Abstract
HIV-1 Nef is necessary and may be sufficient for HIV-1-associated AIDS pathogenicity, in that knockout of Nef alone can protect HIV-infected patients from AIDS. We therefore investigated the feasibility of physical knockout of Nef, using the host ubiquitin proteasome system in HIV-1-infected cells. Our co-immunoprecipitation analysis demonstrated that Nef interacted with ubiquitin specific protease 15 (USP15), and that USP15, which is known to stabilize cellular proteins, degraded Nef. Nef could also cause decay of USP15, although Nef-mediated degradation of USP15 was weaker than USP15-mediated Nef degradation. Direct interaction between Nef and USP15 was essential for the observed reciprocal decay of the proteins. Further, USP15 degraded not only Nef but also HIV-1 structural protein, Gag, thereby substantially inhibiting HIV-1 replication. However, Gag did not degrade USP15, indicating that the Nef and USP15 complex, in distinction to other viral proteins, play an integral role in coordinating viral protein degradation and hence HIV-1 replication. Moreover, Nef and USP15 globally suppressed ubiquitylation of cellular proteins, indicating that these proteins are major determinants for the stability of cellular as well as viral proteins. Taken together, these data indicate that Nef and USP15 are vital in regulating degradation of viral and cellular proteins and thus HIV-1 replication, and specific degradation of viral, not cellular proteins, by USP15 points to USP15 as a candidate therapeutic agent to combat AIDS by eliminating viral proteins from the infected cells via USP15-mediated proteosomal degradation.
Keywords: HIV-1 Nef, Viral Protein Degradation, USP15, Ubiquitin Proteasome System (UPS), Ubiquitylation
1. Introduction
Post-translational modification of proteins by ubiquitin (Ub) and their degradation by the ubiquitin proteasome system (UPS) has emerged as a major regulatory process in virtually all aspects of cell biology (Glickman and Ciechanover, 2002). Given the importance of Ub attachment in regulating the fate and function of proteins, it is not surprising that HIV-1 takes advantage of the UPS to influence the interplay between HIV-1 and its host cells through modulating proteostasis of viral and cellular proteins. Previous studies report that various HIV-1 viral proteins are engaged in regulation of degradation of cellular counterparts at every step of the HIV-1 life cycle – from entry into host cells to release of the infectious progeny viruses to the extracellular milieu to initiate next round of virus life cycle (Cucchiarini et al., 1995; Dang et al., 2006; Dube et al., 2010; Kutluay et al., 2013; Lindwasser et al., 2007; Neil et al., 2008; Sakuma et al., 2007). However, it is completely unknown how these individual events taking place at different stages of virus life cycle are coordinated and what viral and cellular molecules dictate these molecular events.
Nef is essential for HIV-1 pathogenesis (Churchill et al., 2004; Churchill et al., 2006; Kestler et al., 1991; Kirchhoff et al., 1995). However, molecular details on how the Nef protein determines HIV-1-mediated AIDS progression are still unclear. One of the key molecular features of Nef is that, unlike other viral proteins whose functions are phase- (or stage-) specific in the virus life cycle, Nef is the only known viral element which acts throughout the whole virus life cycle within the host cell: it is translated from the early viral messages (Cullen, 1991; Ferguson et al., 2002; Haseltine, 1991) and stays in the infected cells until the protein is packaged into virion particles (Bukovsky et al., 1997), and thus Nef has temporal advantages to determine the fate of not only viral but also cellular proteins throughout the HIV-1 infectious life cycle. Another notable feature is that subcellular localization of Nef, compared with other viral proteins which localize to specific subcellular compartments, is not confined to the cytoplasmic membrane by myristoylation (Allan et al., 1985; Franchini et al., 1986), but occurs in diverse subcellular organelles, such as endosomes (Dikeakos et al., 2012; Kueck and Neil, 2012; Singh et al., 2009), ER (Park et al., 2014), mitochondria, and even the perinuclear membrane by interacting with cellular protein partners (Kammula et al., 2012). Nef is also known to play a pivotal role in localization of Env glycoprotein and Gag in late endosomes (Sandrin and Cosset, 2006) and in regulation of various cellular proteins by inducing endocytosis (Hanna et al., 1998; Sandrin and Cosset, 2006), suggesting that Nef may have spatial advantages in governing viral and cellular protein fates. These reports strongly suggest that the described molecular properties confer multifarious Nef functions in disease progression.
It is recently reported that Nef degrades Tat protein by the UPS (Sugiyama et al., 2011), which is crucial for an efficient virus replication, and counteracts the tetherin protein in SIVs, whose genomes lack vpu (Jia et al., 2009; Sauter et al., 2009; Serra-Moreno et al., 2013; Zhang et al., 2009). Further, it is reported that K144 in HIV-1 Nef is di-ubiquitinated for downregulation of CD4 and MHC-I (Cai et al., 2011). However, little is known as to whether Nef indeed plays an essential role in regulating the stability of HIV-1 viral and cellular proteins and consequent HIV-1 replication and survival of HIV-infected cells, and if so, how and to what degree Nef orchestrates overall HIV-1 and host cell protein fates during pathogenesis. To investigate Nef role in protein degradation, we seek to identify cellular proteins involved in the UPS-mediated protein degradation through association with Nef and found ubiquitin specific protease 15 (USP15) which stabilizes proteins by deubiquitylation and by preventing autoubiquitylation of substrates (Aggarwal and Massague, 2012; Cayli et al., 2009; Inui et al., 2011; Soboleva et al., 2005). Further investigation demonstrated the significance of Nef- and USP15-mediated viral and cellular protein degradation with respect to the regulation of the virus life cycle and HIV-1/host cell competition that is essential for AIDS progression.
2. Materials and methods
2.1. Cells and reagents
Jurkat and 293T cells were cultured in RPMI1640 and Dulbecco modified Eagle medium (DMEM), respectively, supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin/streptomycin. Antibodies (Ab) and reagents: Anti-Myc (9E10), -USP15 (2D5), -EEA (H-300), -HA (F-7), and -GFP antibodies were purchased from Santa Cruz (Santa Cruz, California) and from Clonetech (Takara. Clontech, Mountain View, CA), respectively, anti-β-actin (O61M4808) antibody was obtained from Sigma (St. Louis, MO), and Alexa Fluor 488 or 555 conjugated to a secondary antibodies were purchased from Life technologies (Grand Island, NY). The reagents employed for these experiments are Cycloheximide (Sigma), Bafilomycin A1 (Sigma), MG132 (Cayman, Ann Arbor, MI).
2.2. Plasmids
USP15-expressing plasmid (pUSP15) was constructed by inserting the open reading frame of USP15 in the pCR-BluntII-topo-USP15 (clone ID #40118994, Open Biosystems) between NotI and SalI restriction sites of pCMV-Myc (Company). N- and C-terminal deletion mutants of USP15, pUSP15ΔN and pUSP15ΔC, respectively, were generated, using restriction endonucleases. Similarly, the coding regions of nef of HXBc2 and YU2 strains of HIV-1 were cloned into EcoRI and BamHI sites of pCDNA3.1(−)-Myc. His (Agilent, Santa Clara, CA) to generate pHNef and pYNef, respectively. pHN.GFP plasmid encoding Nef.GFP fusion protein was constructed by placing nef-coding region in pHNef into pEGFP-N3 (Takara. Clontech, Mountain View, CA), using EcoRI and BamHI restriction endonucleases. pDsRed2-ER was purchased from Clontech (Mountain View, CA), and the ubiquitin (Ub)-expressing plasmid, pUb-HA and Gag-expressing plasmid, psPax2 are described in detail elsewhere (Timani et al., 2014) and (Zufferey et al., 1997), respectively.
2.3. Western blot (WB) and immunoprecipitation analysis
Cells were washed twice in ice-cold PBS, suspended in the lysis buffer containing 50 mM Tris-HCl pH 7.4, 300 mM NaCl, 1% NP-40, 50 mM NaF, 1 mM NaVO4, 1mM PMSF and 1x protease inhibitor cocktail (Calbiochem, La Jolla, CA), and incubated on ice for 20 min. After centrifugation at 20,000 g at 4°C for 20 min, the supernatants were collected and saved as cell lysates. The lysates were then employed for immunoprecipitation and Western blot analysis, as described (Park et al., 2013).
2.4. Confocal microscopic analysis
Cells grown on polylysine-coated cover slips were transfected with the indicated plasmids and cultured for 48 hr. Cells were then washed twice with PBS, fixed in 4% paraformaldehyde, permeabilized with 0.3% Triton X-100 for 5 min, and blocked in 2.5% BSA for 30 min. Cells were incubated with the primary antibody followed by Alexa Fluor 488 or 555 conjugated to secondary antibodies at room temperature for 1 h each and washed three times with PBS to visualize subcellular localization of the indicated proteins with an confocal microscope, as described (Park et al., 2014).
2.5. Cycloheximide determination of protein half-life
To investigate whether the observed reductions in the amount of USP15 and Nef were due to the degradation of the expressed proteins, 293T cells transfected with pUSP15 and/or pHNef were treated with 40 μg/ml of cycloheximide (CHX) (Sigma Aldrich, St. Louis, MO) at 48 h post-transfection for the indicated time periods, and changes to protein levels were determined by WB analyses, as described above.
2.5. Reverse transcriptase (RT) assay and preparation of virions
For RT assay, virions in the supernatant were pelleted by centrifugation at 12,000 g for 1 h, and the RT activity was determined, as described (Pyeon and Park, 2015). For virion preparation for WB, cells and cell debris in the culture supernatant were removed by centrifugation at 800 g for 100 min, and the cleared supernatant was passed through a 0.22 μm filter (Corning, NY) to ensure complete removal of smaller cell debris. Virions in the filtrate were precipitated in 20% sucrose in PBS by centrifugation at 238,000 g for 90 min, as described (Pyeon and Park, 2015), and virion proteins were analyzed by WB, as described above.
2.6. Data analysis
All values are expressed as means +/− SD of triplicate experiments. All comparisons were by a controlled two-tailed Student’s t-test. A p value of <0.05 was considered statistically significant (*), and p<0.01 highly significant (**).
3. Results
3.1. Significant amounts of the viral proteins remained in the infected cells without being packaged into virions
Our WB analysis using anti-p24 antibody showed that only a fraction of Gag protein was assembled into virion particles, while significant amount of the viral proteins remained in the infected Jurkat cells (Fig. 1B), even when the amount of HIV-1 virion in the culture supernatants reached to the peak (9 day post-infection) (Fig. 1A). Why, then, does HIV-1 infection yield such large amounts of viral protein, even if only a small fraction of the protein produced is employed in generating infectious progeny, and what is the role and fate of the intracellular viral proteins remaining in the infected cells? We hypothesized that this high percentage of the intracellular viral proteins, specifically Nef, within the cells is required for competing with the infected host in regulating stabilities of cellular restriction factors.
Figure 1.
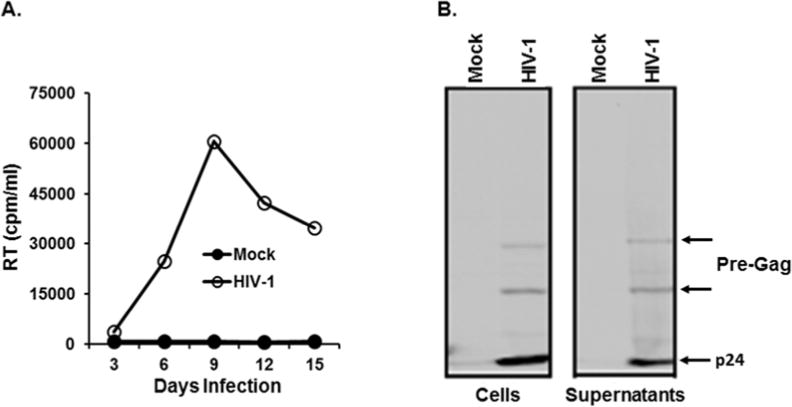
Replication of HIV-1 in Jurkat and WB analysis of viral proteins. (A) HIV-1 corresponding to 10,000 cpm/ml was infected into Jurkat cells (1×106 cells), and replication kinetics of HIV-1 was determined by measuring RT activity in the culture supernatants every 3 days. (B) HIV-1 was infected into Jurkat cells, and cell (left) and virus (right) lysates were prepared at peak virus replication. 1/5 and 1/2 of total lysates of cells and virus (supernatants), respectively, were employed for the analysis. Band intensity of p24 (arrow) in the supernatant was approximately 1/3 of that in the cells.
Since Nef is clearly vital to HIV-1 pathobiology in the clinic, but comprehensive in vitro modeling of the gene has been restricted by its dispensibility to continuous virus replication in the susceptible cells (Joseph et al., 2005; Neri et al., 2011; Schindler et al., 2007), we adopted an alternative strategy that opens with single-cycle analysis of molecular events occurring within 293T cells to answer the above questions.
3.2. HIV-1 Nef and USP15 interacted and reciprocally regulated their stability
To investigate the involvement of Nef in degradation of viral and cellular proteins, we performed the yeast two-hybrid analysis to screen for cellular proteins associating with Nef, as described (Kalpana et al., 1994), and identified USP15. To confirm association of USP15 with Nef, 293T cells were transfected with pUSP15 (lane 1) or with pHNef and pUSP15 (lane 2), and proteins from the transfected cells were precipitated with anti-USP15 Ab followed by WB with anti-USP15 first and the same blot with anti-Myc Ab again. Therefore, lane 1 should detect only USP15, while lane 2 in Fig. 2A could detect both USP15 and Nef. Our data showed that Nef was detected in lane 2 together with USP15, but not in lane 1 (Fig. 2A), indicating that Nef associated with USP15 and suggesting that Nef could be important to UPS-mediated proteosomal degradation processes. We next examined the effect of Nef expression on intracellular USP15 levels by WB analysis of transfecting increasing concentration of pHNef into 293T cells. Our data showed that as the amount of pHNef was increased from 0 to 4 μg, the USP15 levels gradually decreased in a dose-dependent manner (Fig. 2B) without changes to the amount of β-actin (Fig. 2B), indicating that Nef expression caused the intracellular USP15 diminution. Paralleling the pHNef expression (lane 2, Fig. 2C), transfection of pYNef expressing R-tropic nef (YU2 strain) of HIV-1 reduced the amount of USP15 (lane 3, Fig. 2C), establishing that the observed decreases of USP15 were not just strain specific but were common to different strains of HIV-1 Nef. We then examined the effect of USP15 expression on the Nef levels. As shown in Fig. 2D, the amount of Nef gradually declined, as the amount of pUSP15 transfected into 293T was increased from 0 to 4 μg, demonstrating that USP15 can also decrease the amount of the expressed intracellular Nef. These findings indicated that the amounts of intracellular Nef and USP15 were mutually regulated.
Figure 2.
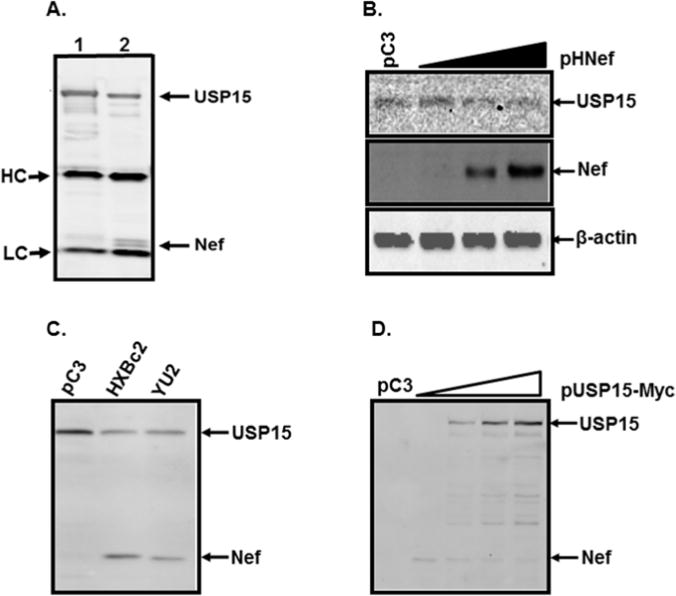
Reciprocal regulation of USP15 and Nef stability. (A) Association between Nef and USP15. 293T cells were transfected with pUSP15 (lane 1) or with pHNef and pUSP15 (lane 2), and proteins from the transfected cells were precipitated with anti-USP15 Ab followed by WB with anti-USP15 first and the same blot with anti-Myc Ab again which can detect both Nef and USP15 tagged with Myc (lane 2), indicating that Nef was co-precipitated with USP15. HC and LC represent immunoglobulin heavy and light chain, respectively. (B) Changes of the amount of endogenous USP15 by Nef. 293T cells were transfected with increasing amounts of nef-expressing plasmid (0, 1, 2, and 4 μg from left to right), and changes of the amount of endogenous USP15 was determined by WB analysis, using anti-USP15 antibody. Protein loading in each lane was determined to be equal by reprobing the membrane with anti-actin antibody (lower panel). The total transfected DNA was adjusted for equivalence by adding control plasmid pC3 (pCDNA3) in each transfection hereafter to eliminate discrepancies which might be arisen by the promoter competition. (C) Nef-mediated degradation of USP15. USP15-expressing plasmid alone (lane 1) or together with nef-expressing plasmids of the HXBc2 (lane 2) and the YU2 (lane 3) was transfected into 293T cells, and Western blot analysis was performed with the transfected cell lysates. (D) USP15-mediated degradation of Nef. Increasing concentration of USP15 (0, 1, 2, and 4 μg from lanes 2 to 5, respectively) reduced the amount of Nef in 293T cells. The data were representative of three independent experiments.
3.3. Reduction of USP15 and Nef was due to the reciprocal degradation of the expressed protein
Next, we investigated whether the observed reduction was due to the reciprocal degradation of the expressed USP15 and Nef. To this end, 293T cells transfected with pUSP15 (4 μg) and pHNef (2 μg) were treated with cycloheximide (CHX) for the indicated time period at 48 h post-transfection, and changes in the amounts of Nef and USP15 were determined by WB (Fig. 3, above) with Bio-Rad image quantitation based on β-actin changes (Fig. 3 line graph). The results showed that the amount of the intracellular USP15 slightly increased in the absence of Nef, while the expressed USP15 levels was reduced in the presence of Nef, indicating that USP15 diminution in the presence of Nef was due to the degradation of the expressed USP15 (Fig. 3). Under the same condition, the amount of Nef protein also increased in the absence of USP15 (Fig. 3A and 3B), while Nef levels significantly declined when the cells were co-transfected with pUSP15 – and the reduction was more dramatic in Nef than in USP15 (Fig. 3) – establishing that the lowered intensity of the protein bands was due to the degradation of the expressed proteins. Taken together, the results show that Nef and USP15 declines were due to expressed protein degradation, and USP15-mediated degradation of Nef was much stronger than Nef-mediated USP15 degradation (Fig. 3).
Figure 3.
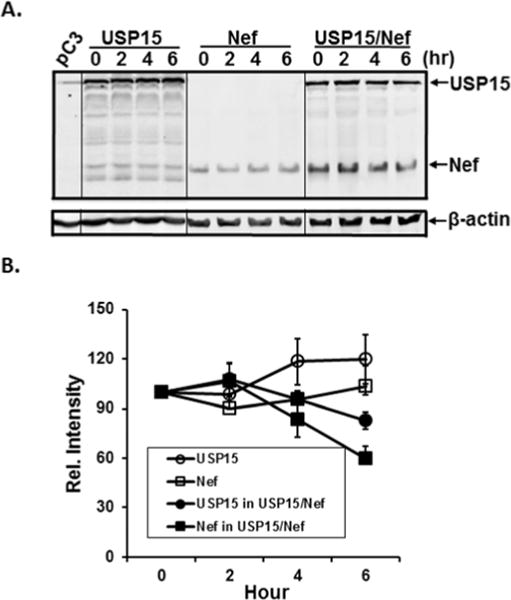
Effect of CHX. USP15- and/or Nef-expressing plasmids were transfected into 293T cells, and at forty-eight hours pos-transfection, the transfected cells were treated with CHX for the indicated time. Changes in the amount of USP15 and Nef were then determined in the cells transfected with USP15- (left), Nef- (middle), or USP15- and Nef-expressing plasmids (right) by WB analysis (above) followed by quantification based on the amount of β-actin. The data were representative of three independent experiments.
3.4. Association of Nef with USP15 is critical for the reciprocal regulation of decays of the proteins
We next investigated whether direct interactions between Nef and USP15 are essential for the observed reciprocal decay of the proteins. Accordingly, we constructed two deletion mutants of USP15, USP15ΔN and USP15ΔC (Fig. 4A), and examined binding of Nef to the mutant USP15 and its consequence to the reciprocal decay of mutant USP15 and Nef. WB analysis showed that GFP and Nef.GFP fusion protein were expressed efficiently (Fig. 4B, top panel), and USP15 and its mutant proteins were also expressed with the expected molecular weights, even if the expression level of USP15ΔN was relatively poor (Fig. 4B, middle panel). Immunoprecipitation of USP15 and its mutant proteins with anti-Myc antibody, followed by WB with anti-GFP antibody for the detection of Nef.GFP and GFP, showed that Nef.GFP, not GFP, was detected only in the cells transfected with wild type- or USPΔN-, but not with USPΔC-expressing plasmids (Fig. 4B bottom panel), indicating that Nef binds USP15 through the encoded protein motif(s) from amino acid 385 to the end of the USP15 gene. We then investigated whether the interaction between USP15 and Nef is required for the observed mutual degradation. Figure 3C showed that the amounts of the expressed USP15 and USP15ΔN which interact with Nef gradually declined as the amount of Nef was increased. However, we did not observe any appreciable reduction in USP15ΔC, which failed to interact with Nef (Fig. 4C), evincing that Nef-triggered decay of USP15 requires the deleted segment. Similarly, the amount of Nef was least in the cells transfected with pUSP15, as well as less in the cells transfected with pUSP15ΔN than in the cells transfected with pUSP15ΔC (Fig. 4C), establishing that interaction of USP15 and Nef is essential for the reciprocal protein decay.
Figure 4.
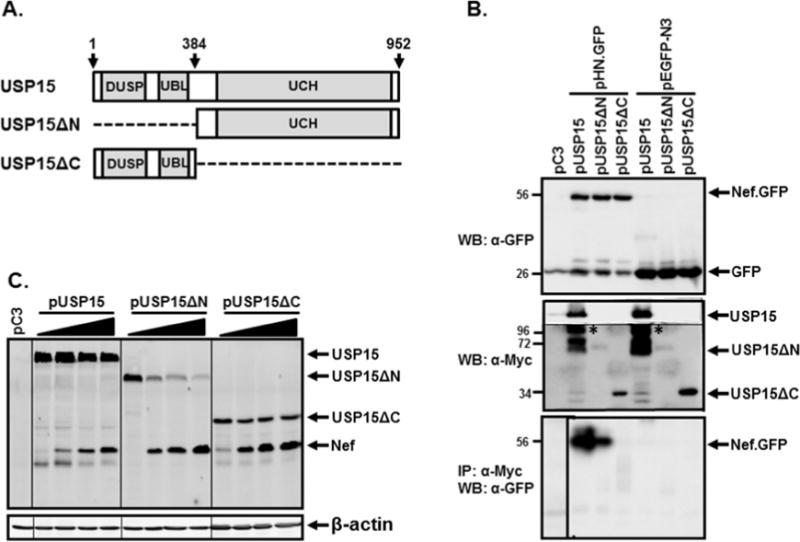
Association and degradation of USP15 and Nef. (A) Schematic representation of USP15 and its mutants. Arabic numbers indicate amino acid number of USP15, and dashed lines in USP15ΔN and USP15ΔC denote deletions. DUSP, UBL, and UCH in the gray boxes are abbreviations of domain present in ubiquitin-specific protease, ubiquitin-like fold, and ubiquitin carboxyl-terminal hydrolase, respectively. (B) Binding of Nef with USP15. pUSP15 or its mutant plasmids (pUSP15ΔN and pUSP15ΔC) was co-transfected with either pHN.GFP or pEGFP-N3 into 293T cells, and association of Nef with the wild type- or mutant USP15 was investigated by IP followed by WB. Expression of Nef.GFP or GFP (top panel) was determined by WB with rabbit anti-GFP antibody, and that of USP15 and its mutant proteins (middle panel) was detected with mouse anti-Myc antibody, wherein low exposed USP15 band was cut and pasted to the same image (*). The lower panel shows association of Nef with USP15, by IP with anti-Myc antibody for USP15 and its mutant proteins, followed by WB with anti-GFP antibody. Arabic numbers on the left of each image indicate positions of pre-stained protein size marker (kDa) (Fisher Scientific, Pittsburgh, PA). (C) Degradation of USP15 and its mutant proteins by Nef. Increasing concentrations of Nef (0, 1, 2, and 4 μg from lanes 1 to 4, respectively, in each bracket) were co-transfected with 2 μg of pUSP15, pUSP15ΔN, or pUSP15ΔC into 293T cells, and changes in the amount of each protein were analyzed by WB analysis. Protein loading in each lane was determined to be equal by reprobing the membrane with anti-actin antibody (lower panel). The data were representative of three independent experiments.
3.5. Both USP15 and Nef were ubiquitylated
Since Ub attachment to proteins followed by degradation of the proteins by UPS plays an integral role in regulation of the fate of proteins, we investigated the possibility of ubiquitylation of Nef and USP15. To this end, pUb-HA together with pHNef or pUSP15 were transfected into 293T cells, and expression and ubiquitylation of Nef and USP15 were determined by WB analysis (lane 1, Fig. 5A) and by immunoprecipitation with anti-HA antibody followed by WB analysis with anti-Myc antibody for the detection of Nef and USP15 (lane 2, Fig. 5A), respectively. Our data showed that both Nef and USP15 were expressed with the expected sizes of molecules, and the expressed proteins were ubiquitylated, indicating that the observed decay of the proteins was achieved by the UPS. Next, we studied whether expression of USP15 and Nef affects ubiquitylation of cellular protein in general. Accordingly, pHNef and pUSP15 were transfected into 293T cells, and effects of Nef or USP15 on ubiquinitylation of cellular proteins were determined by WB analysis with anti-HA Ab. As shown in Fig. 5B and 5C, the amount of the ubiquitylated cellular proteins was significantly diminished with increasing concentration of pUSP15 or pHNef plasmid, showing that both Nef and USP15 play an important role in governing cellular protein stability. In light of the innate function of USP15 to block protein ubiquitylation, the inhibition by USP15 with respect to ubiquitylation, as shown in Fig. 5, was expected. However, it is remarkable that Nef degraded USP15 but reduced ubiquitylation to thereby stabilize cellular proteins in general. These data collectively demonstrate that USP15 and Nef could be degraded by the UPS and that these two proteins are involved in regulating degradation of both viral and cellular proteins.
Figure 5.
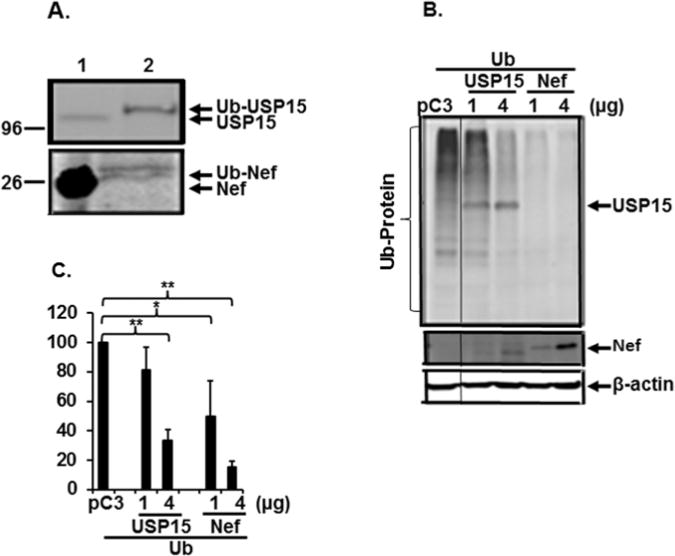
Ubiquitylation of viral and cellular proteins. (A) Ubiquitylation of USP15 and Nef. Ub-expressing plasmid (pUb) was transfected with USP15- (Top) or with nef-expressing plasmid (Bottom), and ubiquitylation of these proteins was determined by immunoprecipitation with anti-HA Ab for Ub followed by Western blot analysis with anti-USP15 (Top) or anti-Myc antibody for Nef (lane 2). Lane 1 indicates WB analysis of USP15 (Top) and Nef (Bottom). Arabic numbers on the left indicate positions of pre-stained protein size marker (kDa) (Fisher Scientific). (B and C) Ubiquitylation of cellular proteins. Total cell lysates generated from 293T cells transfected with pUb and 1 or 4 μg of USP15- or Nef-expressors were analyzed by WB with anti-HA Ab for ubiquitylated cellular proteins, followed with anti-Myc Ab for USP15 and Nef, and anti-β-actin Ab for β-actin protein, and (C) the relative amount of ubiquitylated proteins was determined by scanning the entire area of each lane, using ChemiDoc Touch Imaging System (Bio-Rad, Hercules, CA) and depicted as mean +/− SD of triplicates, shown in (B). The data were representative of three independent experiments.
3.6. Nef and USP15 co-localized to the same subcellular compartments
Nef is known to reside at the cytoplasmic membrane (Fackler et al., 1997; Kaminchik et al., 1994; Kammula et al., 2012; Yu and Felsted, 1992), while subcellular localization of USP15 is poorly understood. Since our data indicated that the stability of Nef and USP15 was affected bilaterally, we examined the possibility that Nef and USP15 co-localize to the same subcellular compartments of 293T cells transfected with pNef.GFP and pUSP15, using confocal microscopic analysis. Our data showed that a significant portion of Nef co-localized with USP15 in the cytosolic compartment (Fig. 6A). Expression of USP15 was also detected in the ER (Fig. 6B) where Nef protein was detected (Park et al., 2014). Further, USP15 was located in the early endosome (Fig. 6C) where endosomal protein degradation takes place, and Nef was also observed in the endosomes together with the early endosomal antigen (EEA) (Fig. 6D), suggesting that USP15 and Nef could regulate their own and other proteins’ destiny cooperatively in the same subcellular organelles. Interestingly, USP15 is known to contain a putative nucleolus localization signal (Soboleva et al., 2005). However, we did not detect significant amounts of USP15 in the nucleus.
Figure 6.
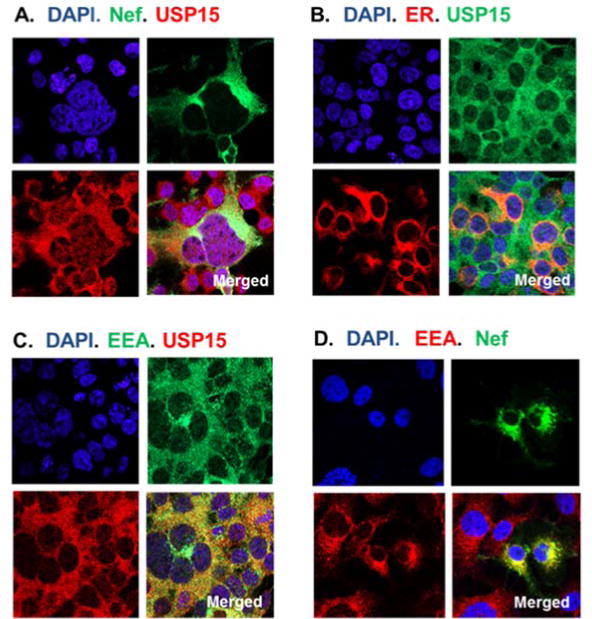
Confocal microscopy. Subcellular localization was determined by confocal microscopic analysis after transfecting USP15- and/or nef.GFP-expressing plasmids into 293T cells. USP15 was detected by anti-USP15 Ab followed by Alexa Fluor 488 (Fig. 6B) or Alexa Fluor 555 (Fig. 6A and C), Nef showed as green by fusion of the protein with GFP (Fig. 6A and D), and early endosomal antigen (EEA) was identified by anti-EEA Ab followed by Alexa Fluor 488 (Fig. 6C) and by Alexa Fluor 555 (Fig. 6D). Expression of pDsRed2-ER encodes a fusion consisting of red fluorescence protein, showing red color in the endoplasmic reticulum (ER) of the transfected cells (Fig. 6B).
3.7. USP15 also degraded viral structural protein and thus impaired HIV-1 replication
The above data demonstrated that Nef and USP15 degraded reciprocally and that USP15-mediated degradation of Nef was very pronounced, compared with Nef-mediated decay of USP15. We thus investigated whether USP15 impairs stability of other viral proteins, such as Gag, and thus alters HIV-1 replication. Accordingly, we transfected 293T cells with different amounts of wt- and Δnef-HIV-1 proviral DNA, and changes in the amount of intracellular viral proteins, Gag, and in virus replication in the presence of USP15 were monitored by WB analysis and RT assay, respectively. Our WB analyses showed that transfection of increasing USP15-expressing plasmid levels reduced the amount of p24 in a dose-dependent fashion in both wt-and Δnef-HIV-1 (Fig. 7A and B, respectively), and decreases of p24 were more noticeable with Δnef-HIV-1, as depicted in Fig. 7C. In parallel, replication of wt- and Δnef-HIV-1 (Fig. 7, below A and B, respectively) was significantly impaired by USP15, indicating that USP15 degraded not only Nef but also Gag and thus hampered HIV-1 replication. Our ensuing study indicated that these changes in the amount of viral proteins were due to the intracellular degradation, not to the inhibition of viral gene expression (data not shown).
Figure 7.
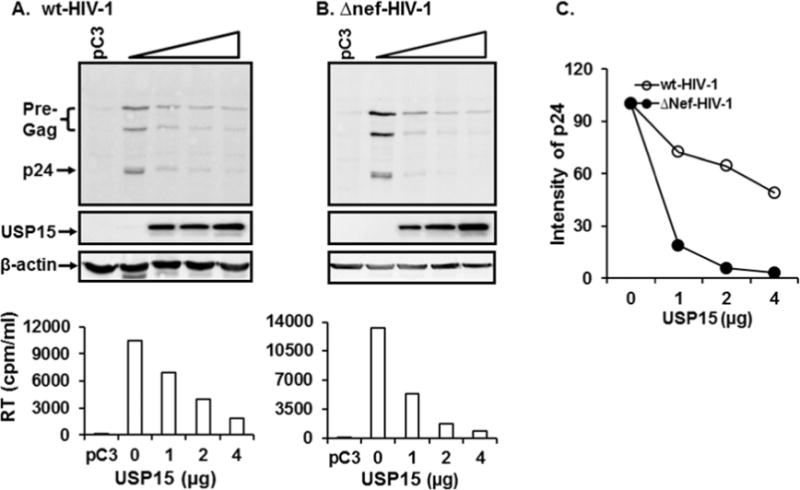
Effect of USP15 on degradation of Gag and HIV-1 replication. (A) and (B) Western blot analysis from cells transfected with wt- and Δnef-HIV-1. Lane 1 indicates 293T cells transfected with isotype plasmids, while lanes 2 to 5 in (A) and (B) represent 293T cells transfected with 4 μg of wt- (A) or Δnef-HIV-proviral DNA together with 0, 1, 2, and 4 μg of USP15-expressing plasmid, respectively. Bar graphs below (A) and (B) represent replication activity of wt- and Δnef-HIV-1 in 293T cells measured by RT activity in the presence of different doses of USP15. (C) depicts relative amount of p24 in panel (A) and (B) normalized based on β-actin. The data were representative of three independent experiments.
3.7. Both endosomal and proteosomal degradation pathways were critical for USP15-triggered degradation of viral proteins
We next investigated how USP15 degraded Gag. In light of the previous reports that Nef drives Gag to endosomes (Sandrin and Cosset, 2006) and that USP15 is essentially ubiquitous in the cytosolic compartments, as shown above (Fig. 6), it is reasonable to hypothesize that USP15 could degrade the structural protein via both enodosomal and proteosomal degradation pathways. To test this possibility, we investigated the effects of endosomal and proteosomal degradation inhibitors, using MG132 (Fig. 8B) and Bafilomycin A1 (Fig. 8C), respectively, on degradation of Gag. Our data showed that in the absence of MG132 (Fig. 8A), p24 was degraded, as the amount of USP15 was increased. However, the observed degradation of p24 by USP15 was effectively abrogated, when the transfected cells were treated with MG132 (Fig. 8B), indicating that the proteosomal degradation pathway plays a key role in USP15- mediated degradation of Gag protein. Remarkably, the band intensity of p24 by USP15 was visibly weaker, when the cells were treated with Bafilomycin A1 (compare p24 intensity in Fig. 8C with A), indicating that endosomal degradation inhibitor actually accelerated p24 degradation by unknown mechanism. Taken together these data showed that USP15-mediated degradation of Gag was modulated via both endosomal and proteosomal degradation cascades.
Figure 8.
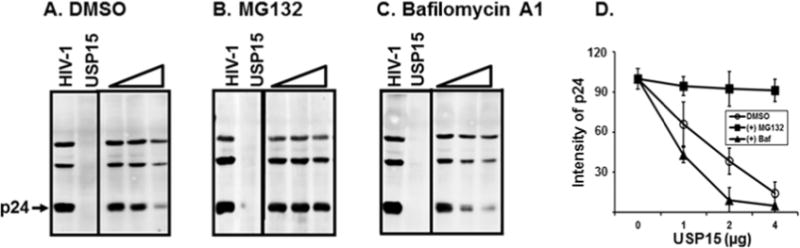
Effect of proteosomal (B) or endosomal (C) protein degradation inhibitor on USP15- (open triangle) mediated degradation of Gag. 293T cells were transfected with 4 μg HIV-1 proviral DNA together with 1, 2, and 4 μg of USP15-expressing plasmid, and at 48 hour post-transfection, cells were treated with DMSO (A) or with either 2 μM MG132 (B) or 100 nM Bafilomycin A1 (C) for 6 hours. WB analysis was then performed with each cell lysate with anti-p24 Ab for p24, and the data were representative of three independent experiments. (D) The amount of p24 in the cells treated with DMSO, MG132, or Bafilomycin A1 (Baf.) of three independent experiments was determined by scanning the corresponding bands, using ChemiDoc Touch Imaging System (Bio-Rad, Hercules, CA), and depicted as mean +/− SD of triplicates.
4. Discussion
Our data demonstrated that cellular protein, USP15, regulates degradation of the intracellular viral proteins in HIV-1 replicating cells using the UPS, and that HIV-1 Nef interacts with and determines the fate of USP15, suggesting that Nef and USP15 play a pivotal role for inter-regulation of HIV-1/host cell competition in the infected cells, which may be integral to AIDS progression. Further, while the amount of intracellular Nef and USP15 was mutually regulated, USP15-mediated degradation of Nef was stronger than Nef-mediated USP15 degradation, implying that USP15 could be employed to knock out Nef, a molecule essential to pathogenicity, within HIV-1 infected cells.
Our WB analysis using anti-p24 antibody showed that significantly more viral proteins remained in the infected Jurkat cells than in the virion particles at peak virus replication (Fig. 1). These data suggest that this large percentage of intracellular viral proteins in the host is critical for competition between the infecting HIV-1 and the infected host cells. Accordingly, USP15 degraded key intracellular viral proteins, such as Nef and Gag in the HIV-1-replicating cells and thereby significantly impaired HIV-1 replication, and Nef induced decay of USP15 which is known to stabilize cellular proteins (Aggarwal and Massague, 2012; Cayli et al., 2009; Inui et al., 2011; Soboleva et al., 2005). These data evinced that Nef and USP15 are key players in governing intracellular viral and cellular protein stability and thus the HIV-1/host cell competition, determining the course of disease.
Our data indicated that USP15 clearly induced degradation of Nef and other viral structural protein, Gag, in the repeated experiments. How, then, can USP15 specifically target viral proteins for decay, while stabilizing cellular proteins in the HIV-1-infected cells? One conceivable mechanism is that USP15 comprises two distinct functional domains, wherein one domain is critical for blocking degradation of cellular proteins, while another motif is important for degradation of foreign pathogenic molecules to restrict replication of the invading pathogen, e.g. HIV-1. This possibility can be tested by introducing mutations in the USP15 gene and by assessing the effect of the mutations on specific viral protein degradation. Alternatively, USP15 could activate other cellular protein(s) in the USP15/Nef complex, and the activated protein(s) in turn degrades Nef protein. We accordingly screened for a potential cellular component involved in the UPS-mediated protein degradation by interacting with Nef, using the yeast- followed by the mammalian-two-hybrid assays (Kalpana et al., 1994; Pyeon and Park, 2015). We found that Nef binds to ubiquitin-protein ligase E3A (UBE3A/E6AP) which induces protein degradation by attaching Ub to substrates (Bernassola et al., 2008; Vande Pol and Klingelhutz, 2013), i.e. Nef associated with two functionally antagonistic proteins in the UPS-mediated protein degradation processes, suggesting that UBE3A could be a major cellular component in regulating USP15-mediated viral protein degradation by interacting with Nef and USP15 simultaneously or independently. These possibilities remain to be studied.
USP15 degraded not only Nef but also Gag, and the USP15-mediated inhibitory effect on wt-HIV-1 replication was more pronounced than on Δnef-HIV-1 replication (Fig. 7). These data indicate that Nef expressed from the wt-HIV-1-, but not from Δnef-HIV-1-replicating cells, degraded USP15, lowering the amount of intracellular USP15, which in turn vitiated USP15-induced degradation of viral proteins and thereby HIV-1 replication, where Nef degrades USP15, but USP15-mediated Nef degradation is more potent (Fig. 3). The observed changes in the amount of viral proteins were not due to the inhibition of viral gene expression but to the intracellular degradation, since luciferase activity of HIV-1 LTR FLuc was not diminished by expression of USP15 (data not shown).
Our observations raise the question of how USP15 would mediate degradation of Gag. From earlier findings that Nef directs Gag to endosomes (Sandrin and Cosset, 2006), and USP15 and Nef are largely omnipresent in the cytosolic compartments (Fig. 6), one may postulate that USP15 alone or together with Nef degrades the structural protein via both the enodosomal and proteosomal degradation routes. It would be interesting to investigate on whether and how Nef regulates USP15-triggerred degradation of Gag. In corroboration, the observed degradation of p24 by USP15 (Fig. 8A) was thoroughly abrogated, when the transfected cells were treated with MG132 (Fig. 8B), indicating that the proteosomal degradation pathway acts on the USP15-mediated degradation of Gag protein. Remarkably, the band intensity of p24 degraded by USP15 was visibly weaker (Fig. 8C vs A), when the cells were treated with Bafilomycin A1. These data establish that USP15-mediated p24 degradation is achieved via both endosomal and proteosomal degradation.
These data collectively demonstrated that stability of Nef and USP15 is regulated reciprocally, and that USP15-mediated degradation of Nef was more pronounced than Nef-mediated degradation of USP15. Further, Nef, but not Gag, degraded USP15, suggesting that reciprocal degradation of Nef and USP15 could play a central role in coordinating decay of viral proteins and hence HIV-1 replication, underlining the dynamic competition between the molecular determinants of the infecting HIV-1 and the infected host cells. Going forward, to maximize biomedical relevance, these experiments should also be conducted in HIV-1-susceptible primary and established CD+4 T cells, though there could be drawbacks as well as advantages to multifarious host-dependent Nef behaviors that may arise in different cell lines. Additionally, our results suggest the need to examine whether blockage of endogenous expression of USP15 reverses USP15-mediated degradation of viral proteins and thus the impairment of HIV-1 replication. We believe that by elucidating the detailed structural basis for Nef/USP15-corrdinated protein degradation, we can improve our understanding of the competing viral pathogenicity and cellular defense strategies setting the course of disease, and we can retrieve key molecular targets for enhancing therapeutics against AIDS.
Highlights.
HIV-1 Nef and USP15 interacted and reciprocally regulated their stability.
Association of Nef with USP15 is critical for the reciprocal regulation of decays of the Proteins.
USP15 also degraded not only Nef but also viral structural protein and thus impaired HIV-1 replication.
Acknowledgments
We thank Lenore Price for editing this manuscript. This work was supported by the NIH/NIDDK R01 DK099055 (I.-W. P).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors have declared that no competing interests exist.
Ethical approval: This article does not contain any studies with human participants performed by any of the authors.
References
- Aggarwal K, Massague J. Ubiquitin removal in the TGF-beta pathway. Nat Cell Biol. 2012;14(7):656–657. doi: 10.1038/ncb2534. [DOI] [PubMed] [Google Scholar]
- Allan JS, Coligan JE, Lee TH, McLane MF, Kanki PJ, Groopman JE, Essex M. A new HTLV-III/LAV encoded antigen detected by antibodies from AIDS patients. Science. 1985;230(4727):810–813. doi: 10.1126/science.2997921. [DOI] [PubMed] [Google Scholar]
- Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14(1):10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Bukovsky AA, Dorfman T, Weimann A, Gottlinger HG. Nef association with human immunodeficiency virus type 1 virions and cleavage by the viral protease. J Virol. 1997;71(2):1013–1018. doi: 10.1128/jvi.71.2.1013-1018.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CY, Zhang X, Sinko PJ, Burakoff SJ, Jin YJ. Two sorting motifs, a ubiquitination motif and a tyrosine motif, are involved in HIV-1 and simian immunodeficiency virus Nef-mediated receptor endocytosis. J Immunol. 2011;186(10):5807–5814. doi: 10.4049/jimmunol.1003506. [DOI] [PubMed] [Google Scholar]
- Cayli S, Klug J, Chapiro J, Frohlich S, Krasteva G, Orel L, Meinhardt A. COP9 signalosome interacts ATP-dependently with p97/valosin-containing protein (VCP) and controls the ubiquitination status of proteins bound to p97/VCP. J Biol Chem. 2009;284(50):34944–34953. doi: 10.1074/jbc.M109.037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill M, Sterjovski J, Gray L, Cowley D, Chatfield C, Learmont J, Sullivan JS, Crowe SM, Mills J, Brew BJ, Wesselingh SL, McPhee DA, Gorry PR. Longitudinal analysis of nef/long terminal repeat-deleted HIV-1 in blood and cerebrospinal fluid of a long-term survivor who developed HIV-associated dementia. J Infect Dis. 2004;190(12):2181–2186. doi: 10.1086/425585. [DOI] [PubMed] [Google Scholar]
- Churchill MJ, Rhodes DI, Learmont JC, Sullivan JS, Wesselingh SL, Cooke IR, Deacon NJ, Gorry PR. Longitudinal analysis of human immunodeficiency virus type 1 nef/long terminal repeat sequences in a cohort of long-term survivors infected from a single source. J Virol. 2006;80(2):1047–1052. doi: 10.1128/JVI.80.2.1047-1052.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucchiarini M, Cagnon L, Giordanengo V, Doglio A, Lefebvre JC. Induction by human immunodeficiency viruses types 1 and 2 of degradation of CD4 but not of a CD4 mutant unable to bind viral envelope glycoproteins. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;8(5):427–436. [PubMed] [Google Scholar]
- Cullen BR. Regulation of HIV-1 gene expression. FASEB J. 1991;5(10):2361–2368. doi: 10.1096/fasebj.5.10.1712325. [DOI] [PubMed] [Google Scholar]
- Dang Y, Wang X, Esselman WJ, Zheng YH. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol. 2006;80(21):10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikeakos JD, Thomas L, Kwon G, Elferich J, Shinde U, Thomas G. An interdomain binding site on HIV-1 Nef interacts with PACS-1 and PACS-2 on endosomes to down-regulate MHC-I. Mol Biol Cell. 2012;23(11):2184–2197. doi: 10.1091/mbc.E11-11-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube M, Bego MG, Paquay C, Cohen EA. Modulation of HIV-1-host interaction: role of the Vpu accessory protein. Retrovirology. 2010;7:114. doi: 10.1186/1742-4690-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fackler OT, Kienzle N, Kremmer E, Boese A, Schramm B, Klimkait T, Kucherer C, Mueller-Lantzsch N. Association of human immunodeficiency virus Nef protein with actin is myristoylation dependent and influences its subcellular localization. Eur J Biochem. 1997;247(3):843–851. doi: 10.1111/j.1432-1033.1997.00843.x. [DOI] [PubMed] [Google Scholar]
- Ferguson MR, Rojo DR, von Lindern JJ, O’Brien WA. HIV-1 replication cycle. Clin Lab Med. 2002;22(3):611–635. doi: 10.1016/s0272-2712(02)00015-x. [DOI] [PubMed] [Google Scholar]
- Franchini G, Robert-Guroff M, Ghrayeb J, Chang NT, Wong-Staal F. Cytoplasmic localization of the HTLV-III 3′ orf protein in cultured T cells. Virology. 1986;155(2):593–599. doi: 10.1016/0042-6822(86)90219-9. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Hanna Z, Kay DG, Rebai N, Guimond A, Jothy S, Jolicoeur P. Nef harbors a major determinant of pathogenicity for an AIDS-like disease induced by HIV-1 in transgenic mice. Cell. 1998;95(2):163–175. doi: 10.1016/s0092-8674(00)81748-1. [DOI] [PubMed] [Google Scholar]
- Haseltine WA. Molecular biology of the human immunodeficiency virus type 1. FASEB J. 1991;5(10):2349–2360. doi: 10.1096/fasebj.5.10.1829694. [DOI] [PubMed] [Google Scholar]
- Inui M, Manfrin A, Mamidi A, Martello G, Morsut L, Soligo S, Enzo E, Moro S, Polo S, Dupont S, Cordenonsi M, Piccolo S. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat Cell Biol. 2011;13(11):1368–1375. doi: 10.1038/ncb2346. [DOI] [PubMed] [Google Scholar]
- Jia B, Serra-Moreno R, Neidermyer W, Rahmberg A, Mackey J, Fofana IB, Johnson WE, Westmoreland S, Evans DT. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 2009;5(5):e1000429. doi: 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph AM, Kumar M, Mitra D. Nef: “necessary and enforcing factor” in HIV infection. Curr HIV Res. 2005;3(1):87–94. doi: 10.2174/1570162052773013. [DOI] [PubMed] [Google Scholar]
- Kalpana GV, Marmon S, Wang W, Crabtree GR, Goff SP. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266(5193):2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- Kaminchik J, Margalit R, Yaish S, Drummer H, Amit B, Sarver N, Gorecki M, Panet A. Cellular distribution of HIV type 1 Nef protein: identification of domains in Nef required for association with membrane and detergent-insoluble cellular matrix. AIDS Res Hum Retroviruses. 1994;10(8):1003–1010. doi: 10.1089/aid.1994.10.1003. [DOI] [PubMed] [Google Scholar]
- Kammula EC, Motter J, Gorgels A, Jonas E, Hoffmann S, Willbold D. Brain transcriptomewide screen for HIV-1 Nef protein interaction partners reveals various membrane-associated proteins. PLoS One. 2012;7(12):e51578. doi: 10.1371/journal.pone.0051578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kestler HW, 3rd, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, Desrosiers RC. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65(4):651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332(4):228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- Kueck T, Neil SJ. A cytoplasmic tail determinant in HIV-1 Vpu mediates targeting of tetherin for endosomal degradation and counteracts interferon-induced restriction. PLoS Pathog. 2012;8(3):e1002609. doi: 10.1371/journal.ppat.1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutluay SB, Perez-Caballero D, Bieniasz PD. Fates of retroviral core components during unrestricted and TRIM5-restricted infection. PLoS Pathog. 2013;9(3):e1003214. doi: 10.1371/journal.ppat.1003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindwasser OW, Chaudhuri R, Bonifacino JS. Mechanisms of CD4 downregulation by the Nef and Vpu proteins of primate immunodeficiency viruses. Curr Mol Med. 2007;7(2):171–184. doi: 10.2174/156652407780059177. [DOI] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451(7177):425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Neri F, Giolo G, Potesta M, Petrini S, Doria M. CD4 downregulation by the human immunodeficiency virus type 1 Nef protein is dispensable for optimal output and functionality of viral particles in primary T cells. J Gen Virol. 2011;92(Pt 1):141–150. doi: 10.1099/vir.0.026005-0. [DOI] [PubMed] [Google Scholar]
- Park IW, Fan Y, Luo X, Ryou MG, Liu J, Green L, He JJ. HIV-1 Nef is transferred from expressing T cells to hepatocytic cells through conduits and enhances HCV replication. PLoS One. 2014;9(6):e99545. doi: 10.1371/journal.pone.0099545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IW, Ndjomou J, Wen Y, Liu Z, Ridgway ND, Kao CC, He JJ. Inhibition of HCV replication by oxysterol-binding protein-related protein 4 (ORP4) through interaction with HCV NS5B and alteration of lipid droplet formation. PLoS One. 2013;8(9):e75648. doi: 10.1371/journal.pone.0075648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyeon D, Park IW. Interaction between Nef and INI1/SMARCB1 augments replicability of HIV-1 in resting human peripheral blood mononuclear cells. Arch Virol. 2015;160(3):727–737. doi: 10.1007/s00705-014-2315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma R, Noser JA, Ohmine S, Ikeda Y. Rhesus monkey TRIM5alpha restricts HIV-1 production through rapid degradation of viral Gag polyproteins. Nat Med. 2007;13(5):631–635. doi: 10.1038/nm1562. [DOI] [PubMed] [Google Scholar]
- Sandrin V, Cosset FL. Intracellular versus cell surface assembly of retroviral pseudotypes is determined by the cellular localization of the viral glycoprotein, its capacity to interact with Gag, and the expression of the Nef protein. J Biol Chem. 2006;281(1):528–542. doi: 10.1074/jbc.M506070200. [DOI] [PubMed] [Google Scholar]
- Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, Takehisa J, Ogando Y, Ochsenbauer C, Kappes JC, Ayouba A, Peeters M, Learn GH, Shaw G, Sharp PM, Bieniasz P, Hahn BH, Hatziioannou T, Kirchhoff F. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe. 2009;6(5):409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler M, Rajan D, Specht A, Ritter C, Pulkkinen K, Saksela K, Kirchhoff F. Association of Nef with p21-activated kinase 2 is dispensable for efficient human immunodeficiency virus type 1 replication and cytopathicity in ex vivo-infected human lymphoid tissue. J Virol. 2007;81(23):13005–13014. doi: 10.1128/JVI.01436-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Moreno R, Zimmermann K, Stern LJ, Evans DT. Tetherin/BST-2 Antagonism by Nef Depends on a Direct Physical Interaction between Nef and Tetherin, and on Clathrin-mediated Endocytosis. PLoS Pathog. 2013;9(7):e1003487. doi: 10.1371/journal.ppat.1003487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Lau D, Noviello CM, Ghosh P, Guatelli JC. An MHC-I cytoplasmic domain/HIV-1 Nef fusion protein binds directly to the mu subunit of the AP-1 endosomal coat complex. PLoS One. 2009;4(12):e8364. doi: 10.1371/journal.pone.0008364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboleva TA, Jans DA, Johnson-Saliba M, Baker RT. Nuclear-cytoplasmic shuttling of the oncogenic mouse UNP/USP4 deubiquitylating enzyme. J Biol Chem. 2005;280(1):745–752. doi: 10.1074/jbc.M401394200. [DOI] [PubMed] [Google Scholar]
- Sugiyama R, Naganuma H, Nishitsuji H, Takaku H. Human immunodeficiency virus-1 Nef suppresses Hsp70-mediated Tat activation. FEBS Lett. 2011;585(21):3367–3371. doi: 10.1016/j.febslet.2011.09.029. [DOI] [PubMed] [Google Scholar]
- Timani KA, Liu Y, Suvannasankha A, He JJ. Regulation of ubiquitin-proteasome system-mediated Tip110 protein degradation by USP15. Int J Biochem Cell Biol. 2014;54:10–19. doi: 10.1016/j.biocel.2014.06.017. [DOI] [PubMed] [Google Scholar]
- Vande Pol SB, Klingelhutz AJ. Papillomavirus E6 oncoproteins. Virology. 2013;445(1–2):115–137. doi: 10.1016/j.virol.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Felsted RL. Effect of myristoylation on p27 nef subcellular distribution and suppression of HIV-LTR transcription. Virology. 1992;187(1):46–55. doi: 10.1016/0042-6822(92)90293-x. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, Johnson MC, Munch J, Kirchhoff F, Bieniasz PD, Hatziioannou T. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe. 2009;6(1):54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15(9):871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]


