Abstract
The development of a versatile technique to induce RNA interference (RNAi) without immune stimulation in vivo is of interest as existing approaches to trigger RNAi, such as small interfering RNA (siRNA) and plasmid DNA (pDNA) expressing short hairpin RNA (shRNA), present drawbacks arising from innate immune stimulation. To overcome them, an intelligent shRNA expression device (iRed) designed to induce RNAi was developed. The minimum sequence of iRed encodes only the U6 promoter and shRNA. A series of iRed comprises a polymerase chain reaction (PCR)-amplified 4′-thioDNA in which any one type of adenine (A), guanine (G), cytosine (C), or thymine (T) nucleotide unit was substituted by each cognate 4′-thio derivatives, i.e., dSA iRed, dSG iRed, dSC iRed, and ST iRed respectively. Each modified iRed acted as a template to transcribe shRNA with RNAi activity. The highest shRNA yield was generated using dSC iRed that exerted gene silencing activity in an orthotopic mouse model of mesothelioma. Reducing the minimal structure required to transcribe shRNA and the presence of the 4′-thiomodification synergistically function to abrogate innate immune response induced by dsDNA. The iRed will introduce a new approach to induce RNAi without inducing a detectable innate immune response.
Keywords: 4′-thioDNA, innate immune response, RNA interference, shRNA, TLR9
Introduction
RNA interference (RNAi) is the standard method to suppress gene expression because of its target specificity, potency, and ability to silence the expression of virtually any gene. Using a small interfering RNA (siRNA),1 which comprises a 21-mer, is the general approach used to induce RNAi because it can be easily prepared using a DNA/RNA synthesizer. Synthetic siRNA can be chemically modified to increase the potency of RNAi activity and to abrogate the innate immune stimulation.2,3 Accordingly, siRNAs have been used as biological tools and new drug candidates.4 Plasmid DNA (pDNA) expressing short hairpin RNA (shRNA) can also be used to induce RNAi.5,6,7,8 pDNA produces numerous shRNAs that induce RNAi with potent and long-term RNAi activity, even if only one pDNA molecule is delivered to the nucleus. However, it has some drawbacks in therapeutic use, such as low pDNA transfection efficiency arising from its huge molecular size and innate immune responses induced by extra genes, such as CpG motifs.9,10,11,12 To overcome these drawbacks, minimally-sized plasmid vectors transcribing shRNAs have been developed.13,14,15 These vectors effectively suppress targeted gene expression in vitro. To the best of our knowledge, there are no reports of their use in vivo. Moreover, it is unclear whether pDNA downsizing itself can diminish the innate immune responses leading to systematic adverse effects.
Meanwhile, our groups have explored a series of 4′-thionucleic acids with sulfur atoms at the 4′-position instead of oxygen atoms. We reported that both 4′-thioRNA and 4′-thioDNA exhibit not only increased hybridization and nuclease resistance compared with unmodified molecules16,17,18 but are biologically equivalent to naturally occurring RNA and DNA because sulfur and oxygen atoms belong to the same group in the periodic table. Thus, a series of 4′-thioRNAs were utilized as siRNAs in gene silencing via RNAi19,20,21 and/or anti-microRNA oligonucleotides.22,23 Concerning 4′-thioDNA, we could amplify a highly modified 4′-thioDNA using 2′-deoxy-4′-thionucleoside 5′-triphosphates (dSNTPs),15,24 and we used the polymerase chain reaction (PCR)-amplified 4′-thioDNA products as templates to synthesize natural RNAs in vitro.15,25 These results prompted us to develop a new artificial DNA device to produce RNAi inducers, that we call the intelligent shRNA expression device (iRed), which is composed of 4′-thioDNA (Figure 1). If shRNAs are transcribed from the iRed, similar to natural dsDNA, RNAi activities should be expected. Further, we predicted that the iRed prevents undesired immune responses due to the synergistic effect of downsizing and chemical modifications using 2′-deoxy-4′-thioribonucleoside units as chemically-modified siRNA did. Here we describe the development of the iRed as an alternative producer of shRNA and show that the shRNAs transcribed from iRed in HeLa cells silenced gene expression. We succeeded in inducing RNAi using an iRed in mice without inducing a detectable innate immune response.
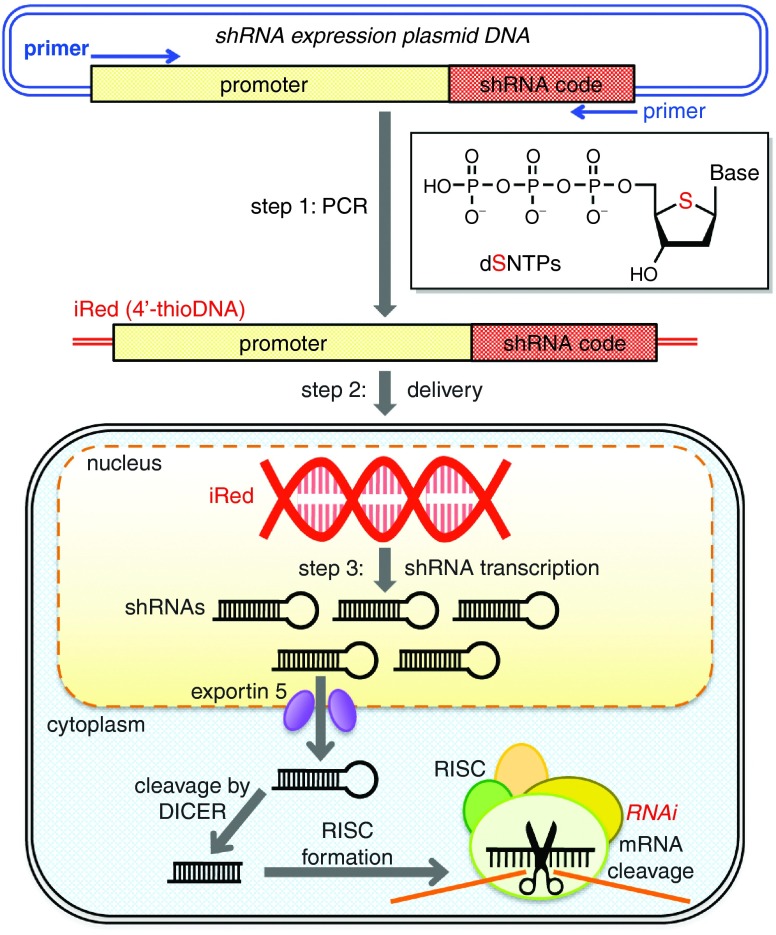
Figure 1.
Schematic of gene silencing using an intelligent shRNA expression device (iRed). Step1: A region of plasmid DNA (pDNA) encoding the U6 promoter and a short hairpin RNA (shRNA) was amplified using PCR in the presence of one type of 2'-deoxy-4'-thionucleoside triphosphate (dSNTP) and three other dNTPs to synthesize an iRed. Step 2: The iRed was delivered into the nucleus. Step 3: Numerous shRNAs were transcribed from the iRed to possess a potent RNA interference (RNAi) activity.
Results
Construction of iRed consisting of 4′-thioDNA
We first attempted the construction of iRed. As shown in Figure 1, the iRed contained only an shRNA transcriptional unit encoded by the pDNA, which comprised the promoter and shRNA coding sequences. In our previous study, we reported PCR amplification in the presence of each dSNTP that generated 104 base pairs (bp) of double-stranded (ds) 4′-thioDNA.24 Using similar PCR conditions and one type of dSNTP to substitute the cognate dNTP, the sequences encoding the U6 promoter and an shRNA specific for pGL2 firefly luciferase (Fluc) were amplified to produce a series of iReds as follows: dSA iRed, dSG iRed, ST iRed, and dSC iRed (Supplementary Figure S1a). The size of the iRed with 4′-thiomodification was 362 bp compared with 6,503 bp of the parental pDNA. The incorporation of 209 residues of 2′-deoxy-4′-thioadenosine (dSA) into dSA iRed and of 212 residues of ST in ST iRed were required in every PCR cycle. In contrast, 113 and 118 residues of dSG in dSG iRed and dSC in dSC iRed were incorporated, respectively. However, there was no correlation between the PCR amplification yields and the number of incorporated 4′-thionucleoside residues (Supplementary Figure S1). Although dSG iRed and dSC iRed contained similar numbers of 4′-thiomodifications, the latter was the most efficient. The preparation of an iRed with greater than two types of 4′-thio units was also attempted but was unsuccessful because the iRed sequence was much longer compared with that of a 104 bp template that we previously used (data not shown).24 Therefore, we decided to use 4′-thioDNAs containing one type of 2′-deoxy-4′-thionucleoside unit to produce the iReds.
The iRed suppressed target gene expression via RNAi machinery in HeLa cells
To evaluate in vitro gene silencing activity, the resulting iReds specific for pGL2 Fluc were cotransfected with pGL2 Fluc and Renilla luciferase (Rluc) pDNAs into HeLa cells. In Figure 2a, the gene silencing activities of each iRed at 24, 48, and 72 hours post-transfection are shown with those of pDNA, synthetic shRNA, and PCR-amplified natural dsDNA having the same sequence as the iRed (called the natural device). pDNA had a high potency and continued to suppress pGL2 Fluc gene expression by 95% from 24 to 72 hours post-transfection into HeLa cells at 1.0 nmol/l. Under equimolar conditions, shRNA reduced target gene expression, although its potency was less than that of pDNA. The iReds suppressed pGL2 Fluc gene expression with different efficiencies depending on the variations in the 4′-thiomodification. The dSA iRed and ST iRed, having more than 200 units of 4′-thionucleotide in their sequences, showed moderate activities, which increased over time post-transfection. In contrast, the dSG iRed and dSC iRed having approximately 100 units of 4′-thionucleotide strongly silenced luciferase gene expression even at 24 hours post-transfection; their activities were maintained up to 72 hours post-transfection. The RNAi activities of the dSG iRed and dSC iRed were more potent than those of synthetic shRNA. The most potent RNAi activity was observed when the dSC iRed was used.
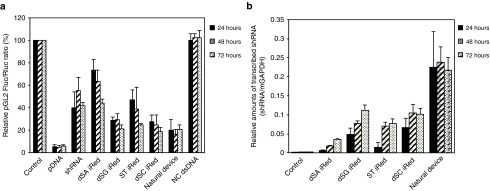
Figure 2.
Evaluation of silencing of pGL2 firefly luciferase (Fluc) activity. (a) RNAi activities of iReds compared with those of pDNA, shRNA, and dsDNA with the same sequence as the iRed (natural device). HeLa cells were transfected with each construct at 1.0 nmol/l. (b) The amounts of shRNAs transcribed from iReds and the natural device. The shRNAs were extracted at the indicated times after transfection (1.0 nmol/l, each construct). Error bars indicate the standard deviations of three independent experiments.
To further validate that iReds reduced target gene expression via RNAi, the amounts of shRNAs transcribed from each iRed were determined using reverse-transcription quantitative PCR (Figure 2b).26 The natural device acted as a genetic template to produce shRNA. Similarly, the iReds consisting of 4′-thioDNA also yielded shRNA, although their amounts were less than those of the natural device at 24 hours post-transfection. However, continued monitoring revealed that the amounts of shRNAs transcribed from the iReds increased over time post-transfection up to 72 hours. In addition, these accretions of shRNA production could result in long-term RNAi activities of iReds (Figure 2a).
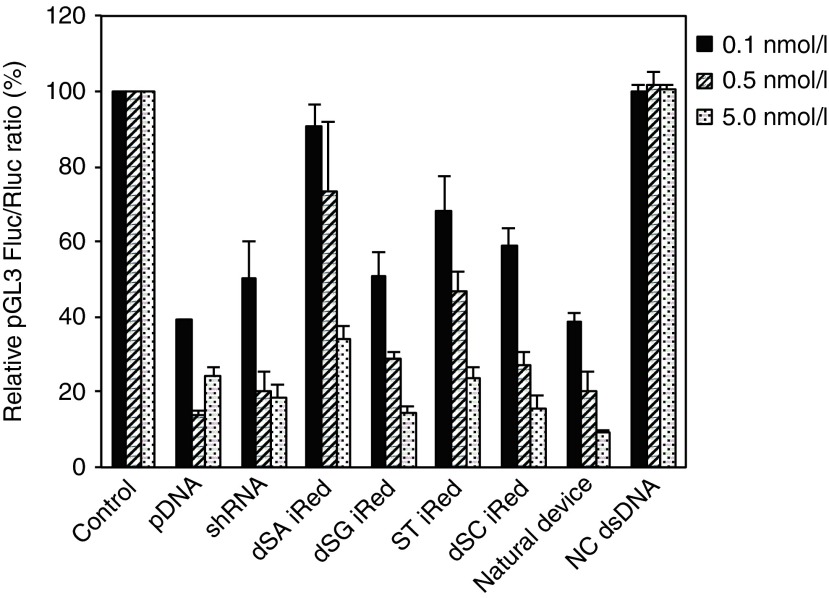
Theoretically, a new artificial DNA used to produce shRNA can suppress the expression of any gene. To validate this, we applied the iRed to the pGL3 Fluc target. The iReds targeting pGL3 Fluc were prepared similarly to those targeting pGL2 Fluc by PCR in the presence of a mixture of one type of dSNTP and the three other dNTPs (Supplementary Figure S2), and their RNAi activities were evaluated in HeLa cells. All iReds containing each modification pattern reduced pGL3 Fluc gene expression in a dose-dependent manner, similar to pDNA and synthetic shRNA (Figure 3). Their relative potencies of RNAi activity, which may depend on the modification patterns, were quite similar to those targeting pGL2 Fluc. To examine the target specificity, the iReds targeting pGL2 Luc were transfected into the HeLa cells expressing pGL3 Luc. The iReds targeting pGL2 Luc did not cause a significant suppression of pGL3 Luc activity at 1.0 nmol/l post-transfection. This indicates that our new, artificial DNA can suppress target gene expression with high specificity, even in a highly homologous gene expression (Supplementary Figure S3).
Figure 3.
Dose-dependent RNAi activities of iReds targeting pGL3 Fluc compared with those of the pDNA, synthetic shRNA, and natural device. HeLa cells were transfected with all samples at the indicated concentrations. Error bars indicate the standard deviations of three independent experiments.
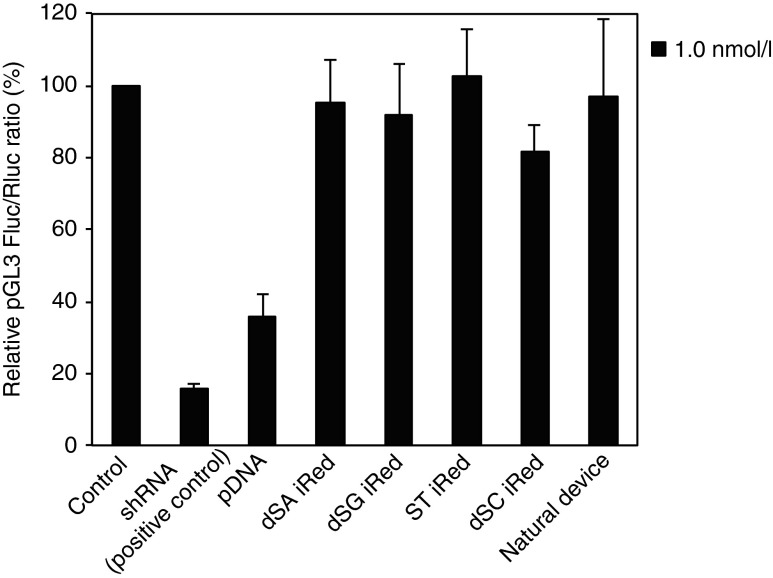
The RNA polymerase III-dependent RNA transcription is initiated at the U6 promoter sequence and terminated at the small polyT transcription terminator.27 Accordingly, shRNAs having poly(U) sequences longer than five nucleotides are not transcribed using U6-promoter transcription system. To ensure that this termination in RNA polymerase III-dependent transcription was conserved even in case of 4′-thioDNA in the iRed, we prepared another series of iReds with a poly(U) stretch in the middle of the shRNA coding regions, which targeted pGL3 Fluc (Supplementary Figure S4). We compared their RNAi activities with those of pDNA, the natural device, and synthetic shRNA (Figure 4) and found that none of the modified iReds reduced Fluc activity. Contrary to our expectations, we found that the pDNA silenced pGL3 Fluc gene expression, similar to shRNA, indicating that a systematic adverse effect caused off-target silencing. Our strategy to induce RNAi using iReds showed highly conserved sequence-specific RNAi activity.
Figure 4.
Evaluation of RNAi activities of the iReds with a transcription terminator in the middle of the shRNA coding region. RNAi activities of each iRed were compared with those of the pDNA, synthetic shRNA and dsDNA with the same sequences as the iRed. Error bars indicate the standard deviations of three independent experiments.
The iRed consisting of 4′-thioDNA acted as genetic template for transcribing shRNA even in vivo
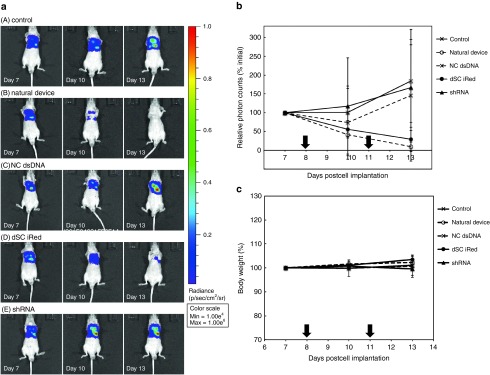
Our in vitro studies showed that the iReds have great potential to induce RNAi silencing compared with synthetic shRNA against both pGL2 and pGL3 Fluc. To determine silencing activity in vivo, the gene silencing efficacy of dSC iRed targeting pGL3 Fluc (Supplementary Figure S2) was evaluated using an orthotopic malignant pleural mesothelioma mouse model. The malignant pleural mesothelioma mouse model was prepared by injecting human pleural mesothelioma cells (MSTO-211H) expressing Fluc (MSTO-211H-Luc) into the left pleural cavity of BALB/c nu/nu mice. Tumor development was monitored using in vivo imaging system (IVIS). On days 8 and 11 after the cells were inoculated, the model mice were intrapleurally injected with equimolecular amounts of the natural device, negative control dsDNA (NC dsDNA), dSC iRed, or synthetic shRNA each specific for pGL3 Fluc as a complex with a transfection reagent designed for use in vivo. On days 7, 10, and 13 after the inoculation, we used IVIS to determine the luciferase activities of pleural tumors.
The bioluminescence of the pleural cavities of untreated control mice was the highest on day 13 after inoculation, indicating aggressive growth of the inoculated cells (Figure 5a,b). Similar luciferase activities were detected that localized with the pleural tumors of mice injected with NC dsDNA or shRNA. In contrast, the dSC iRed as well as natural device markedly suppressed luciferase activity to approximately the same extent, altough a statistical significance of difference was not recognized between a control group/administration groups, because of slightly high value of standard deviation in a control group. Furthermore, as shown in Figure 5c, the mice in all tested groups did not show a significant decrease in body weight, indicating that the treatment samples containing the dSC iRed had less systemic adverse effects following intrapleural administration.
Figure 5.
In vivo evaluation of RNAi activities. Mice that served as an orthotopic model of malignant pleural mesothelioma were intrapleurally administered two doses of either the natural device, negative control dsDNA (NC dsDNA), iRed, or synthetic shRNA once every 3 days. On days 7, 10, and 13 after inoculation, the bioluminescence of the pleural cavities was observed using an in vivo imaging system. (a) Bioluminescence intensities. (b) Time course of RNAi activity of each sample. Data are expressed as the mean of relative photon counts to that of the initial (%). (c) The body weights of the mice were determined from day 7 to 13 after inoculation of the transfected cells. Data are represented as the mean of % initial body weight ± standard deviation. The arrows indicate the administration of samples. The error bars indicate the standard deviation of three or more independent experiments.
Scarcely did the administration of iRed induce innate immune stimulation in vivo
As described in the Introduction, the iReds would have a great advantage not only in inducing RNAi but also in abrogating undesired immune stimulation. The development of a versatile technique for inducing RNAi without immune stimulation in vivo is of great interest because approaches for RNAi therapeutics using pDNA present issues arising from immune stimulation.9,10,11,12
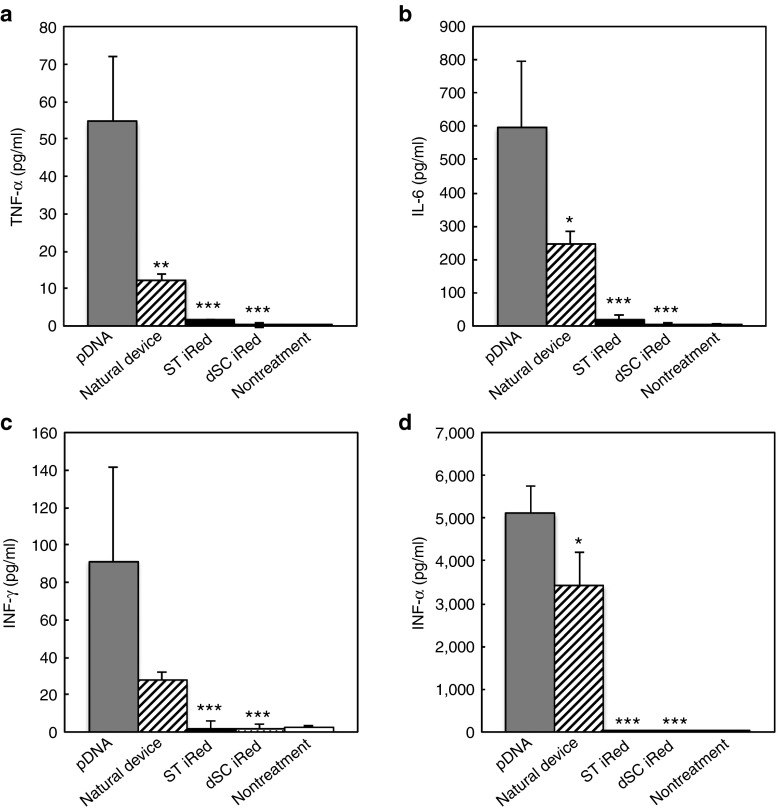
Therefore, we next determined whether the iReds induced an immune response. For this purpose, we used BALB/c mice injected intravenously with of 20 μg of pDNA, natural device, ST iRed, or dSC iRed complexed with PEGylated cationic liposomes (lipoplex) to improve blood circulation time and exposure to cells. The average diameters and ζ-potentials of the PEGylated lipoplexes containing iReds with 4′-thioDNA were not significantly different from those that contained the natural device (Supplementary Table S3). At 4 hours after injection, proinflammatory cytokines (TNF-α, IL-6, and INF-γ) and type-I interferon (INF-α) induction levels in the serum were evaluated. pDNA treatment intensely stimulated immune responses in TNF-α, IL-6, or INF-γ, and INF-α (Figure 6). Similarly, the natural device showed moderate cytokine levels, indicating that only pDNA downsizing could not completely abrogate innate immune stimulation. In particular, similar levels of INF-α were induced by the natural device and pDNA. In contrast, dSC iRed and dST iRed induced lower levels of these cytokines, although they also contained CpG motifs. We therefore conclude that the downsizing and 4′-thiomodification synergized to abrogate the innate immune response to pDNA. Thus, the innate immune systems of these mice distinguished 4′-thioDNA from natural DNA when dC or T nucleotides were substituted by corresponding 2′-deoxy 4′-thionucleotide.
Figure 6.
Evaluation of innate immune response induced by PEGylated lipoplexes each containing 20 μg of the pDNA, natural device, ST iRed, or dSC iRed. Induction levels of (a) TNF-α, (b) IL-6, (c) INF-γ, and (d) INF-α. Each sample was designed to transcribe a shRNA targeting pGL2 Fluc. Four hours after intravenous injection, the serum level of each molecular was determined using an enzyme-linked immunosorbent assay. Error bars indicate the standard deviations of three independent experiments. The P values indicate the statistical difference between the mice treated with pDNA and the mice treated with natural device, ST iRed or dSC iRed. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
The pDNA expressing shRNA in cells, that induce long-term RNAi activity, has become an important and widely used approach in biological studies. However it still presents some drawbacks arising from innate immune response toward the therapeutic use. To overcome them, we explored RNAi gene silencing using iReds consisting of 4′-thioDNA. PCR amplification including one type of dSNTP produced c.a. 360 bp 4′-thioDNAs encoding the U6 promoter and a shRNA corresponding to that of the iRed, indicating that the 4′-thioDNA was a good substrate for a DNA polymerase in vitro. In this way, the downsizing and chemical modification of iRed were achieved in one pot. In our examination, there was no correlation between the PCR amplification yields and number of incorporated 4′-thionucleoside residues (Supplementary Figure S1b,c). Further investigations will be required to determine the superiority of the incorporation of each dSNTP and optimize the PCR conditions in the presence of more than two types of dSNTPs.
The transfection of HeLa cells with equimolar amounts of each Fluc-specific iRed suppressed luciferase expression more potently compared with a synthetic shRNA (Figures 2 and 3). HeLa cells transfected with dSC iReds yielded relatively large quantities of shRNA (Figure 2b). Although the amounts of shRNAs yielded from dSC iReds were less than that of natural device, the RNAi activity of dSC was equal to that of natural device because RNA-induced silencing complexes (RISCs) are recycled and proceed through several rounds of mRNA cleavage events. We considered that the amount of shRNA yielded from dSC iRed was proper, but that from natural device might be surplus. These results indicate that 4′-thioDNA could act as an artificial genetic template to synthesize naturally occurring RNA in organisms. In addition, careful evaluation revealed that the sequences of iRed were exactly recognized by RNA polymerase III of mammalian cells to obtain highly specific RNAi activity (Figure 4). Furthermore, dSC iRed exerted potent RNAi activities even in an orthotopic mouse model of mesothelioma without detectable systematic adverse effects, whereas synthetic shRNA showed no silencing activities under the equimolar conditions (Figure 5).
There are several reports evaluating transcriptional process using chemically-modified DNAs used to transfect cell lines or bacteria. For example Krueger et al.28 used a DNA having some size-expanded unnatural base pair (xDNA) and demonstrated that the endogenous enzymes of Escherichia coli accurately transcribe xDNA. Birts et al. found that DNA with one unnatural triazole linkage directs the transcription of mRNAs in eukaryotic cells.29,30 However, there are no report of a sugar modified DNA template. To the best of our knowledge, the present study is the first to show that a highly chemical-modified DNA, dSC iRed, encoded a naturally occurring RNA that exhibited a high level of gene silencing activity in a mouse model.
Furthremore, the administration of dSC iRed or ST iRed did not induce a detectable innate immune response in vivo, although the natural device with the same sequence as the iRed induced the production of proinflammatory cytokines and a type-I interferon. In the case of siRNAs, the incorporation of chemical modifications, particularly 2′-O-methyl (2′-OMe) derivatives, prevent innate immune responses.31,32 However, it is difficult to modify the pDNA structure. Thus, we reasoned that an iRed with 4′-thioDNA may provide a versatile approach to prepare noninflammatory shRNAs in organisms.
The sequence of the natural device encoding the U6 promoter and shRNA still contained CpG motifs having the general formula RRCGYY (where R and Y represent a purine and a pyrimidine, respectively),33 although the length is approximately 5% of that of pDNA. Therefore, the natural device would intensely induce innate immune response. The toll-like receptor 9 (TLR9) plays a critical role in immune stimulation triggered by dsDNA that harbors CpG motifs.34 The crystal structure of the complex between TLR9 and CpG-DNA reveals the CpG sequence and phosphate backbones that are recognized by TLR9.35 The conformational changes in the sugar ring caused by the substitution of O4′ with sulfur may result in the loss of recognition of the iRed–4′-thioDNA through the interaction of TLR9 with the phosphate backbone. Moreover, we have reported that 4′-thioDNA forms an A-form-like duplex structure, whereas natural DNA forms a B-form duplex structure.18 This higher-order structural change may affect recognition of the DNA by the innate immune systems of mice. It is not clear whether 4′-thioDNA acts as an antagonist of TLR9 or is not recognized by TLR9. However, these results suggest a strategy to avoid the induction of an innate immune response by DNAs.
In conclusion, our study showed that the 4′-thioDNA iRed acted as a bioisostere of the natural DNA template for DNA polymerase and RNA polymerase III to synthesize shRNA that exhibited potent gene-silencing activity. Our study is the first to demonstrate that a highly substituted artificial DNA acts as a template both for replication and transcription. The lack of immunogenicity of an iRed indicates its potential as an RNAi therapeutic. Theoretically, the 4′-thoDNA may serve as an artificial template to synthesize not only shRNA but also other functional RNAs, such as micro-RNA and mRNA for translation. Further studies to address these concerns are currently under way. Details of these results will be published elsewhere.
Materials and methods
General. Natural dNTPs were purchased from GE Healthcare Japan (Tokyo, Japan). 2′-Deoxy-4′-thionucleoside triphosphates (dSNTPs) were prepared according to our previous reports.15,24 Oligonucleotides were purchased from FASMAC (Kanagawa, Japan). 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-n-[methoxy-(PEG)-2000] (mPEG2000-DSPE), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine were generously donated by the NOF Corporation (Tokyo, Japan). The animal and ethics review committee of Tokushima University approved the experiments using mice.
Construction of shRNA expression pDNA. Oligonucleotides having shRNA and terminator coding sequences were ligated into the BamHI and EcoRI sites of an shRNA expression cassette containing the U6 promoter (RNAi-Ready pSIREN-RetroQ, Clontech, Mountain View, CA) according to the manufacturer's instructions. The inserted sequences are summarized in Supplementary Table S1.
Preparation of an intelligent shRNA expression device (iRed) and the natural device. The iReds were prepared using the shRNA expression pDNA as a template for PCR as follows. The sequences encoding the U6 promoter and shRNA in pDNA were amplified in 20 μl of KOD buffer containing KOD Dash DNA polymerase (0.05 unit/μl, TOYOBO, Osaka, Japan), pDNA template (0.1 fmol/μl), 200 μmol/l dNTPs, and 0.5 μmol/l of primers. The reaction mixture was gently vortexed, and the DNA was amplified using a thermal cycler. PCR was performed under the following conditions: initial denaturation at 94 °C for 15 seconds, 15 cycles of denaturation/amplification (94 °C, 30 seconds; 62 °C, 30 seconds; 72 °C, 30 seconds), and final extension at 72 °C for 15 minutes. Amplicons were purified using 1% agarose gel electrophoresis, and the DNAs were further purified using a High Pure PCR Product Purification Kit (Roche, Basel, Switzerland) and used as templates for the next round of PCR. The second PCR amplification was performed in 100 μl of reaction buffer containing KOD Dash DNA polymerase (0.1 unit/μl), a template (0.2 fmol/μl), 200 μmol/l of a nucleoside 5′-triphosphate mixture containing one type of dSNTP and three types of dNTPs, and 1.25 μmol/l of primers. The reaction mixture was gently vortexed, and the DNA was amplified using a thermal cycler. PCR was performed under the following conditions: initial denaturation at 94 °C for 15 seconds, 30 cycles of denaturation/amplification (94 °C, 30 seconds; 62 °C, 30 seconds; 72 °C, 10 minutes), and final extension at 72 °C for 15 minutes. The iRed amplicons were purified using a High Pure PCR Product Purification Kit (Roche).
The natural device was prepared using this same protocol but with unmodified dNTPs.
Cell culture. HeLa cells were cultured at 37 °C in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (BioSource International, Camarillo, CA), 100 units/ml penicillin, and 100 μg/ml streptomycin (ICN Biomedicals, Irvine, CA) in an atmosphere containing 5% CO2.
A human pleural mesothelioma cell line (MSTO-211H) expressing pGL3 firefly luciferase (MSTO-211H-Luc) generated by stable transfection with the firefly luciferase (Fluc) gene (pGL3 Basic plasmid, Promega, Madison, WI) was generously supplied by Dr. Masashi Kobayashi (Department of Thoracic Surgery, Faculty of Medicine, Kyoto University). MSTO-211H-Luc was in RPMI-1640 medium (Wako Pure Chemical, Osaka, Japan) supplemented with 10% of heat-inactivated fetal bovine serum (Corning, Corning, NY), 100 units/ml penicillin, and 100 μg/ml streptomycin in an atmosphere containing 5% CO2 at 37 °C. The cells were regularly passaged to maintain exponential growth.
In vitro Luciferase reporter assay. HeLa cells were seeded in a 96-well plate (1 × 104 cells/100 μl per well) in Dulbecco's modified Eagle's medium. After incubation for 24 hours, the cells were cotransfected with equimolar amounts of reporter pDNAs (0.2 μg per well, pGL2-control or pGL3-control, and pRL-TK; Promega), and iRed, shRNA expression pDNA, synthetic shRNA, or the natural device in equimolar amounts using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Each well contained a total volume of 100 μl of Opti-MEM I (Invitrogen). After incubation for 1 hour at 37 °C, the medium was replaced with 100 μl of flesh culture medium containing 10% serum, and the cells were incubated at 37 °C. The cells were washed with phosphate-buffered saline and lysed with Passive Lysis Buffer (Promega) at the indicated times after transfection, and Fluc and Renilla luciferase (Rluc) activities of the cell lysates were measured using the Dual-Luciferase Reporter Assay System (Promega) with an Infinite 200 PRO (TECAN, Männedorf, Switzerland) according to the manufacturer's instructions. Fluc signals were normalized to those of Rluc. The results were expressed as the Fluc/Rluc ratios compared with those of untreated cells. All experiments were performed in triplicate, and the data represent the mean values from at least three assays.
Quantification of shRNA in HeLa cells. The iRed or natural device (1.0 nmol/l each) was used to transfect the HeLa cells in the presence of Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. At the indicated times after transfection, the cells were washed with phosphate-buffered saline, and small RNAs were isolated and resuspended in 50 μl of elution buffer using a mirVana miRNA Isolation Kit (Ambion, Foster City, CA) according to the manufacturer's protocol. Reverse transcription reactions were performed in 25 μl of First Standard Buffer containing an annealed specific primer (0.05 μmol/l) and extracted small RNAs (2.0 μmol/l/total 50 μl) mixture, dNTPs (1.0 mmol/l), Super Script III (2.5 unit/μl, Invitrogen), dithiothreitol (8.5 μmol/l), and RNaseOUT (0.1 unit/μl, Invitrogen). The cDNAs were synthesized as follows: 16 °C for 30 minutes; 60 cycles of synthesis (30 °C, 30 seconds; 42 °C, 30 seconds; 50 °C, 1 minute); and 85 °C for 15 minutes.26 Next, quantitative PCR reactions were performed in 20 μl of reaction mixture containing FASTStart Universal Probe Master (Rox) (Roche), Universal Probe Library (#21) (Roche), and primers (each 900 nmol/l) using a StepOnePlus Real-Time PCR System (Applied Biosystems). The reaction was performed under the following conditions: initial denaturation at 95 °C for 10 minutes, 40 cycles at 95 °C for 10 seconds, and 60 °C for 1 minute. The data are expressed as shRNA/mGAPDH. All experiments were performed in triplicate, and the data were analyzed using StepOne Software v2.1 (Applied Biosystems, Waltham, MA). Primers sequences are summarized in Supplementary Table S2.
Preparation of an orthotopic mouse model of mesothelioma. BALB/c nu/nu mice (male, 5 weeks old), which were purchased from Japan SLC (Shizuoka, Japan) were allowed free access to water and mouse chow and were housed under a constant temperature, humidity, and 12-hour dark–light cycle. An orthotopic mouse model of mesothelioma was prepared by the direct injection of MSTO-211H-Luc cells (1 × 106 cells per mouse) into the left pleural cavity of the mice. The tumor was monitored using an in vivo imaging system (Xenogen, Advanced Molecular Vision, Lincolnshire, UK). For in vivo imaging, the mice were intraperitoneally injected with 100 μl of 7.5 mg/ml D-luciferin potassium salt and were subsequently anesthetized using isoflurane inhalation. Three minutes after injection, bioluminescence was recorded using a charge-coupled device camera (1-minute exposure). The bioluminescent region of interest (ROI) was calculated and shown as photon counts (photons/s/cm2/steradian).
In vivo luciferase reporter assay. An orthotopic model mouse of mesothelioma was prepared by the inoculation of MSTO-211H-Luc cells into the pleural cavity. On days 8 and 11 after inoculation, the tumor-bearing mice (four mice per group) were intrapleurally injected with two doses of either the natural device (1 mg/kg), NC dsDNA (1 mg/kg), dSC iRed (1 mg/kg), or synthetic shRNA (0.077 mg/kg). Each sample was complexed with a transfection reagent designed for use in vivo (TurboFect Transfection Reagents, Life Technologies, Carlsbad, CA) and used according to the manufacturer's instructions. On days 7, 10, and 13 after the inoculation, the luciferase activity of the pleural tumor was observed using IVIS as described in the section “Preparation of an orthotopic mouse model of mesothelioma”.
Preparation of PEGylated lipoplex of iRed, pDNA, and the natural device. Briefly, the lipids 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, cholesterol (Wako Pure Chemical), and O,O'-ditetradecanoyl-N-(α-trimethylammonioacetyl)diethanolamine chloride (DC-6–14, Sogo Pharmaceutical, Tokyo, Japan) were dissolved in chloroform and mixed at molar ratios of 3:2:3:2. After evaporating chloroform, the thin lipid films were rehydrated with 9% sucrose at 37 °C, and the suspensions were sized by repeated extrusion through polycarbonate membrane filters (Nucleopore) with consecutive pore sizes of 400, 200, and 100 nm and mixed with 5 mol% of mPEG2000-DSPE at 37 °C. The phospholipid concentrations of the resulting PEGylated cationic liposomes were quantified using the Fiske and Subbarow phosphate assay.36 To form a lipoplex, equal volumes of PEGylated cationic liposomes (1.2 μmol total lipids) and DNA solution (20 μg) were mixed and vortexed for 15 minutes at room temperature.
Particle size and ζ-potential measurement. Size and ζ-potentials were measured using a NICOMP 370 HPL submicron particle analyzer (Particle Sizing System, Port Richey, FL). Lipoplexes were diluted with 9% sucrose, and measurements were performed at 25 °C and repeated independently three times.
Cytokine assays. BALB/c mice (male, 5 weeks old, Japan SLC) were intravenously injected with 20 μg each DNA sample formulated with a lipoplex. Four hours after the injection, peripheral blood was collected from the retro-orbital plexus while the mice were anesthetized. To obtain serum, the blood was stored for 30 minutes at room temperature and then centrifuged at 3,000 rpm at 4 °C for 15 minutes. The serum levels of IL-6, INF-γ, and TNF-α were quantified using Quantikine Immunoassay Kits (R&D Systems, Minneapolis, MN), and INF-α was quantified using a VeriKine Mouse Interferon Alpha ELISA Kit (PBL interferon source, Piscataway, NJ). Experiments were performed in triplicate at room temperature.
Statistical analysis. Differences in a group were evaluated via analysis of variance testing using the Tukey post-hoc test.
SUPPLEMENTARY MATERIAL Table S1. Sequences of ligated ODNs for construction of pDNAs expressing shRNAs. Table S2. Sequences of primers used for the quantification of transcribed shRNA in HeLa cells. Table S3. Physicochemical properties of PEGylated iRed-lipoplex, pDNA-lipoplex and natural device-lipoplex. Figure S1. PCR preparation of an iRed targeting pGL2 Fluc. Figure S2. Preparation of iRed targeting pGL3 Fluc. Figure S3. Evaluation of RNAi activities of iReds targeting pGL2 against pGL3 Fluc. Figure S4. Preparation of iRed targeting pGL3 Fluc but having terminator sequence in the middle of shRNA coding region.
Acknowledgments
This work was supported by Grant-in-Aid for Scientific Research (B) (Grant Number; 24390027 and 15H04656). The authors would like to thank Masashi Kobayashi (Department of Thoracic Surgery, Faculty of Medicine, Kyoto University) for kindly providing MSTO-211H-Luc and Shuji Kitaike (Center for Instrumental Analysis, Tokushima University) for providing technical assistance. N.T. thanks the Japanese Society for the Promotion of Science (JSPS) for a research fellowship and the research program for the development of intelligent Tokushima artificial exosome (iTEX) from Tokushima University. The authors declare no competing financial interest.
Supplementary Material
References
- Elbashir, SM, Harborth, J, Lendeckel, W, Yalcin, A, Weber, K and Tuschl, T (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498. [DOI] [PubMed] [Google Scholar]
- Watts, JK, Deleavey, GF and Damha, MJ (2008). Chemically modified siRNA: tools and applications. Drug Discov Today 13: 842–855. [DOI] [PubMed] [Google Scholar]
- Shukla, S, Sumaria, CS and Pradeepkumar, PI (2010). Exploring chemical modifications for siRNA therapeutics: a structural and functional outlook. ChemMedChem 5: 328–349. [DOI] [PubMed] [Google Scholar]
- Davidson, BL and McCray, PB Jr (2011). Current prospects for RNA interference-based therapies. Nat Rev Genet 12: 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp, TR, Bernards, R and Agami, R (2002). A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553. [DOI] [PubMed] [Google Scholar]
- Paddison, PJ, Caudy, AA, Bernstein, E, Hannon, GJ and Conklin, DS (2002). Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev 16: 948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, CP, Good, PD, Winer, I and Engelke, DR (2002). Effective expression of small interfering RNA in human cells. Nat Biotechnol 20: 505–508. [DOI] [PubMed] [Google Scholar]
- Yu, JY, DeRuiter, SL and Turner, DL (2002). RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci USA 99: 6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessis, N, GarciaCozar, FJ and Boissier, MC (2004). Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther 11 Suppl 1: S10–S17. [DOI] [PubMed] [Google Scholar]
- Khalil, IA, Kogure, K, Akita, H and Harashima, H (2006). Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol Rev 58: 32–45. [DOI] [PubMed] [Google Scholar]
- Seow, Y and Wood, MJ (2009). Biological gene delivery vehicles: beyond viral vectors. Mol Ther 17: 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, DR, Pringle, IA and Hyde, SC (2009). Progress and prospects: the design and production of plasmid vectors. Gene Ther 16: 165–171. [DOI] [PubMed] [Google Scholar]
- Taki, M, Kato, Y, Miyagishi, M, Takagi, Y and Taira, K (2004). Small-interfering-RNA expression in cells based on an efficiently constructed dumbbell-shaped DNA. Angew Chem Int Ed Engl 43: 3160–3163. [DOI] [PubMed] [Google Scholar]
- Castanotto, D and Scherer, L (2005). Targeting cellular genes with PCR cassettes expressing short interfering RNAs. Methods Enzymol 392: 173–185. [DOI] [PubMed] [Google Scholar]
- Inoue, N, Shionoya, A, Minakawa, N, Kawakami, A, Ogawa, N and Matsuda, A (2007). Amplification of 4′-thioDNA in the presence of 4′-thio-dTTP and 4′-thio-dCTP, and 4′-thioDNA-directed transcription in vitro and in mammalian cells. J Am Chem Soc 129: 15424–15425. [DOI] [PubMed] [Google Scholar]
- Hoshika, S, Minakawa, N and Matsuda, A (2004). Synthesis and physical and physiological properties of 4′-thioRNA: application to post-modification of RNA aptamer toward NF-kappaB. Nucleic Acids Res 32: 3815–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, N, Minakawa, N and Matsuda, A (2006). Synthesis and properties of 4′-ThioDNA: unexpected RNA-like behavior of 4′-ThioDNA. Nucleic Acids Res 34: 3476–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsugami, A, Ohyama, T, Inada, M, Inoue, N, Minakawa, N, Matsuda, A et al. (2008). Unexpected A-form formation of 4′-thioDNA in solution, revealed by NMR, and the implications as to the mechanism of nuclease resistance. Nucleic Acids Res 36: 1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshika, S, Minakawa, N, Kamiya, H, Harashima, H and Matsuda, A (2005). RNA interference induced by siRNAs modified with 4′-thioribonucleosides in cultured mammalian cells. FEBS Lett 579: 3115–3118. [DOI] [PubMed] [Google Scholar]
- Hoshika, S, Minakawa, N, Shionoya, A, Imada, K, Ogawa, N and Matsuda, A (2007). Study of modification pattern-RNAi activity relationships by using siRNAs modified with 4′-thioribonucleosides. Chembiochem 8: 2133–2138. [DOI] [PubMed] [Google Scholar]
- Takahashi, M, Nagai, C, Hatakeyama, H, Minakawa, N, Harashima, H and Matsuda, A (2012). Intracellular stability of 2′-OMe-4′-thioribonucleoside modified siRNA leads to long-term RNAi effect. Nucleic Acids Res 40: 5787–5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, M, Yamada, N, Hatakeyama, H, Murata, M, Sato, Y, Minakawa, N et al. (2013). In vitro optimization of 2′-OMe-4′-thioribonucleoside-modified anti-microRNA oligonucleotides and its targeting delivery to mouse liver using a liposomal nanoparticle. Nucleic Acids Res 41: 10659–10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, Y, Hashimoto, Y, Arai, M, Tarashima, N, Miyazawa, T, Miki, K et al. (2014). Chemistry, properties, and in vitro and in vivo applications of 2′-O-methoxyethyl-4′-thioRNA, a novel hybrid type of chemically modified RNA. Chembiochem 15: 2535–2540. [DOI] [PubMed] [Google Scholar]
- Kojima, T, Furukawa, K, Maruyama, H, Inoue, N, Tarashima, N, Matsuda, A et al. (2013). PCR amplification of 4′-thioDNA using 2′-deoxy-4′-thionucleoside 5′-triphosphates. ACS Synth Biol 2: 529–536. [DOI] [PubMed] [Google Scholar]
- Maruyama, H, Furukawa, K, Kamiya, H, Minakawa, N and Matsuda, A (2015). Transcription of 4′-thioDNA templates to natural RNA in vitro and in mammalian cells. Chem Commun (Camb) 51: 7887–7890. [DOI] [PubMed] [Google Scholar]
- Chen, C, Ridzon, DA, Broomer, AJ, Zhou, Z, Lee, DH, Nguyen, JT et al. (2005). Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, S, Yuzenkova, Y and Zenkin, N (2013). Mechanism of eukaryotic RNA polymerase III transcription termination. Science 340: 1577–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger, AT, Peterson, LW, Chelliserry, J, Kleinbaum, DJ and Kool, ET (2011). Encoding phenotype in bacteria with an alternative genetic set. J Am Chem Soc 133: 18447–18451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanzone, AP, El-Sagheer, AH, Brown, T and Tavassoli, A (2012). Assessing the biocompatibility of click-linked DNA in Escherichia coli. Nucleic Acids Res 40: 10567–10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birts, CN, Sanzone, AP, El-Sagheer, AH, Blaydes, JP, Brown, T and Tavassoli, A (2014). Transcription of click-linked DNA in human cells. Angew Chem Int Ed Engl 53: 2362–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, M, Judge, A and MacLachlan, I (2009). siRNA and innate immunity. Oligonucleotides 19: 89–102. [DOI] [PubMed] [Google Scholar]
- Judge, AD, Bola, G, Lee, AC and MacLachlan, I (2006). Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther 13: 494–505. [DOI] [PubMed] [Google Scholar]
- Krieg, AM, Yi, AK, Matson, S, Waldschmidt, TJ, Bishop, GA, Teasdale, R et al. (1995). CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 374: 546–549. [DOI] [PubMed] [Google Scholar]
- Kawai, T and Akira, S (2010). The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11: 373–384. [DOI] [PubMed] [Google Scholar]
- Ohto, U, Shibata, T, Tanji, H, Ishida, H, Krayukhina, E, Uchiyama, S et al. (2015). Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9. Nature 520: 702–705. [DOI] [PubMed] [Google Scholar]
- Bartlett, GR (1959). Colorimetric assay methods for free and phosphorylated glyceric acids. J Biol Chem 234: 469–471. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.