Abstract
The conserved RNA-binding protein Musashi1 (MSI1) has been characterized as a stem cell marker, controlling the balance between self-renewal and differentiation and as a key oncogenic factor in numerous solid tumors, including glioblastoma. To explore the potential use of MSI1 targeting in therapy, we studied MSI1 in the context of radiation sensitivity. Knockdown of MSI1 led to a decrease in cell survival and an increase in DNA damage compared to control in cells treated with ionizing radiation. We subsequently examined mechanisms of double-strand break repair and found that loss of MSI1 reduces the frequency of nonhomologous end-joining. This phenomenon could be attributed to the decreased expression of DNA–protein kinase catalytic subunit, which we have previously identified as a target of MSI1. Collectively, our results suggest a role for MSI1 in double-strand break repair and that its inhibition may enhance the effect of radiotherapy.
Glioblastoma multiforme is the most aggressive form of glioma and is the most common among primary brain tumors. The current standard of care for newly diagnosed glioblastoma is surgical resection, followed by concurrent temozolomide and radiotherapy, then by maintenance chemotherapy with temozolomide. Unfortunately, this route offers a median survival of only approximately 14 months.1, 2 Recently, the addition of tumor-treating fields to maintenance temozolomide has extended overall survival to 20.5 months.3 Local recurrence is a problem that has been observed for many years, suggesting that tumors may harbor a subset of radio-resistant cells that can repopulate a tumor, even after aggressive treatment. Different molecular factors are thought to play a role in radiation resistance, such as altered expression of molecules involved in growth factor receptor signaling pathways, DNA damage and repair, and angiogenesis; consequentially, multipathway-targeted therapies have been proposed as a more effective way to treat cancer.4 In the past decade, much progress has been made in understanding the cellular and molecular heterogeneity in glioblastoma, and its links to clinical aggressiveness, differential response to chemotherapy and radiation treatments, and different patient outcomes. Despite the advances, it is clear that many alterations implicated in glioblastoma initiation and development remain unknown. In particular, the role of aberrant post-transcriptional regulation in gliomagenesis, mediated primarily by RNA-binding proteins, is underexplored and deserving of extra attention.
The stem cell–related RNA-binding protein Musashi1 (MSI1) is emerging as an important oncogenic factor in numerous tumor types, including glioblastoma multiforme, in which it is frequently up-regulated.5 MSI1 is evolutionarily conserved, displays important functions during nervous system development and embryogenesis, and serves as an important regulatory molecule in neural stem cells by balancing self-renewal and differentiation.6 Widespread gene regulatory activities both as a repressor and activator of translation7, 8, 9, 10, 11, 12 suggest that MSI1 promotes and potentiates tumorigenesis in multiple ways. In glioblastoma multiforme, MSI1 influences numerous cancer-relevant processes. Our functional genomic analysis showed that MSI1 controls hundreds of targets that are preferentially located in pathways such as focal adhesion, adherens junction, Wnt signalling pathway, Janus kinase/signal transducers and activators of transcription, p53, mitogen-activated protein kinase, and ErbB and suggests that MSI1 could be an interesting therapeutic target.13 However, it remains to be determined how high levels of MSI1 impact glioblastoma therapy, as its overexpression correlates with poor prognosis.14 Here, we elucidate the role that MSI1 has on DNA damage repair and radio-resistance. We demonstrate that MSI1 mediates glioblastoma cell survival after radiation through increased DNA damage repair by end-joining (EJ), presumably via its stabilization of DNA-dependent protein kinase catalytic subunit (PKcs).
Materials and Methods
Cell Lines, Transfection, and Reagents
U251 and U343 glioblastoma cells were obtained from ATCC (Rockville, MD), and U2OS cells with stably integrated EJ5–Green fluorescent protein (GFP)15 were obtained from Dr. Jeremy Stark (City of Hope, Duarte, CA). Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% Pen/Strep (Life Technologies, Carlsbad, CA). glioblastoma transcriptome data performed in the study by Uren et al13 were used for authenticating the U251 and U343 cell lines.
Cells were transiently transfected with control siRNA or MSI1 siRNA using Lipofectamine RNAiMAX (MSI1HSS106732, MSI1HSS106733, MSI1HSS106734; Life Technologies). MSI1 or DNA-PKcs transgenic expression was achieved after transfecting U251 cells with pcDNA3.1-MSI1 or pCMV-F2_k_DNA-PKcs (obtained from Dr. David Chen, University of Texas Southwestern, Dallas, TX) vectors and GeneJammer transfection reagent (Agilent Technologies, Santa Clara, CA). For sustained suppression of MSI1 expression, short hairpin RNA (shRNAmir) GIPZ lentiviral vector carrying shRNA targeting MSI1 (shMSI1) and a nonspecific shRNA control (shCtrl) (Thermo Fisher Scientific, Rochester, NY) were used for transducing glioblastoma cells, according to the manufacturer's protocol.
Clustered Regularly Interspaced Short Palindromic Repeat
Two MSI1 target sequences (5′-CACCGTGGGGCGCGTCAGTCTCCAT-3′/5′-CACCGCGAATACTTCGGCCAGTTCG-3′) were cloned into lenti–clustered regularly interspaced short palindromic repeat v2 plasmid (catalog number 52961; Addgene, Cambridge, MA) and used for co-transfecting U251 cells. After selection using puromycin, cells were submitted to cell cloning by serial dilution in 96-well plates. Single colonies were transferred to a 12-well plate and allowed to grow. Three different clones were analyzed in this work.
Clonogenic Assay
Cells were plated after appropriate dilution and ionizing radiation (IR) treatment was performed on the next day at a dose ranging from 0.15 to 5 Gy. A cabinet X-ray system (CP-160 Cabinet X-Radiator; Faxitron X-Ray Corp., Tucson, AZ) was used for all treatments. After IR, cells were cultured for 10 to 14 days. Then, cells were stained with crystal violet and all colonies of 50 or more cells were manually counted. The survival fraction, expressed as a function of IR, was estimated according to the following formula: Survival fraction = Colonies formed/(Cells seeded × Plating efficiency of the control group/100).
Alternatively, crystal violet was dissolved from stained plates, and optic density was measured with a microplate reader at 570 nm. All experiments were performed in triplicate.
Alkaline Comet Assay
Cells were plated, subjected to IR, and collected at different time points. The alkaline comet assay was performed according to Singh et al,16 with some modifications. Briefly, after cell lysis, the slides were washed three times (5 minutes each) with electrophoresis buffer (300 mmol/L NaOH/1 mmol/L EDTA, pH ≥13.0) and placed in horizontal electrophoresis tank filled with electrophoresis buffer to allow for DNA unwinding for 20 minutes. Electrophoresis was performed for 20 minutes at 25 V (300 mA). Subsequently, the slides were neutralized three times (5 minutes each) with 400 mmol/L Tris–HCl (pH 7.5), fixed with 100% ethanol, and dried at room temperature. All steps were conducted under dim light to prevent the occurrence of additional DNA damage. Each slide was stained with 20 μg/mL ethidium bromide and covered with a coverslip before analysis. Images were captured on an Eclipse TE2000-U microscope (Nikon Instruments, Melville, NY) at ×40 magnification. Data are presented as the mean values of tail moment ± SD. Tail moment values of 50 randomly selected cells per slide from duplicate slides were scored using the Scion Image software package version 4.0.3.2 (NIH, Bethesda, MD) and used as the index of DNA damage.
Quantitative Reverse Transcription and PCR
Total RNA was extracted using the TRIzol reagent (Life Technologies), following the manufacturer's instructions. Reverse transcription was performed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) with random priming. After reverse transcription, quantitative PCR was performed using the Gene Expression Assay (Applied Biosystems) in TaqMan Gene Expression Master Mix (Applied Biosystems) for mRNA analysis. Real-time PCRs were performed on a 7500 Real-Time PCR System (Applied Biosystems). Data were acquired using the SDS software package version 2.0.1 (Applied Biosystems), and analyzed using the 2-ΔΔCt method using β-actin as an endogenous control.
Immunoblot Analysis
Cells were lysed in lysis buffer and subjected to Western blot analysis with the following primary antibodies: anti-MSI1 (1:1000; EMD Millipore, Billerica, MA), anti–replication protein A 32 (1:1000; Bethyl Laboratories, Montgomery, TX), anti–phospho-replication protein A 32(S4/S8) (1:1000; Bethyl), anti–CCCTC-binding factor (1:4000; Abcam, Cambridge, UK), anti–DNA-PKcs (1 μg/mL; Abcam), and anti–α-tubulin (1:5000; Sigma-Aldrich St. Louis, MO). Densitometry was performed using ImageJ analysis software version 1.47v (NIH; http://imagej.nih.gov/ij). Samples were normalized to α-tubulin content and results are expressed as the means ± SD values of integrated optical density.
Immunofluorescence
U251 cells were grown on fibronectin-coated coverslips. After MSI1 knockdown, cells were treated with 5 Gy IR. Thirty minutes after IR, cells were fixed and permeabilized. Cells were then blocked with 1% bovine serum albumin/4% goat serum followed by an overnight incubation with a 1:3000 dilution of 53BP1 primary antibody (Bethyl). Anti-rabbit IgG, Alexa Fluor 568–conjugated was used as secondary antibody (InvitroGen, Carlsbad, CA). Cells were then stained with DAPI and coverslips were mounted on slides using Vectashield (Vector Laboratories, Burlingame, CA). Images were captured on an Axiovert 200 M microscope (Carl Zeiss, Oberkochen, Germany) at ×40 magnification. A minimum of 100 nuclei were counted for each condition performed in triplicate.
End-Joining Repair Assay
Briefly, EJ5-GFP U2OS cells were seeded in a 24-well plate and transfected with relevant transfections (siCtrl, siMSI1). Twenty-four hours later, cells were transfected with either ISceI expression vector alone or in combination with DNA-PKcs. After 72 hours, cells were harvested and GFP-positive cells were evaluated by flow cytometry on a BD flow cytometer (Becton, Dickinson and Company, Franklin Lakes, NJ). Appropriate controls were used, and all experiments were performed in triplicate. Data were analyzed with the t-test and are presented as means ± SD.
Results
MSI1 Levels Increase on Ionizing Radiation
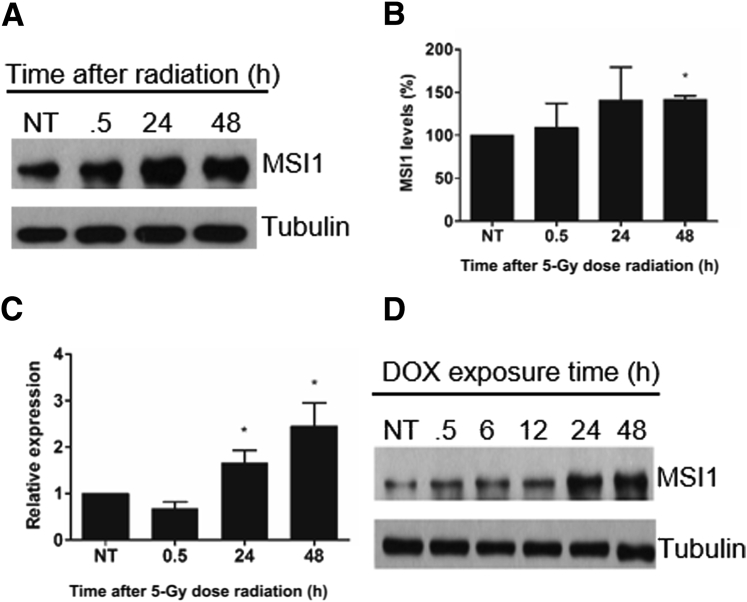
Several studies have described elevated MSI1 expression in different cancer/tumor types and subsequent association with a poor prognosis.5 However, whether a targeted reduction in MSI1 expression contributes to treatment outcome remains unknown. We looked into the connection between high MSI1 expression and radio-resistance. Initially, to determine the impact of IR on MSI1 expression, U251 glioblastoma cells were treated with a single dose of IR (5 Gy). After treatment, the cells showed an increase in MSI1 expression at both the protein and RNA levels (Figure 1, A–C). To confirm the stress-induced expression of MSI1, cells were also treated with doxorubicin, an intercalating chemotherapeutic agent that blocks topoisomerase II activity, thereby causing double-strand breaks in the DNA. Protein levels of MSI1 were evaluated and, similar to the radiation results, we observed an increase in MSI1 expression caused by doxorubicin in an exposure time–dependent manner (Figure 1D). Thus, our data indicate that MSI1 expression is induced in response to DNA damage.
Figure 1.
Increased expression of MSI1 after ionizing radiation and doxorubicin (DOX) treatment in U251 glioblastoma cells. A: Expression of MSI1 at different times after radiation was verified by immunoblot analysis. B: Protein bands of three independent Western blots were quantified by densitometry using ImageJ. MSI1 levels were normalized by α-tubulin levels. C: Quantification of MSI1 mRNAs levels after IR. D: Analysis of MSI1 protein expression levels by Western blot after different time points of 50 nmol/L doxorubicin treatment. Data are presented as means ± SD. n = 3 (B and C). ∗P < 0.05 (t-test). h, hours; NT, no treatment.
MSI1 Impacts Response to Genotoxic Stress
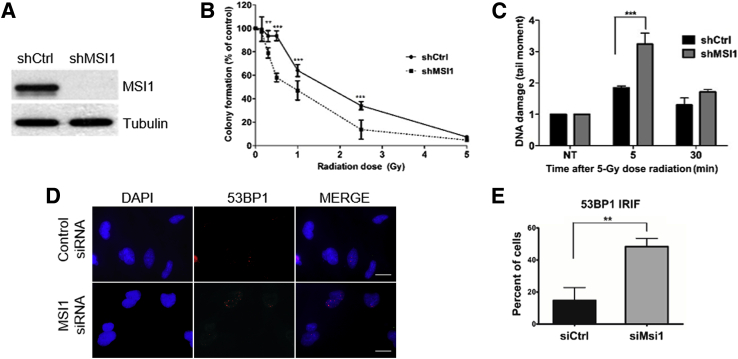
Next, we assessed MSI1 impact on cellular response to radiotherapy. To determine the survival rate after IR, U251 shMSI1 and shCtrl cells were treated with different doses of IR and evaluated in a clonogenic assay. MSI1 knockdown cells (Figure 2A) showed increased sensitivity to IR (Figure 2B). Similar results were obtained with U343 cells (Supplemental Figure S1). We also directly measured the presence of broken DNA using the alkaline comet assay and observed that MSI1 knockdown led to a significant increase in DNA damage levels at early time points after radiation (Figure 2C). Finally, we evaluated IR-induced DNA damage levels in MSI1-silenced and control cells by measuring levels of 53BP1 foci, an established marker of double-strand breaks. Based on spontaneous levels of 53BP1 foci in U251 cells, a frequency of <15 foci/cell was considered as background. MSI1 knockdown resulted in a significant increase in cells with >15 foci, indicating higher levels of DNA damage (Figure 2, D and E).
Figure 2.
Decreased cell survival and increased DNA damage levels after knockdown (KD) of MSI1. A: Analysis of MSI1 expression in U251 cells transfected with control (shCtrl) or shMSI1 by Western blot. B: Clonogenic assay of U251 MSI1-KD after exposure to X-rays. C: DNA damage and repair assessed by the alkaline comet assay at different times after radiation. D: Representative field with U251 cells with 53BP1 foci after IR treatment. E: Quantification of 53BP1 ionizing radiation–induced foci (IRIF) levels in U251 cells after MSI1 knockdown. Data are presented as means ± SD. Data was collected from 3 sets of 100 cells each per condition. ∗∗P < 0.01, ∗∗∗P < 0.001 (t-test). Scale bars = 25 μm. NT, no treatment; sh, small hairpin; si, small interfering.
MSI1 Targets DNA-PKcs
Cross-linking immunoprecipitation analysis of U251 cells, previously performed by our group,13, 17, 18, 19 identified PRKDC (protein kinase, DNA-activated, catalytic polypeptide) or DNA-PKcs as a potential target of MSI1, with binding sites present in the 3′-untranslated region (Supplemental Figure S2A). Protein kinase, DNA-activated, catalytic polypeptide encodes DNA-PKcs, which is the key enzyme involved in the classic nonhomologous (NH) EJ pathway of DNA double-strand break repair in mammalian cells. DNA-PKcs promotes NHEJ while suppressing homologous recombination20 and facilitates repair of genotoxic and replication stress associated damage via phosphorylation of the single-stranded DNA-binding protein replication protein A 2.21
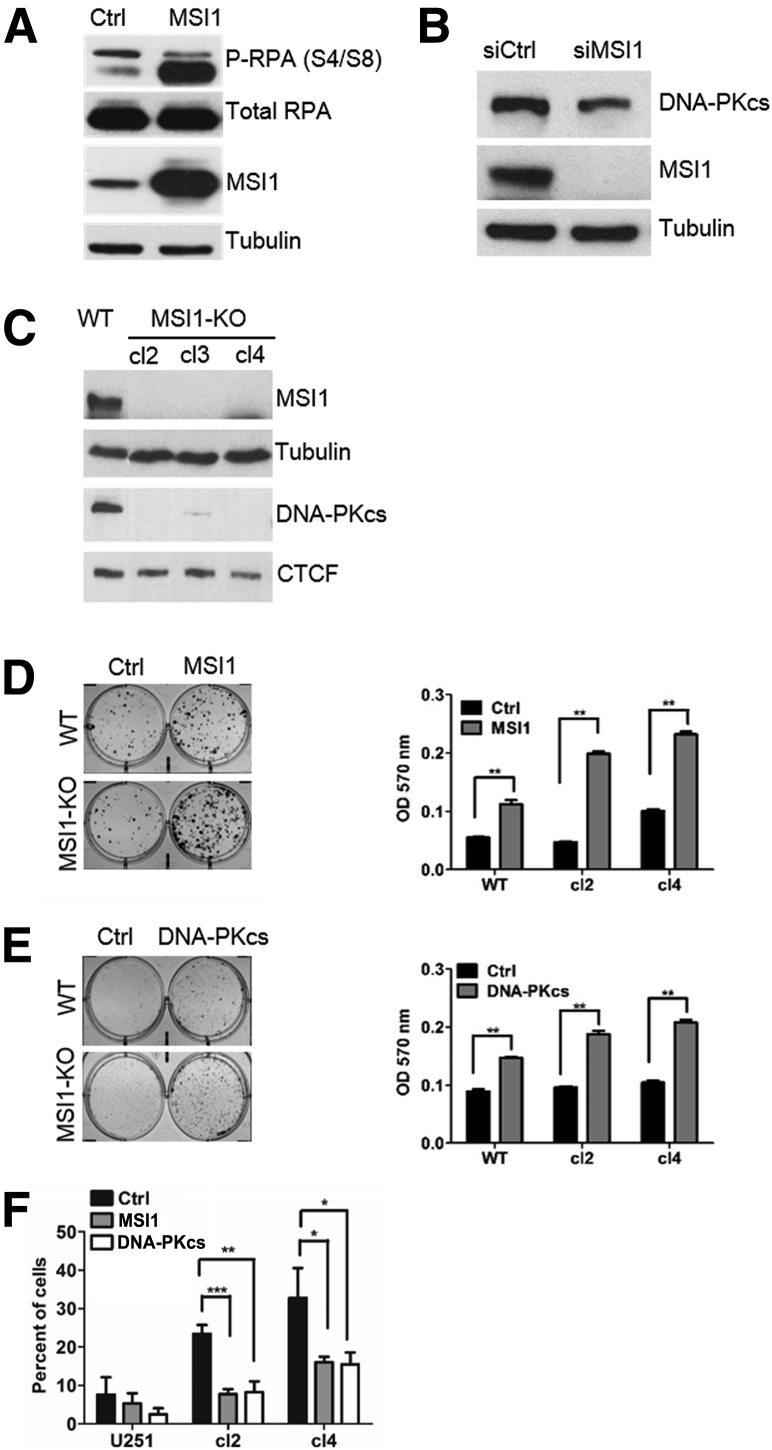
Similar to MSI1, DNA-PKcs gene expression is increased on radiation (Supplemental Figure S2B). Moreover, we observed a replication protein A 2 hyperphosphorylation pattern after MSI1 overexpression in U251 cells (Figure 3A) concordant with the induction of DNA damage response in a DNA-PK–dependent manner.22 Most importantly, MSI1 knockdown via siRNA decreased the expression of DNA-PKcs in U251 glioblastoma cells (Figure 3B). Due to the high molecular weight of DNA-PKcs protein (approximately 460 kDa), we used CCCTC-binding factor protein as loading control (83 kDa).
Figure 3.
Regulation of protein kinase, DNA-activated, catalytic polypeptide (PRKDC). A: Immunoblot analysis showing replication protein A 2 hyperphosphorylation (P-RPA) after MSI1 overexpression. B: Immunoblot analysis showing decreased levels of DNA-PKcs after MSI1 knockdown. C: MSI1 knockout (KO) cells show reduced DNA-PKcs expression. D: Colony formation after MSI1 rescuing. Right panel: Representative clonogenic assay of MSI1 knockout–rescued cells after exposure to 2.5 Gy. Left panel: Stain absorbed by the cells was dissolved in 10% SDS solution and quantified by absorbance at 595 nm. Results are representative of three independent experiments. E: Colony formation after DNA-PKcs rescuing. Right panel: representative clonogenic assay of MSI1 knockout cells transfected with DNA-PKcs expression vector after exposure to 2.5 Gy. Left panel: stain absorbed by the cells was dissolved in 1% SDS solution and quantified by absorbance at 570 nm. Results are representative of three independent experiments. F: Quantification of endogenous 53BP1 foci levels in MSI1 knockout cells after MSI1 or DNA-PKcs overexpression. A minimum of 100 nuclei were counted for each condition performed in triplicate. Data are presented as means ± SD. n = 3 (D–F). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 (t-test). CTCF, CCCTC-binding factor; Ctrl, control; si, small interfering; WT, wild-type.
To ascertain that DNA-PKcs is a critical MSI1 target in the context of DNA repair, we generated MSI1-knockout cells using the clustered regularly interspaced short palindromic repeat/Cas9 system23 and performed rescue experiments. MSI1 knockout was confirmed by Western blot (Figure 3C). DNA-PKcs levels were dramatically reduced in the clones analyzed, confirming that DNA-PKcs expression is indeed controlled by MSI1 (Figure 3C). Then, MSI1-knockout glioblastoma lines were transfected with vectors containing empty control, MSI1, or DNA-PKcs coding sequence, subjected to a single dose of IR (2.5 Gy), and evaluated in clonogenic assays. MSI1 (Figure 3D) and DNA-PKcs (Figure 3E) knockout-rescued cells showed higher radio-resistance compared to cells transfected with an empty vector (Ctrl). MSI1 rescue was more efficient compared to DNA-PKcs, possibly because MSI1 might regulate other genes involved in DNA repair. Finally, the number of endogenous 53BP1 foci was quantified in MSI1-knockout cells after transfection with MSI1 or DNA-PKcs coding sequences (Figure 3F). Endogenous 53BP1 foci levels were higher in MSI1-knockout compared to wild-type cells. Most importantly, the percentage of cells containing multiple 53BP1 foci decreased after overexpression of MSI1 or DNA-PKcs. These results suggest the association of MSI1 expression with lowering levels of DNA damage caused by IR and its role in the double-strand break repair, most likely by regulating DNA-PKcs expression and activity.
MSI1 Regulates Double-Strand Break Repair
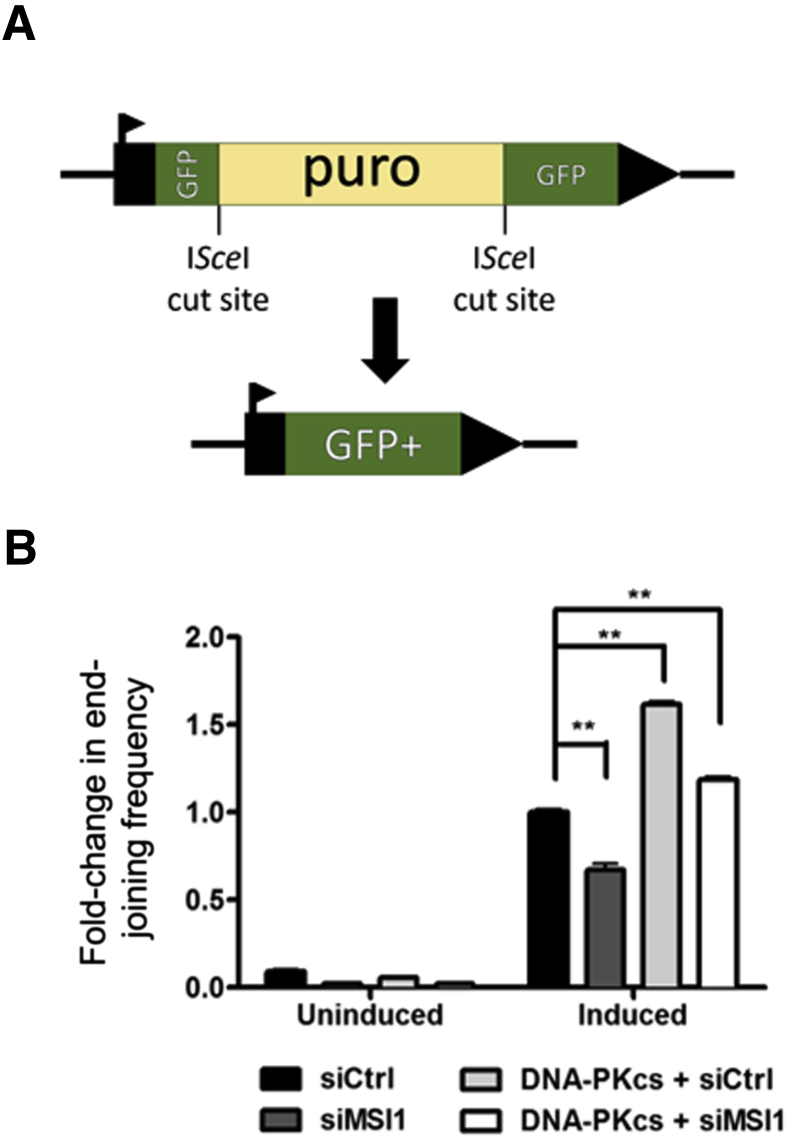
Given the high levels of persistent DNA damage with MSI1 knockdown and its regulation of DNA-PKcs, we asked whether MSI1 plays a role in the repair of double-strand breaks. DNA-PKcs is a key player in the NHEJ pathway to repair double-strand breaks.24 We therefore asked whether MSI1 modulated EJ repair via its control of DNA-PKcs expression. To evaluate the frequency of EJ, we used the EJ5-GFP reporter assay integrated into U2OS cells.15, 25 This reporter (Figure 4A) consists of a promoter and a GFP-coding cassette that is interrupted by a puromycin (puro) gene. The puro gene is flanked by ISceI homing endonuclease recognition sites in the same orientation. The expression of ISceI allows for site-specific double-strand breaks, resulting in excision of the puro gene and in the presence of functional NHEJ, leading to restoration of wild-type GFP gene. Interestingly, we observed a reduction in NHEJ frequency in MSI1-knockdown cells (Figure 4B), and this phenotype was rescued with the overexpression of DNA-PKcs (Figure 4B).
Figure 4.
Repair analysis after MSI1 alterations. A: Schematic representation of vector EJ5-GFP, which is designed to measure frequency of total end-joining events. Expression of ISceI introduces double-strand breaks at corresponding recognition sites, excision of the intervening puro cassette, and restoration of functional Green fluorescent protein (GFP) gene. B: Analysis of change in end-joining frequency with MSI1 knockdown and DNA–PKcs overexpression in the absence of MSI1 using EJ5-GFP assay. Data are presented as means ± SD. n = 3 (B). ∗∗P < 0.01. Ctrl, control; si, small interfering.
Discussion
Few recent studies have reported on the biological consequence of MSI1 in cancer. In colon cancer, the silencing of MSI1 induced apoptosis, mitosis, G2/M arrest, and tumor regression.26 In breast cancer, MSI1 knockdown resulted in decreased tumor mammosphere formation, decreased proliferation, and reduced breast cancer xenograft growth.27 In lung cancer, MSI1 silencing reduced spheroid colony formation with inhibition of the Wnt and Notch pathways.28 Silencing of MSI1 in DAOY medulloblastoma cells decreased proliferation and neurosphere formation, and induced differentiation and apoptosis.29 In the particular case of glioblastoma, we observed that a reduction in MSI1 expression increased apoptosis, decreased proliferation, affected cell cycle regulation, and interfered with adhesion-related functions such as invasion and migration.13 However, the impact of MSI1 expression on treatment outcome is poorly understood. Here, we demonstrated the induced expression of MSI1 by radiation in glioblastoma cell lines, which correlates with increased cell survival. Knockout of MSI1 increased radiosensitivity by affecting DNA damage repair through a mechanism involving protein kinase, DNA-activated, catalytic polypeptide expression.
Consistent with the role of cancer stem cells in radio-resistance, different studies have reported that radiation was able to induce the expansion of glioma cells that express stem cell markers, such as MSI1.30 Increased DNA repair capacity of cancer stem cells has been associated with resistance to radiotherapy.31 We suggest in this work that the functional involvement of MSI1 in the efficient repair of double-strand breaks by the NHEJ pathway is a possible mechanism of the observed chemoresistance. NHEJ is the predominant repair pathway in the mammalian system during the G1 and M phases, although it is active throughout the cell cycle. Increased protein expression of DNA-PKcs has been reported in a variety of tumor types, such as nasopharyngeal cancer, colorectal cancer, and non–small cell lung cancer, and overexpression has been correlated with tumor grade and poor survival.32 In glioma specimens, hyperactivity of this protein was associated with tumor grading33 and radio-resistance,34 and its up-regulation after radiation treatment has been associated with radio-resistance in recurrent oral squamous cell carcinomas.35 Using a DNA-PK kinase assay and assessing the phosphorylation status of replication protein A 2, a previous study demonstrated IR-induced DNA-PK activity in glioblastoma cell lines.36 This increased activity was correlated with the survival fraction of those cells after a 5-Gy dose of irradiation, which suggests the role of DNA-PK in radiation resistance.36 Furthermore, different studies have demonstrated the efficacy of its inhibition in promoting radio-sensitization of glioblastoma by inducing autophagy.37
In conclusion, considering the important impact of MSI1 on DNA repair, radio-resistance, and other cancer-relevant processes,13 MSI1 targeting can prove to be an important avenue for the treatment of glioblastoma patients when combined with radiotherapy. Moreover, MSI1 is specifically expressed in tumor cells and not in adjacent and differentiated cells such as neurons and astrocytes, leaving collateral damage from local inhibition of MSI1 to a minimum. Therapeutic targeting of MSI1 could be accomplished using a myriad of methodologies that could include small-molecule inhibitors (in a similar strategy for drug discovery for DNA-binding proteins),38 RNA-based decoys mimicking MSI1 binding motifs,39, 40 and miRNA mimics.41, 42 There are several known elements that control its expression,43, 44, 45, 46 and a recent report shows that MSI1 is a potential pharmacotherapeutic target.47 Because MSI1 has been connected to numerous solid tumors,5 strategies to modulate MSI1 function or levels of expression could have a large impact on cancer treatment.
Acknowledgment
We thank Dr. Pei Wang for the lenti-CRISPR v2 vector used for generating MSI1 knockout cell lines by CRISPR technology.
Footnotes
Supported by Cancer Prevention Research Institute of Texas (CPRIT) fellowship RP140105 (P.R.d.A.), CPRIT Greehey fellowship RP101491 (A.G.), a Translational Science Training Across Disciplines Scholarship from the University of Texas System Graduate Programs Initiative (A.G.), National Cancer Institute T32 postdoctoral training fellowship T32CA148724 (A.G.) and training grant 5T32CA148724-2 (S.S.T.), a scholarship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brazil (A.E.d.S.). Work performed in the Bishop laboratory was supported by NIH grants K22ES012264, 1R15ES019128, and 1R01CA152063. Work performed in the Penalva laboratory was supported by The Max and Minnie Tomerlin Voelcker Fund grants 1R01HG006015-01A and 1R21CA175875-01A1, The Institute for Integration of Medicine and Sciences (IIMS)-Cancer Therapy & Research Center (CTRC) (University of Texas Health Sciences Center at San Antonio), and Voices Against Brain Cancer (VABC; L.O.F.P.).
Disclosures: None declared.
P.R.d.A., A.G., and A.E.d.S. contributed equally to this work.
Current address of D.T.V., Department of Radiation Oncology, University of Texas Southwestern Medical Center, Dallas, Texas.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2016.05.020.
Contributor Information
Alexander J.R. Bishop, Email: bishopa@uthscsa.edu.
Luiz O.F. Penalva, Email: penalva@uthscsa.edu.
Supplemental Data
Decreased U343 cell survival after knockdown (KD) of Musashi1 (MSI1). A: Immunoblot analysis of MSI1 expression in U343 cells after transfection with sort-hairpin control (shCtrl) or shMSI1. B: Clonogenic assay of U343 MSI1-KD after exposure to increasing doses of X-rays. ∗P < 0.05, ∗∗∗P < 0.001.
A: The University of California at Santa Cruz (UCSC) genome browser (https://genome.ucsc.edu, accessed October 10, 2015) tracks showing individual-nucleotide resolution UV cross-linking immunoprecipitation (iCLIP) read coverage in the 3′ untranslated region (UTR) of protein kinase, DNA-activated, catalytic polypeptide (PRKDC). Vertical red lines indicate occurrences of the trimer UAG, which forms the core of the Musashi1 (MSI1) binding site. The density of iCLIP reads, which mark MSI1 binding activity, correlates with regions showing high occurrence of UAG trimers. Reads were mapped to hg19 using RMAP.17 Read-coverage wiggle tracks were produced using a combination of in-house scripts and the UCSC genome browser toolchain. Visualization of read coverage and identification of UAG trimers were performed using the UCSC genome browser. B: Immunoblot analysis showing increased DNA–protein kinase catalytic subunit (PKcs) expression in different times after radiation (5 Gy). CTCF, CCCTC-binding factor.
References
- 1.Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J., Belanger K., Brandes A.A., Marosi C., Bogdahn U., Curschmann J., Janzer R.C., Ludwin S.K., Gorlia T., Allgeier A., Lacombe D., Cairncross J.G., Eisenhauer E., Mirimanoff R.O. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Van Meir E.G., Hadjipanayis C.G., Norden A.D., Shu H.K., Wen P.Y., Olson J.J. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stupp R., Taillibert S., Kanner A.A., Kesari S., Steinberg D.M., Toms S.A., Taylor L.P., Lieberman F., Silvani A., Fink K.L., Barnett G.H., Zhu J.J., Henson J.W., Engelhard H.H., Chen T.C., Tran D.D., Sroubek J., Tran N.D., Hottinger A.F., Landolfi J., Desai R., Caroli M., Kew Y., Honnorat J., Idbaih A., Kirson E.D., Weinberg U., Palti Y., Hegi M.E., Ram Z. Maintenance therapy with tumor-treating fields plus temozolomide vs temozolomide alone for glioblastoma: a randomized clinical trial. JAMA. 2015;314:2535–2543. doi: 10.1001/jama.2015.16669. [DOI] [PubMed] [Google Scholar]
- 4.Scaringi C., Enrici R.M., Minniti G. Combining molecular targeted agents with radiation therapy for malignant gliomas. Onco Targets Ther. 2013;6:1079–1095. doi: 10.2147/OTT.S48224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glazer R.I., Vo D.T., Penalva L.O. Musashi1: an RBP with versatile functions in normal and cancer stem cells. Front Biosci. 2012;17:54–64. doi: 10.2741/3915. [DOI] [PubMed] [Google Scholar]
- 6.Siddall N.A., McLaughlin E.A., Marriner N.L., Hime G.R. The RNA-binding protein Musashi is required intrinsically to maintain stem cell identity. Proc Natl Acad Sci U S A. 2006;103:8402–8407. doi: 10.1073/pnas.0600906103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imai T., Tokunaga A., Yoshida T., Hashimoto M., Mikoshiba K., Weinmaster G., Nakafuku M., Okano H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Sousa Abreu R., Sanchez-Diaz P.C., Vogel C., Burns S.C., Ko D., Burton T.L., Vo D.T., Chennasamudaram S., Le S.Y., Shapiro B.A., Penalva L.O. Genomic analyses of musashi1 downstream targets show a strong association with cancer-related processes. J Biol Chem. 2009;284:12125–12135. doi: 10.1074/jbc.M809605200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charlesworth A., Wilczynska A., Thampi P., Cox L.L., MacNicol A.M. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 2006;25:2792–2801. doi: 10.1038/sj.emboj.7601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Battelli C., Nikopoulos G.N., Mitchell J.G., Verdi J.M. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol Cell Neurosci. 2006;31:85–96. doi: 10.1016/j.mcn.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 11.MacNicol M.C., Cragle C.E., MacNicol A.M. Context-dependent regulation of Musashi-mediated mRNA translation and cell cycle regulation. Cell Cycle. 2011;10:39–44. doi: 10.4161/cc.10.1.14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuwako K., Kakumoto K., Imai T., Igarashi M., Hamakubo T., Sakakibara S., Tessier-Lavigne M., Okano H.J., Okano H. Neural RNA-binding protein Musashi1 controls midline crossing of precerebellar neurons through posttranscriptional regulation of Robo3/Rig-1 expression. Neuron. 2010;67:407–421. doi: 10.1016/j.neuron.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Uren P.J., Vo D.T., de Araujo P.R., Potschke R., Burns S.C., Bahrami-Samani E., Qiao M., de Sousa Abreu R., Nakaya H.I., Correa B.R., Kuhnol C., Ule J., Martindale J.L., Abdelmohsen K., Gorospe M., Smith A.D., Penalva L.O. RNA-Binding Protein Musashi1 Is a Central Regulator of Adhesion Pathways in Glioblastoma. Mol Cell Biol. 2015;35:2965–2978. doi: 10.1128/MCB.00410-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahlrot R.H., Hansen S., Herrstedt J., Schroder H.D., Hjelmborg J., Kristensen B.W. Prognostic value of Musashi-1 in gliomas. J Neurooncol. 2013;115:453–461. doi: 10.1007/s11060-013-1246-8. [DOI] [PubMed] [Google Scholar]
- 15.Gunn A., Bennardo N., Cheng A., Stark J.M. Correct end use during end joining of multiple chromosomal double strand breaks is influenced by repair protein RAD50, DNA-dependent protein kinase DNA-PKcs, and transcription context. J Biol Chem. 2011;286:42470–42482. doi: 10.1074/jbc.M111.309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh N.P., McCoy M.T., Tice R.R., Schneider E.L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 17.Smith A.D., Chung W.Y., Hodges E., Kendall J., Hannon G., Hicks J., Xuan Z., Zhang M.Q. Updates to the RMAP short-read mapping software. Bioinformatics. 2009;25:2841–2842. doi: 10.1093/bioinformatics/btp533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen C., Kurimasa A., Brenneman M.A., Chen D.J., Nickoloff J.A. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc Natl Acad Sci U S A. 2002;99:3758–3763. doi: 10.1073/pnas.052545899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serrano M.A., Li Z., Dangeti M., Musich P.R., Patrick S., Roginskaya M., Cartwright B., Zou Y. DNA-PK, ATM and ATR collaboratively regulate p53-RPA interaction to facilitate homologous recombination DNA repair. Oncogene. 2013;32:2452–2462. doi: 10.1038/onc.2012.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liaw H., Lee D., Myung K. DNA-PK-dependent RPA2 hyperphosphorylation facilitates DNA repair and suppresses sister chromatid exchange. PLoS One. 2011;6:e21424. doi: 10.1371/journal.pone.0021424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 24.Uematsu N., Weterings E., Yano K., Morotomi-Yano K., Jakob B., Taucher-Scholz G., Mari P.O., van Gent D.C., Chen B.P., Chen D.J. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao Z., Bozzella M., Seluanov A., Gorbunova V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst) 2008;7:1765–1771. doi: 10.1016/j.dnarep.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sureban S.M., May R., George R.J., Dieckgraefe B.K., McLeod H.L., Ramalingam S., Bishnupuri K.S., Natarajan G., Anant S., Houchen C.W. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134:1448–1458. doi: 10.1053/j.gastro.2008.02.057. [DOI] [PubMed] [Google Scholar]
- 27.Wang X.Y., Penalva L.O., Yuan H., Linnoila R.I., Lu J., Okano H., Glazer R.I. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Mol Cancer. 2010;9:221. doi: 10.1186/1476-4598-9-221. doi: 10.1186/1476-4598-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X.Y., Yu H., Linnoila R.I., Li L., Li D., Mo B., Okano H., Penalva L.O., Glazer R.I. Musashi1 as a potential therapeutic target and diagnostic marker for lung cancer. Oncotarget. 2013;4:739–750. doi: 10.18632/oncotarget.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez-Diaz P.C., Burton T.L., Burns S.C., Hung J.Y., Penalva L.O. Musashi1 modulates cell proliferation genes in the medulloblastoma cell line Daoy. BMC Cancer. 2008;8:280–292. doi: 10.1186/1471-2407-8-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bao S., Wu Q., McLendon R.E., Hao Y., Shi Q., Hjelmeland A.B., Dewhirst M.W., Bigner D.D., Rich J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 31.Hussein D., Punjaruk W., Storer L.C., Shaw L., Othman R., Peet A., Miller S., Bandopadhyay G., Heath R., Kumari R., Bowman K.J., Braker P., Rahman R., Jones G.D., Watson S., Lowe J., Kerr I.D., Grundy R.G., Coyle B. Pediatric brain tumor cancer stem cells: cell cycle dynamics, DNA repair, and etoposide extrusion. Neuro Oncol. 2011;13:70–83. doi: 10.1093/neuonc/noq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu F.M., Zhang S., Chen B.P. Role of DNA-dependent protein kinase catalytic subunit in cancer development and treatment. Transl Cancer Res. 2012;1:22–34. doi: 10.3978/j.issn.2218-676X.2012.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao C.J., Fu J., Shi H.L., Mu Y.G., Chen Z.P. Activities of DNA-PK and Ku86, but not Ku70, may predict sensitivity to cisplatin in human gliomas. J Neurooncol. 2008;89:27–35. doi: 10.1007/s11060-008-9592-7. [DOI] [PubMed] [Google Scholar]
- 34.Mukherjee B., McEllin B., Camacho C.V., Tomimatsu N., Sirasanagandala S., Nannepaga S., Hatanpaa K.J., Mickey B., Madden C., Maher E., Boothman D.A., Furnari F., Cavenee W.K., Bachoo R.M., Burma S. EGFRvIII and DNA double-strand break repair: a molecular mechanism for radioresistance in glioblastoma. Cancer Res. 2009;69:4252–4259. doi: 10.1158/0008-5472.CAN-08-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shintani S., Mihara M., Li C., Nakahara Y., Hino S., Nakashiro K., Hamakawa H. Up-regulation of DNA-dependent protein kinase correlates with radiation resistance in oral squamous cell carcinoma. Cancer Sci. 2003;94:894–900. doi: 10.1111/j.1349-7006.2003.tb01372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen Y., Wang Y., Sheng K., Fei X., Guo Q., Larner J., Kong X., Qiu Y., Mi J. Serine/threonine protein phosphatase 6 modulates the radiation sensitivity of glioblastoma. Cell Death Dis. 2011;2:e241. doi: 10.1038/cddis.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhuang W., Li B., Long L., Chen L., Huang Q., Liang Z.Q. Knockdown of the DNA-dependent protein kinase catalytic subunit radiosensitizes glioma-initiating cells by inducing autophagy. Brain Res. 2011;1371:7–15. doi: 10.1016/j.brainres.2010.11.044. [DOI] [PubMed] [Google Scholar]
- 38.Berg T. Inhibition of transcription factors with small organic molecules. Curr Opin Chem Biol. 2008;12:464–471. doi: 10.1016/j.cbpa.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 39.Keefe A.D., Pai S., Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Symensma T.L., Baskerville S., Yan A., Ellington A.D. Polyvalent Rev decoys act as artificial Rev-responsive elements. J Virol. 1999;73:4341–4349. doi: 10.1128/jvi.73.5.4341-4349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bader A.G., Brown D., Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027–7030. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henry J.C., Azevedo-Pouly A.C., Schmittgen T.D. MicroRNA replacement therapy for cancer. Pharm Res. 2011;28:3030–3042. doi: 10.1007/s11095-011-0548-9. [DOI] [PubMed] [Google Scholar]
- 43.Vo D.T., Qiao M., Smith A.D., Burns S.C., Brenner A.J., Penalva L.O. The oncogenic RNA-binding protein Musashi1 is regulated by tumor suppressor miRNAs. RNA Biol. 2011;8:817–828. doi: 10.4161/rna.8.5.16041. [DOI] [PubMed] [Google Scholar]
- 44.Vo D.T., Abdelmohsen K., Martindale J.L., Qiao M., Tominaga K., Burton T.L., Gelfond J.A., Brenner A.J., Patel V., Trageser D., Scheffler B., Gorospe M., Penalva L.O. The oncogenic RNA-binding protein Musashi1 is regulated by HuR via mRNA translation and stability in glioblastoma cells. Mol Cancer Res. 2012;10:143–155. doi: 10.1158/1541-7786.MCR-11-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rezza A., Skah S., Roche C., Nadjar J., Samarut J., Plateroti M. The overexpression of the putative gut stem cell marker Musashi-1 induces tumorigenesis through Wnt and Notch activation. J Cell Sci. 2010;123:3256–3265. doi: 10.1242/jcs.065284. [DOI] [PubMed] [Google Scholar]
- 46.Clingman C.C., Deveau L.M., Hay S.A., Genga R.M., Shandilya S.M., Massi F., Ryder S.P. Allosteric inhibition of a stem cell RNA-binding protein by an intermediary metabolite. Elife. 2014;3 doi: 10.7554/eLife.02848. doi: 10.7554/eLife.02848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lan L., Appelman C., Smith A.R., Yu J., Larsen S., Marquez R.T., Liu H., Wu X., Gao P., Roy A., Anbanandam A., Gowthaman R., Karanicolas J., De Guzman R.N., Rogers S., Aube J., Ji M., Cohen R.S., Neufeld K.L., Xu L. Natural product (-)-gossypol inhibits colon cancer cell growth by targeting RNA-binding protein Musashi-1. Mol Oncol. 2015;9:1406–1420. doi: 10.1016/j.molonc.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Decreased U343 cell survival after knockdown (KD) of Musashi1 (MSI1). A: Immunoblot analysis of MSI1 expression in U343 cells after transfection with sort-hairpin control (shCtrl) or shMSI1. B: Clonogenic assay of U343 MSI1-KD after exposure to increasing doses of X-rays. ∗P < 0.05, ∗∗∗P < 0.001.
A: The University of California at Santa Cruz (UCSC) genome browser (https://genome.ucsc.edu, accessed October 10, 2015) tracks showing individual-nucleotide resolution UV cross-linking immunoprecipitation (iCLIP) read coverage in the 3′ untranslated region (UTR) of protein kinase, DNA-activated, catalytic polypeptide (PRKDC). Vertical red lines indicate occurrences of the trimer UAG, which forms the core of the Musashi1 (MSI1) binding site. The density of iCLIP reads, which mark MSI1 binding activity, correlates with regions showing high occurrence of UAG trimers. Reads were mapped to hg19 using RMAP.17 Read-coverage wiggle tracks were produced using a combination of in-house scripts and the UCSC genome browser toolchain. Visualization of read coverage and identification of UAG trimers were performed using the UCSC genome browser. B: Immunoblot analysis showing increased DNA–protein kinase catalytic subunit (PKcs) expression in different times after radiation (5 Gy). CTCF, CCCTC-binding factor.