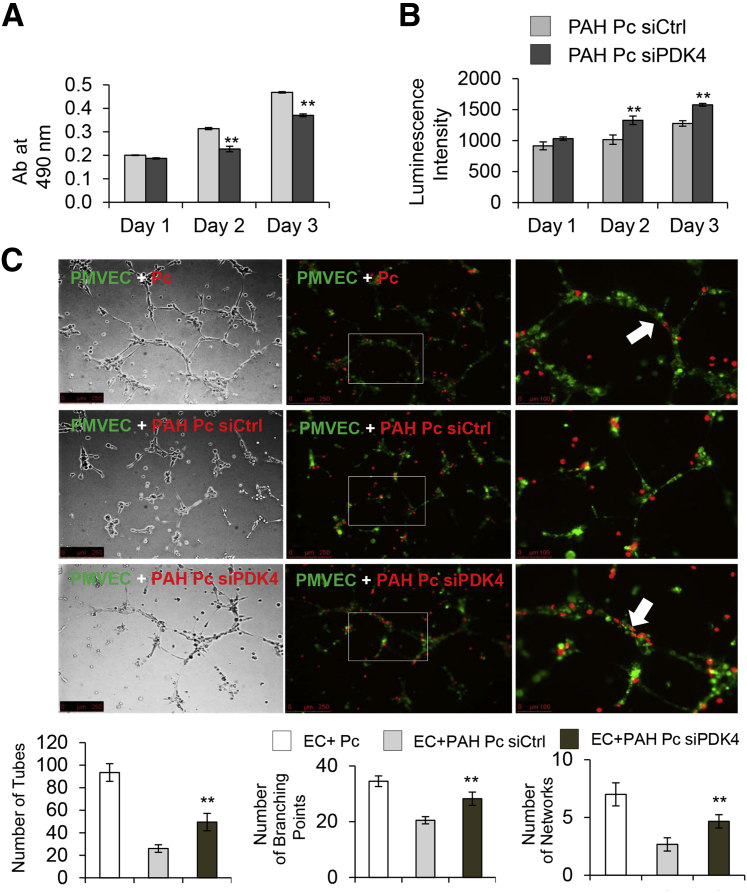
Given that restoration of mitochondria activity has been associated with reductions in cell proliferation and survival in PAH, we proceeded to measure these parameters in PDK4 siRNA–treated PAH pericytes. As anticipated, siPDK4 transfected PAH pericytes had significant reductions in proliferation (Figure 6A) and survival (Figure 6B) as measured by MTS and caspase 3/7 activity assays, respectively. As before, we did not see any differences in donor control pericytes transfected with either siCtrl or PDK4 siRNA (Supplemental Figure S2).
Figure 6.
Knockdown of PDK4 reduces proliferation and survival of pulmonary arterial hypertension (PAH) pericytes and restores capacity to enhance endothelial tube formation. Cell growth by MTS (A) and cell apoptosis by caspase 3/7 (B) were evaluated on days 1, 2, and 3 of culture after 48 hours siRNA posttransfection. C: Pericytes (Pcs) were transfected with nontargeting siRNA control (siCtrl) or PDK4-specific (siPDK4) for 48 hours when compared with healthy pericytes. Before seeded on Matrigel, cells were stained with PKHs. Pulmonary microvascular endothelial cells (PMVECs) were stained in PKH67 (green) and PAH Pcs in PKH26 (red). The white boxes indicate the enlarged areas of magnification in the right column. The white arrows indicate EC-Pc interaction. The numbers of tubes, branching points, and networks were assessed after six hours. ∗∗P < 0.01 compared with control (unpaired t-test, one-way analysis of variance with Bonferroni post-test). Scale bars: 250 μm (C, left and middle columns); 100 μm (C, right column). EC, endothelial cell.
PDK4 Knockdown Restores Capacity of PAH Pericytes to Associate with Donor Control PMVECs and Establish Vascular Tube Networks
Co-culture of healthy lung pericytes with PMVECs results in formation of vascular networks characterized by thick tubes composed of endothelial cells covered by pericytes (Figure 6C). Our previously published studies established that PAH pericytes have an inherent inability to properly associate with endothelial cells, resulting in formation of small and disorganized vascular networks.18 Given that steady ATP production is required to support the cellular bioenergetics associated with motility,38 we tested whether reduction of PDK4 can improve the ability of PAH pericytes to associate with PMVECs during angiogenesis. Compared with cells transfected with siCtrl (Figure 6C), co-culture of PDK4 siRNA–transfected PAH pericytes with healthy PMVECs resulted in networks characterized by greater tube number, branching points, and loops (Figure 6C).
Discussion
A major contributor to the clinical deterioration of patients with PAH is progressive small vessel loss and lack of proper compensatory pulmonary angiogenesis, which contributes to increased pulmonary vascular resistance and decompensated right-sided heart failure.41 A major determinant of donor control angiogenesis is the capacity of newly formed blood vessels to recruit neighboring pericytes to provide structural support and induce normalization.42 To date, studies focused on the role of pericytes in PAH have identified an abnormal cellular phenotype characterized by hyperproliferation and an impaired ability to associate with developing microvessels.18, 43 In an effort to increase our knowledge of the genomic landscape of PAH pericytes and identify possible genetic sources for their abnormal behavior, we conducted a transcriptome analysis, which revealed significant enrichment for genes involved in cell signaling and the regulation of processes essential for preservation of cellular homeostasis. In particular, our GO and KEGG enrichment analysis revealed that PAH pericytes have a significant enrichment for genes involved in various aspects of cell metabolism (Figure 1). Available studies strongly support metabolic dysregulation as a major pathogenic mechanism in both human and rodent models of PAH and stress the concept that dysregulation of cell metabolism is a common finding in PAH pathogenesis and could explain recent associations between PAH and disorders, such as insulin resistance, metabolic syndrome, and obesity.44, 45, 46, 47 The relevance of this finding is further supported by the efficacy of agents used to treat these metabolic disorders to reverse PAH in experimental animal models45, 48, 49 and several ongoing clinical trials testing their use as treatment for PAH. Targeting cell metabolism is an attractive concept because most of the cells involved in vascular remodeling have been reported to exhibit similar altered metabolic responses, and their restoration via lifestyle modifications and pharmacologic therapy could provide a powerful opportunity to alter disease progression.
PAH is a disease state characterized by uncoupling of glycolysis and glucose oxidation. In this pathologic state, pulmonary vascular cells, such as fibroblasts, endothelial, smooth muscle cells, and cardiomyocytes,24, 50, 51 increase glucose uptake to drive ATP production through glycolysis, a less efficient metabolic strategy compared with mitochondrial-based glucose oxidation. Evidence of increased glucose uptake has been documented in the right ventricle of patients with PAH by measurements of fludeoxyglucose (a glucose analog) uptake via positron emission tomography.52, 53 A feature of this abnormal metabolic switch is up-regulation of PDKs, which phosphorylate and inhibit PDH in the mitochondria.38 We chose to validate PDK4 in our cell model not only because it had the highest fold increase among the genes selected in our analysis but also because of previous studies documenting similar alterations in this and other PDK isoforms in both animal models and human PAH samples.19, 21, 34 PDK4 is highly expressed in the liver, heart, and skeletal muscle,24, 54 and its expression is regulated by various transcription factors associated with metabolic pathways, such as hypoxia-inducible factor-1α, FOXO1, estrogen, and peroxisome proliferator-activated receptor gamma, among others.24, 55, 56 Skeletal-muscle PDK4 protein expression is selectively increased in insulin-deficient and -resistant states in both disease models and humans.57 In studies looking at right ventricular function in fawn-hooded rats and samples from patients with PAH, PDK4 was elevated in right ventricle hypertrophy and correlated with reduced glucose oxidation and increased glycolysis.24 Our studies support a similar model in which elevated PDK4 correlated with a switch to glycolysis-based metabolism in PAH pericytes that could be partially reversed by reducing PDK4 levels to improve endothelial-pericyte interactions and support pulmonary angiogenesis (Figure 7). In contrast to mitochondrial extracts, Western blotting of whole lysates failed to reveal major differences in PDK4 protein levels; this was further evident in our siPDK4 studies, in which the magnitude of gene knockdown was substantially higher in mitochondrial compared with whole cell extracts. The explanation for this is not completely clear, but there is evidence that some PDK4 can be found in the cytoplasm, where it can interact with signaling mediators of the BMP signaling in vascular smooth muscle cells.58 We speculate that there is a pool of PDK4 that is distributed between cytoplasm and that the latter is more susceptible to alterations in mRNA transcription and could be influenced by altered protein translocation to mitochondria.
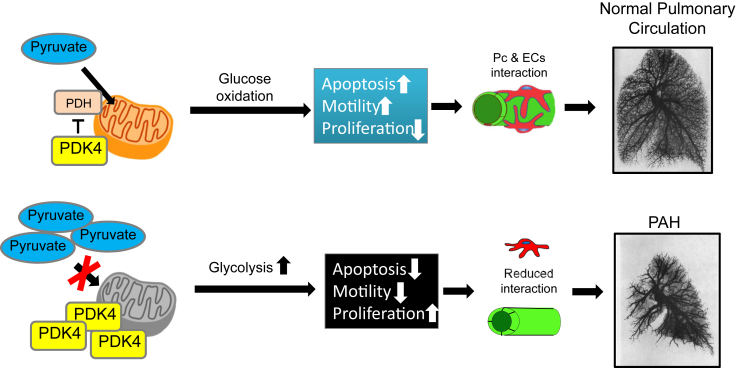
Figure 7.
Proposed model. Elevated PDK4 in pulmonary arterial hypertension (PAH) pericytes (Pcs) reduces mitochondrial metabolism in favor of glycolysis. Although pericytes exhibit heightened proliferation and survival, they have limited capacity to establish communication with pulmonary microvascular endothelial cells (ECs), resulting in small vessel loss and impaired angiogenesis.
The salutary effects of reducing PDK4 in PAH pericytes are supported by multiple studies that have targeted this and other PDKs to reduce the burden of pulmonary lesions and right ventricular failure in various models of PAH using the classic PDK inhibitor DCA,23, 24, 34, 59, 60 an agent that is currently at the center of a multicenter clinical trial in PAH (NCT01083524). Of note, we tested DCA in our cells and found mild improvement in angiogenesis with increasing doses, but the effects were less pronounced compared with PDK4 siRNA (data not shown). We speculate that this could be because DCA has greater inhibitory binding capacity for some PDK isoforms compared with PDK4 and that other PDK inhibitors, such as AZD7545 and radicicol,61 could have a greater biological effect, which could be tested in cell and animal models of PAH. Furthermore, PDK4 sits in a GO term node that interacts with multiple genes, such as CYP1A1, LPL, and PDE3B, based on gene-concept network analysis (Supplemental Figures S3 and S4); overactivity could create ripples through this network that could result in alteration of other processes, which may not be properly targeted by treatment with DCA alone. One possible approach to study this would be to perform metabolomic studies to capture the profile of lipid and organic molecules present in healthy and PAH pericyte cytosol at baseline in response to exogenous injury, which would serve to prioritize metabolic genes and pathways relevant to abnormal cellular phenotype. We anticipate that our data will likely continue to yield candidate genes that could be further characterized to elucidate their contribution to aberrant pericyte behavior in PAH.
The similarity of cellular and metabolic phenotype of pericytes and PASMCs is significant and bears special mention given the common origin of these cells from mesenchymal stem cells. Pericytes are considered multipotent cells with mesenchymal stem cell–like properties that can give rise to different cell lineages when exposed to specific environmental cues.62, 63, 64, 65 The concept that pericytes may differentiate into smooth muscle cells had been proposed a decade ago.66 Recently, a study looked at pericyte distribution in the pulmonary circulation of patients with PAH and reported increased pericyte numbers in close proximity to pulmonary microvessels, along with a pro-proliferative and promigratory phenotype when the cells were purified and cultured in vitro.43 Although these observations seem to support a potential contribution of these pericytes to PAH pathology, it is still unclear whether smooth muscle cells s around the distal pulmonary arteries originate from pericytes. On the basis of our current data, we are tempted to speculate that lung pericytes could serve as a niche for PASMCs in PAH. Availability of microarray data sets from PAH PASMCs could be used to compare similarities with pericytes to determine whether gene enrichment for PASMC genes are found in PAH pericytes to detect lineage similarities. Should pericytes be a significant source of PASMCs, it is possible that strategies that can reverse PASMCs to a pericyte-like phenotype may help reduce vascular remodeling and encourage vessel stability in patients with PAH.
Although targeting PDK4 improves cellular phenotype and pericyte function, it is clearly not enough to restore normal function. We speculate that this could explain the lack of a significant effect of PDK4 siRNA in donor control cells because other family proteins could compensate for down-regulation of PDK4. Furthermore, given the strong metabolic signature in our gene set, it is very likely that alteration in other gene modifiers are likely present and may not be properly corrected with targeted knockdown of PDK4. To address these challenges, we have initiated network analysis combined with metabolomics to study how alteration in metabolic networks affects the production of relevant metabolites that could serve to produce the altered PAH phenotype. It is also attractive to speculate that such studies could identify common patterns that could be expanded to other cell types involved in PAH vascular remodeling. It is worth pointing out that our treatment of healthy control pericytes with siPDK4 did not result in significant changes in mitochondrial ROS production and glycolytic activity. We speculate that this may be because of the capacity of other PDK isoforms and related molecules to be up-regulated and compensate for PDK4 deficiency; in the case of PAH pericytes, PDK4 up-regulation may overshadow the effect of these other genes, leading to the salutary effect of its reduction in these cells.
Our study has some important limitations. Our analysis could have missed other relevant genes with differential expression in PAH versus donor control pericytes as a result of small sample size and different binding affinity of probes contained in the chip. These are well-documented issues that must be taken into account when analyzing microarray data sets and have been addressed in our studies by our choice of a well-tailored statistical method that takes into account these limitations. However, current next-generation sequencing technologies, such as RNA sequencing.67 have become more accessible and provide a viable alternative to microarray studies because of greater sensitivity, depth of coverage, and capacity to capture temporal changes in a large cohort of gene expression. As a next step, we plan to perform RNA-sequencing studies to compare differential gene expression in pericytes cultured alone versus in co-culture with endothelial cells. We anticipate that these experiments will provide information on genes whose temporal expression during the establishment of endothelial-pericyte interaction is critical and whose dysregulation could compromise vessel stability in PAH. Finally, it is important to point out that the genotype of the patients with PAH who served as tissue donors is unknown, and we cannot rule out whether mutations in genes known to be associated with PAH are present in our samples.68
In conclusion, we present evidence of a differential gene expression profile between PAH pericytes and donor control cells linked to impaired establishment of endothelial-pericyte interactions during pulmonary angiogenesis. Although targeting PDK4 appears to be a promising strategy to partially restore pericyte function, a more ambitious approach incorporating comprehensive knowledge of cellular metabolism together with PDK4 network analysis will be required to identify drugs capable of restoring mitochondrial metabolism with minimal effect on other genes within the network whose function remains unaffected in PAH. The ultimate goal is to improve the ability of pericytes to associate with endothelial cells during pulmonary angiogenesis and preserve blood flow to alveoli for proper oxygenation, which would lead to improved clinical outcomes in PAH.
Acknowledgments
Lung tissues from patients with PAH and controls were provided by the Pulmonary Hypertension Breakthrough Initiative, which is funded by the Cardiovascular Medical Research and Education Fund. We thank Drs. Marlene Rabinovitch and Mark Nicolls for their careful review and thoughtful comments.
Footnotes
Supported by an American Heart Association Scientist Development grant 15SDG25710448 (K.Y.) and Pulmonary Hypertension Association Proof of Concept grant SPO121940 (K.Y.). J.K.H. was supported by German Research Foundation fellowship He 6855/1-1. V.A.d.J.P. was supported by a career development award from the Robert Wood Johnson Foundation, NIH grant K08 HL105884-01, a Pulmonary Hypertension Association Award, a Biomedical Research Award from the American Lung Association, and a Translational Research and Applied Medicine Award from Stanford University.
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2016.05.016.
Supplemental Data
Additional comparison of gene expression patterns of healthy donor and pulmonary arterial hypertension (PAH) pericytes by microarray analysis. A: Multidimensional scaling (MDS) plot of gene expression levels based on microarrays of all samples. B: The line plot is based on log2 (log2 gene expression) to make the genes comparable to others, using the normalized untransformed gene expression levels. The normalized gene expression level = 2 superscript (the log 2 expression levels). Log2 expression levels for controls 1, 2, and 3 (Ctrl1, Ctrl2, and Ctrl3, respectively) and patients with PAH 1, 2, and 3 (PAH1, PAH2, and PAH3, respectively) are 10.19, 6.85, and 7.95 and 11.97, 9.85, and 12.06, respectively. Expression of PDK4 is the top highest among all up-regulated genes.
Knockdown of PDK4 in donor pericytes does not change mitochondrial activity. A: Reactive oxygen species (ROS) production in donor control with PDK4 siRNA treatment. Fluorescence intensity was normalized by MitoTracker (MitoT). Aerobic glycolysis and glycolytic capacity (B) in donor control with PDK4 siRNA treatment as measured by Seahorse assay. Cell growth by MTS assay (C) and cell apoptosis by caspase 3/7 assay (D) were evaluated on days 1, 2, and 3 of culture in donor control with PDK4 siRNA treatment. Scale bar = 25 μm. ECAR, extracellular acidification rate; Pc, pericyte; siCtrl, siRNA control.
Gene Ontology concept–gene network analysis for all significantly up-regulated genes in pulmonary arterial hypertension pericytes.
Gene Ontology concept–gene network analysis for all significantly down-regulated genes in pulmonary arterial hypertension pericytes.
References
- 1.Humbert M., Morrell N.W., Archer S.L., Stenmark K.R., MacLean M.R., Lang I.M., Christman B.W., Weir E.K., Eickelberg O., Voelkel N.F., Rabinovitch M. Cellular and molecular pathobiology of pulmonary arterial hypertension. J Am Coll Cardiol. 2004;43:13S–24S. doi: 10.1016/j.jacc.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 2.Rich S., Dantzker D.R., Ayres S.M., Bergofsky E.H., Brundage B.H., Detre K.M. Primary pulmonary hypertension: a national prospective study. Ann Intern Med. 1987;107:216–223. doi: 10.7326/0003-4819-107-2-216. [DOI] [PubMed] [Google Scholar]
- 3.Tuder R.M., Voelkel N.F. Angiogenesis and pulmonary hypertension: a unique process in a unique disease. Antioxid Redox Signal. 2002;4:833–843. doi: 10.1089/152308602760598990. [DOI] [PubMed] [Google Scholar]
- 4.Tuder R.M., Cool C.D., Yeager M., Taraseviciene-Stewart L., Bull T.M., Voelkel N.F. The pathobiology of pulmonary hypertension: endothelium. Clin Chest Med. 2001;22:405–418. doi: 10.1016/s0272-5231(05)70280-x. [DOI] [PubMed] [Google Scholar]
- 5.Voelkel N.F., Gomez-Arroyo J. The role of vascular endothelial growth factor in pulmonary arterial hypertension: the angiogenesis paradox. Am J Respir Cell Mol Biol. 2014;51:474–484. doi: 10.1165/rcmb.2014-0045TR. [DOI] [PubMed] [Google Scholar]
- 6.Eilken H.M., Adams R.H. Dynamics of endothelial cell behavior in sprouting angiogenesis. Curr Opin Cell Biol. 2010;22:617–625. doi: 10.1016/j.ceb.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 7.Birbrair A., Zhang T., Wang Z.M., Messi M.L., Mintz A., Delbono O. Pericytes: multitasking cells in the regeneration of injured, diseased, and aged skeletal muscle. Front Aging Neurosci. 2014;6:245. doi: 10.3389/fnagi.2014.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Dijk C.G., Nieuweboer F.E., Pei J.Y., Xu Y.J., Burgisser P., van Mulligen E., el Azzouzi H., Duncker D.J., Verhaar M.C., Cheng C. The complex mural cell: pericyte function in health and disease. Int J Cardiol. 2015;190:75–89. doi: 10.1016/j.ijcard.2015.03.258. [DOI] [PubMed] [Google Scholar]
- 9.de Jesus Perez V.A., Alastalo T.P., Wu J.C., Axelrod J.D., Cooke J.P., Amieva M., Rabinovitch M. Bone morphogenetic protein 2 induces pulmonary angiogenesis via Wnt-beta-catenin and Wnt-RhoA-Rac1 pathways. J Cell Biol. 2009;184:83–99. doi: 10.1083/jcb.200806049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jesus Perez V.A., Yuan K., Orcholski M.E., Sawada H., Zhao M., Li C.G., Tojais N.F., Nickel N., Rajagopalan V., Spiekerkoetter E., Wang L., Dutta R., Bernstein D., Rabinovitch M. Loss of adenomatous poliposis coli-alpha3 integrin interaction promotes endothelial apoptosis in mice and humans. Circ Res. 2012;111:1551–1564. doi: 10.1161/CIRCRESAHA.112.267849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fessel J.P., Chen X., Frump A., Gladson S., Blackwell T., Kang C., Johnson J., Loyd J.E., Hemnes A., Austin E., West J. Interaction between bone morphogenetic protein receptor type 2 and estrogenic compounds in pulmonary arterial hypertension. Pulm Circ. 2013;3:564–577. doi: 10.1086/674312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nickel N.P., Spiekerkoetter E., Gu M., Li C.G., Li H., Kaschwich M., Diebold I., Hennigs J.K., Kim K.Y., Miyagawa K., Wang L., Cao A., Sa S., Jiang X., Stockstill R.W., Nicolls M.R., Zamanian R.T., Bland R.D., Rabinovitch M. Elafin reverses pulmonary hypertension via caveolin-1-dependent bone morphogenetic protein signaling. Am J Respir Crit Care Med. 2015;191:1273–1286. doi: 10.1164/rccm.201412-2291OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu W., Erzurum S.C. Endothelial cell energy metabolism, proliferation, and apoptosis in pulmonary hypertension. Compr Physiol. 2011;1:357–372. doi: 10.1002/cphy.c090005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalkara T., Gursoy-Ozdemir Y., Yemisci M. Brain microvascular pericytes in health and disease. Acta Neuropathol. 2011;122:1–9. doi: 10.1007/s00401-011-0847-6. [DOI] [PubMed] [Google Scholar]
- 15.Kaur C., Foulds W.S., Ling E.A. Blood-retinal barrier in hypoxic ischaemic conditions: basic concepts, clinical features and management. Prog Retin Eye Res. 2008;27:622–647. doi: 10.1016/j.preteyeres.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Motiejunaite R., Kazlauskas A. Pericytes and ocular diseases. Exp Eye Res. 2008;86:171–177. doi: 10.1016/j.exer.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell T.S., Bradley J., Robinson G.S., Shima D.T., Ng Y.S. RGS5 expression is a quantitative measure of pericyte coverage of blood vessels. Angiogenesis. 2008;11:141–151. doi: 10.1007/s10456-007-9085-x. [DOI] [PubMed] [Google Scholar]
- 18.Yuan K., Orcholski M.E., Panaroni C., Shuffle E.M., Huang N.F., Jiang X., Tian W., Vladar E.K., Wang L., Nicolls M.R., Wu J.Y., de Jesus Perez V.A. Activation of the Wnt/planar cell polarity pathway is required for pericyte recruitment during pulmonary angiogenesis. Am J Pathol. 2015;185:69–84. doi: 10.1016/j.ajpath.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fillmore N., Lopaschuk G.D. Targeting mitochondrial oxidative metabolism as an approach to treat heart failure. Biochim Biophys Acta. 2013;1833:857–865. doi: 10.1016/j.bbamcr.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 20.Hur H., Xuan Y., Kim Y.B., Lee G., Shim W., Yun J., Ham I.H., Han S.U. Expression of pyruvate dehydrogenase kinase-1 in gastric cancer as a potential therapeutic target. Int J Oncol. 2013;42:44–54. doi: 10.3892/ijo.2012.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sutendra G., Michelakis E.D. Pyruvate dehydrogenase kinase as a novel therapeutic target in oncology. Front Oncol. 2013;3:38. doi: 10.3389/fonc.2013.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S.L., Hu X., Zhang W., Yao H., Tam K.Y. Development of pyruvate dehydrogenase kinase inhibitors in medicinal chemistry with particular emphasis as anticancer agents. Drug Discov Today. 2015;20:1112–1119. doi: 10.1016/j.drudis.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 23.McMurtry M.S., Bonnet S., Wu X., Dyck J.R., Haromy A., Hashimoto K., Michelakis E.D. Dichloroacetate prevents and reverses pulmonary hypertension by inducing pulmonary artery smooth muscle cell apoptosis. Circ Res. 2004;95:830–840. doi: 10.1161/01.RES.0000145360.16770.9f. [DOI] [PubMed] [Google Scholar]
- 24.Piao L., Sidhu V.K., Fang Y.H., Ryan J.J., Parikh K.S., Hong Z., Toth P.T., Morrow E., Kutty S., Lopaschuk G.D., Archer S.L. FOXO1-mediated upregulation of pyruvate dehydrogenase kinase-4 (PDK4) decreases glucose oxidation and impairs right ventricular function in pulmonary hypertension: therapeutic benefits of dichloroacetate. J Mol Med. 2013;91:333–346. doi: 10.1007/s00109-012-0982-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng G., Shaw P., Rosen S.T., Lin S.M., Kibbe W.A. Using the bioconductor GeneAnswers package to interpret gene lists. Methods Mol Biol. 2012;802:101–112. doi: 10.1007/978-1-61779-400-1_7. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro H.M., Natale P.J., Kamentsky L.A. Estimation of membrane potentials of individual lymphocytes by flow cytometry. Proc Natl Acad Sci U S A. 1979;76:5728–5730. doi: 10.1073/pnas.76.11.5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diebold I., Hennigs J.K., Miyagawa K., Li C.G., Nickel N.P., Kaschwich M., Cao A., Wang L., Reddy S., Chen P.I., Nakahira K., Alcazar M.A., Hopper R.K., Ji L., Feldman B.J., Rabinovitch M. BMPR2 preserves mitochondrial function and DNA during reoxygenation to promote endothelial cell survival and reverse pulmonary hypertension. Cell Metab. 2015;21:596–608. doi: 10.1016/j.cmet.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho N., Coomber B.L. Pyruvate dehydrogenase kinase expression and metabolic changes following dichloroacetate exposure in anoxic human colorectal cancer cells. Exp Cell Res. 2015;331:73–81. doi: 10.1016/j.yexcr.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Sutendra G., Dromparis P., Kinnaird A., Stenson T.H., Haromy A., Parker J.M., McMurtry M.S., Michelakis E.D. Mitochondrial activation by inhibition of PDKII suppresses HIF1a signaling and angiogenesis in cancer. Oncogene. 2013;32:1638–1650. doi: 10.1038/onc.2012.198. [DOI] [PubMed] [Google Scholar]
- 31.Dando I., Dalla Pozza E., Biondani G., Cordani M., Palmieri M., Donadelli M. The metabolic landscape of cancer stem cells. IUBMB Life. 2015;67:687–693. doi: 10.1002/iub.1426. [DOI] [PubMed] [Google Scholar]
- 32.Vaitheesvaran B., Xu J., Yee J., Q-Y L., Go V.L., Xiao G.G., Lee W.N. The Warburg effect: a balance of flux analysis. Metabolomics. 2015;11:787–796. doi: 10.1007/s11306-014-0760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu X.D., Shao S.X., Jiang H.P., Cao Y.W., Wang Y.H., Yang X.C., Wang Y.L., Wang X.S., Niu H.T. Warburg effect or reverse Warburg effect? a review of cancer metabolism. Oncol Res Treat. 2015;38:117–122. doi: 10.1159/000375435. [DOI] [PubMed] [Google Scholar]
- 34.Guignabert C., Tu L., Izikki M., Dewachter L., Zadigue P., Humbert M., Adnot S., Fadel E., Eddahibi S. Dichloroacetate treatment partially regresses established pulmonary hypertension in mice with SM22alpha-targeted overexpression of the serotonin transporter. FASEB J. 2009;23:4135–4147. doi: 10.1096/fj.09-131664. [DOI] [PubMed] [Google Scholar]
- 35.Li B., Yan J., Shen Y., Liu Y., Ma Z. Dichloroacetate prevents but not reverses the formation of neointimal lesions in a rat model of severe pulmonary arterial hypertension. Mol Med Rep. 2014;10:2144–2152. doi: 10.3892/mmr.2014.2432. [DOI] [PubMed] [Google Scholar]
- 36.Palsson-McDermott E.M., O'Neill L.A. The Warburg effect then and now: from cancer to inflammatory diseases. Bioessays. 2013;35:965–973. doi: 10.1002/bies.201300084. [DOI] [PubMed] [Google Scholar]
- 37.Cereijo-Santalo R. Aerobic glycolysis and mitochondrial swelling. Biochem Biophys Res Commun. 1966;24:650–655. doi: 10.1016/0006-291x(66)90373-1. [DOI] [PubMed] [Google Scholar]
- 38.Cohen P.J. Oxygen and intracellular metabolism. Int Anesthesiol Clin. 1981;19:9–19. doi: 10.1097/00004311-198119030-00004. [DOI] [PubMed] [Google Scholar]
- 39.Marsboom G., Toth P.T., Ryan J.J., Hong Z., Wu X., Fang Y.H., Thenappan T., Piao L., Zhang H.J., Pogoriler J., Chen Y., Morrow E., Weir E.K., Rehman J., Archer S.L. Dynamin-related protein 1-mediated mitochondrial mitotic fission permits hyperproliferation of vascular smooth muscle cells and offers a novel therapeutic target in pulmonary hypertension. Circ Res. 2012;110:1484–1497. doi: 10.1161/CIRCRESAHA.111.263848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan J., Dasgupta A., Huston J., Chen K.H., Archer S.L. Mitochondrial dynamics in pulmonary arterial hypertension. J Mol Med. 2015;93:229–242. doi: 10.1007/s00109-015-1263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rabinovitch M. Molecular pathogenesis of pulmonary arterial hypertension. J Clin Invest. 2012;122:4306–4313. doi: 10.1172/JCI60658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams R.H., Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol. 2007;8:464–478. doi: 10.1038/nrm2183. [DOI] [PubMed] [Google Scholar]
- 43.Ricard N., Tu L., Le Hiress M., Huertas A., Phan C., Thuillet R., Sattler C., Fadel E., Seferian A., Montani D., Dorfmuller P., Humbert M., Guignabert C. Increased pericyte coverage mediated by endothelial-derived fibroblast growth factor-2 and interleukin-6 is a source of smooth muscle-like cells in pulmonary hypertension. Circulation. 2014;129:1586–1597. doi: 10.1161/CIRCULATIONAHA.113.007469. [DOI] [PubMed] [Google Scholar]
- 44.Assad T.R., Hemnes A.R. Metabolic dysfunction in pulmonary arterial hypertension. Curr Hypertens Rep. 2015;17:20. doi: 10.1007/s11906-014-0524-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansmann G., Wagner R.A., Schellong S., Perez V.A., Urashima T., Wang L., Sheikh A.Y., Suen R.S., Stewart D.J., Rabinovitch M. Pulmonary arterial hypertension is linked to insulin resistance and reversed by peroxisome proliferator-activated receptor-gamma activation. Circulation. 2007;115:1275–1284. doi: 10.1161/CIRCULATIONAHA.106.663120. [DOI] [PubMed] [Google Scholar]
- 46.Moral-Sanz J., Moreno L., Cogolludo A., Perez-Vizcaino F. Pulmonary vascular function in insulin resistance and diabetes. Curr Vasc Pharmacol. 2014;12:473–482. doi: 10.2174/1570161112666140423222808. [DOI] [PubMed] [Google Scholar]
- 47.Rabinovitch M. PPARgamma and the pathobiology of pulmonary arterial hypertension. Adv Exp Med Biol. 2010;661:447–458. doi: 10.1007/978-1-60761-500-2_29. [DOI] [PubMed] [Google Scholar]
- 48.Hemnes A.R., Brittain E.L., Trammell A.W., Fessel J.P., Austin E.D., Penner N., Maynard K.B., Gleaves L., Talati M., Absi T., Disalvo T., West J. Evidence for right ventricular lipotoxicity in heritable pulmonary arterial hypertension. Am J Respir Crit Care Med. 2014;189:325–334. doi: 10.1164/rccm.201306-1086OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansmann G., de Jesus Perez V.A., Alastalo T.P., Alvira C.M., Guignabert C., Bekker J.M., Schellong S., Urashima T., Wang L., Morrell N.W., Rabinovitch M. An antiproliferative BMP-2/PPARgamma/apoE axis in human and murine SMCs and its role in pulmonary hypertension. J Clin Invest. 2008;118:1846–1857. doi: 10.1172/JCI32503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crosswhite P., Sun Z. Molecular mechanisms of pulmonary arterial remodeling. Mol Med. 2014;20:191–201. doi: 10.2119/molmed.2013.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freund-Michel V., Khoyrattee N., Savineau J.P., Muller B., Guibert C. Mitochondria: roles in pulmonary hypertension. Int J Biochem Cell Biol. 2014;55:93–97. doi: 10.1016/j.biocel.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 52.Oikawa M., Kagaya Y., Otani H., Sakuma M., Demachi J., Suzuki J., Takahashi T., Nawata J., Ido T., Watanabe J., Shirato K. Increased [18F]fluorodeoxyglucose accumulation in right ventricular free wall in patients with pulmonary hypertension and the effect of epoprostenol. J Am Coll Cardiol. 2005;45:1849–1855. doi: 10.1016/j.jacc.2005.02.065. [DOI] [PubMed] [Google Scholar]
- 53.Lundgrin E.L., Park M.M., Sharp J., Tang W.H., Thomas J.D., Asosingh K., Comhair S.A., DiFilippo F.P., Neumann D.R., Davis L., Graham B.B., Tuder R.M., Dostanic I., Erzurum S.C. Fasting 2-deoxy-2-[18F]fluoro-D-glucose positron emission tomography to detect metabolic changes in pulmonary arterial hypertension hearts over 1 year. Ann Am Thorac Soc. 2013;10:1–9. doi: 10.1513/AnnalsATS.201206-029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bowker-Kinley M.M., Davis W.I., Wu P., Harris R.A., Popov K.M. Evidence for existence of tissue-specific regulation of the mammalian pyruvate dehydrogenase complex. Biochem J. 1998;329(Pt 1):191–196. doi: 10.1042/bj3290191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim J.W., Tchernyshyov I., Semenza G.L., Dang C.V. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 56.Holness M.J., Bulmer K., Gibbons G.F., Sugden M.C. Up-regulation of pyruvate dehydrogenase kinase isoform 4 (PDK4) protein expression in oxidative skeletal muscle does not require the obligatory participation of peroxisome-proliferator-activated receptor alpha (PPARalpha) Biochem J. 2002;366:839–846. doi: 10.1042/BJ20020754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McAinch A.J., Cornall L.M., Watts R., Hryciw D.H., O'Brien P.E., Cameron-Smith D. Increased pyruvate dehydrogenase kinase expression in cultured myotubes from obese and diabetic individuals. Eur J Nutr. 2015;54:1033–1043. doi: 10.1007/s00394-014-0780-2. [DOI] [PubMed] [Google Scholar]
- 58.Lee S.J., Jeong J.Y., Oh C.J., Park S., Kim J.Y., Kim H.J., Doo Kim N., Choi Y.K., Do J.Y., Go Y., Ha C.M., Choi J.Y., Huh S., Ho Jeoung N., Lee K.U., Choi H.S., Wang Y., Park K.G., Harris R.A., Lee I.K. Pyruvate dehydrogenase kinase 4 promotes vascular calcification via SMAD1/5/8 phosphorylation. Sci Rep. 2015;5:16577. doi: 10.1038/srep16577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Archer S.L., Fang Y.H., Ryan J.J., Piao L. Metabolism and bioenergetics in the right ventricle and pulmonary vasculature in pulmonary hypertension. Pulm Circ. 2013;3:144–152. doi: 10.4103/2045-8932.109960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Michelakis E.D., McMurtry M.S., Wu X.C., Dyck J.R., Moudgil R., Hopkins T.A., Lopaschuk G.D., Puttagunta L., Waite R., Archer S.L. Dichloroacetate, a metabolic modulator, prevents and reverses chronic hypoxic pulmonary hypertension in rats: role of increased expression and activity of voltage-gated potassium channels. Circulation. 2002;105:244–250. doi: 10.1161/hc0202.101974. [DOI] [PubMed] [Google Scholar]
- 61.Kato M., Li J., Chuang J.L., Chuang D.T. Distinct structural mechanisms for inhibition of pyruvate dehydrogenase kinase isoforms by AZD7545, dichloroacetate, and radicicol. Structure. 2007;15:992–1004. doi: 10.1016/j.str.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dore-Duffy P. Pericytes: pluripotent cells of the blood brain barrier. Curr Pharm Des. 2008;14:1581–1593. doi: 10.2174/138161208784705469. [DOI] [PubMed] [Google Scholar]
- 63.Wittig D., Jaszai J., Corbeil D., Funk R.H. Immunohistochemical localization and characterization of putative mesenchymal stem cell markers in the retinal capillary network of rodents. Cells Tissues Organs. 2013;197:344–359. doi: 10.1159/000346661. [DOI] [PubMed] [Google Scholar]
- 64.Dar A., Domev H., Ben-Yosef O., Tzukerman M., Zeevi-Levin N., Novak A., Germanguz I., Amit M., Itskovitz-Eldor J. Multipotent vasculogenic pericytes from human pluripotent stem cells promote recovery of murine ischemic limb. Circulation. 2012;125:87–99. doi: 10.1161/CIRCULATIONAHA.111.048264. [DOI] [PubMed] [Google Scholar]
- 65.Teichert-Kuliszewska K., Kutryk M.J., Kuliszewski M.A., Karoubi G., Courtman D.W., Zucco L., Granton J., Stewart D.J. Bone morphogenetic protein receptor-2 signaling promotes pulmonary arterial endothelial cell survival: implications for loss-of-function mutations in the pathogenesis of pulmonary hypertension. Circ Res. 2006;98:209–217. doi: 10.1161/01.RES.0000200180.01710.e6. [DOI] [PubMed] [Google Scholar]
- 66.Meyrick B., Reid L. Ultrastructural findings in lung biopsy material from children with congenital heart defects. Am J Pathol. 1980;101:527–542. [PMC free article] [PubMed] [Google Scholar]
- 67.Wesolowski S., Birtwistle M.R., Rempala G.A. A comparison of methods for RNA-Seq differential expression analysis and a new empirical bayes approach. Biosensors. 2013;3:238–258. doi: 10.3390/bios3030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Jesus Perez V.A., Yuan K., Lyuksyutova M.A., Dewey F., Orcholski M.E., Shuffle E.M., Mathur M., Yancy L., Jr., Rojas V., Li C.G., Cao A., Alastalo T.P., Khazeni N., Cimprich K.A., Butte A.J., Ashley E., Zamanian R.T. Whole-exome sequencing reveals TopBP1 as a novel gene in idiopathic pulmonary arterial hypertension. Am J Respir Crit Care Med. 2014;189:1260–1272. doi: 10.1164/rccm.201310-1749OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional comparison of gene expression patterns of healthy donor and pulmonary arterial hypertension (PAH) pericytes by microarray analysis. A: Multidimensional scaling (MDS) plot of gene expression levels based on microarrays of all samples. B: The line plot is based on log2 (log2 gene expression) to make the genes comparable to others, using the normalized untransformed gene expression levels. The normalized gene expression level = 2 superscript (the log 2 expression levels). Log2 expression levels for controls 1, 2, and 3 (Ctrl1, Ctrl2, and Ctrl3, respectively) and patients with PAH 1, 2, and 3 (PAH1, PAH2, and PAH3, respectively) are 10.19, 6.85, and 7.95 and 11.97, 9.85, and 12.06, respectively. Expression of PDK4 is the top highest among all up-regulated genes.
Knockdown of PDK4 in donor pericytes does not change mitochondrial activity. A: Reactive oxygen species (ROS) production in donor control with PDK4 siRNA treatment. Fluorescence intensity was normalized by MitoTracker (MitoT). Aerobic glycolysis and glycolytic capacity (B) in donor control with PDK4 siRNA treatment as measured by Seahorse assay. Cell growth by MTS assay (C) and cell apoptosis by caspase 3/7 assay (D) were evaluated on days 1, 2, and 3 of culture in donor control with PDK4 siRNA treatment. Scale bar = 25 μm. ECAR, extracellular acidification rate; Pc, pericyte; siCtrl, siRNA control.
Gene Ontology concept–gene network analysis for all significantly up-regulated genes in pulmonary arterial hypertension pericytes.
Gene Ontology concept–gene network analysis for all significantly down-regulated genes in pulmonary arterial hypertension pericytes.