Abstract
Elevated expression levels of eukaryotic initiation factor 4E (eIF4E) promote cancer development and progression. MAP kinase interacting kinases (MNKs) modulate the function of eIF4E through the phosphorylation that is necessary for oncogenic transformation. Therefore, pharmacologic MNK inhibitors may provide a nontoxic and effective anticancer strategy. MNK1b is a truncated isoform of MNK1a that is active in the absence of stimuli. Using in vitro selection, high-affinity DNA aptamers to MNK1b were selected from a library of ssDNA. Selection was monitored using the enzyme-linked oligonucleotide assay (ELONA), and the selected aptamer population was cloned and sequenced. Four groups of aptamers were identified, and the affinities of one representative for rMNK1b were determined using ELONA and quantitative polymerase chain reaction. Two aptamers, named apMNK2F and apMNK3R, had a lower Kd in the nmol/l range. The secondary structure of the selected aptamers was predicted using mFold, and the QGRS Mapper indicated the presence of potential G-quadruplex structures in both aptamers. The selected aptamers were highly specific against MNK1, showing higher affinity to MNK1b than to MNK1a. Interestingly, both aptamers were able to produce significant translation inhibition and prevent tumor cell proliferation and migration and colony formation in breast cancer cells. These results indicate that MNK1 aptamers have an attractive therapeutic potential.
Keywords: aptamer, cancer, MNK1, SELEX, translation
Introduction
The MAP kinase interacting kinase 1 (MNK1) has been shown to bind to and be a substrate for both the mitogen activated extracellular signal-regulated protein kinases (ERK) 1/2 and the stress-activated p38 MAP kinases (p38 MAPKs) both in vitro and in vivo.1 MNK1b is a truncated isoform lacking the MAP kinase-binding motif present in the C-terminal region of MNK1a that arises from an alternatively spliced transcript of the MNK1 gene.2 Both MNK1 isoforms have a nuclear localization signal in the N-terminal region that allows the kinase to enter into the nucleus.3 MNK1a contains a nuclear export signal that ensures its cytoplasmic localization,3 whereas MNK1b lacks this nuclear export signal, being cytoplasmic and nuclear.2 The different features in the human MNK1 isoforms lead to MNK1b having a higher basal activity than MNK1a. Moreover, MNK1b activity does not correlate with the phosphorylation of the activation loop residues and seems to be independent of the upstream kinases (ERK1/2 and p38 MAP kinase).2
The only well-characterized substrate for MNK1 is the eukaryotic initiation factor 4E (eIF4E).4 In higher eukaryotes, eIF4E is a component of the eukaryotic initiation factor 4F (eIF4F). The other two components are an ATP-dependent RNA helicase, eIF4A, and a scaffold protein, eIF4G. eIF4E was reported as physiologically phosphorylated on the residue Serine 209 following cell treatments with growth factors, hormones and mitogens.5,6,7 Previous results showed that nuclear eIF4E phosphorylation appears to be important for controlling the transport of cyclin D1 mRNA and for the transforming properties of eIF4E.8 Conversely, overexpression of the oncogene HMD2 in cancer cells is regulated by eIF4E; therefore, the overexpression of eIF4E promotes the export of HDM2 mRNA in a MAP kinases and MNK1-dependent manner.9 Recent studies established that the phosphorylation of eIF4E by MNK1/2 plays an important role for the oncogenic action of eIF4E.10 For instance, treatment of human cancer cells with the MNK1 inhibitors reduced colony formation, proliferation, and survival.11,12,13,14 In addition, Wendel et al.15 showed that overexpression of a constitutively active MNK1 diminishes apoptosis and accelerates the development of tumors in an experimental model of mice, while an inactive mutant reduces the development of these tumors. Interestingly, preliminary findings in our lab (unpublished results) demonstrated that the isoform MNK1b, which is active in the absence of stimuli,2 was overexpressed in several types of tumors (breast and colorectal) relative to normal tissue. Finally, it was demonstrated that eIF4E was completely dephosphorylated in knockout mice for MNK1 and MNK2, which do not exhibit any apparent phenotypical consequences that questions the vital role of MNK activity.16 These results suggest that protein kinase MNK1b could be considered as a potential diagnostic/prognostic marker or as a potential therapeutic target by regulating its expression and/or activity.
Aptamers are structured polynucleotide sequences isolated from randomized oligonucleotide libraries by the systematic evolution of ligands by exponential enrichment (SELEX) technology, which selectively binds target molecules with high affinity and specificity.17,18,19 Aptamers form stable and specific complexes with targets that have dissociation constants in the nanomolar range. Furthermore, aptamers have several advantages over antibodies due to the nature of nucleic acids, such as increased stability, easy regeneration, and simple modifications with different reporters during their synthesis. In addition, they are significantly smaller, can be isolated rapidly in vitro, and do not elicit a significant immune response.20,21
In this study, we detail the isolation and characterization of two aptamers that specifically recognize MNK1 with affinity in the low nanomolar range. In addition, both aptamers, more significantly apMNK2F, block protein translation in vitro. We also demonstrated that aptamers inhibit cell proliferation, cell migration, and colony formation in vitro. This opens the possibility to initiate in vivo studies addressing the use of these aptamers in cancer treatments.
Results
Selection and characterization of high-affinity aptamers against MNK1b
Aptamers were selected from libraries of oligonucleotides by iterative cycles of selection (SELEX methodology). We performed a selection of specific DNA aptamers against the MNK1b protein as indicated in the Materials and Methods section. We carried out 10 rounds of selection and 3 counterselection rounds (after rounds 4, 7, and 10) using a Ni-NTA resin that binds the recombinant MNK1b protein fused to the 6xHIS tail at the N-terminal end. To confirm the enrichment of the populations obtained after successive rounds of selection, enzyme-linked oligonucleotide assays (ELONA) were performed as described in the Materials and Methods section. The results showed an increase in the population signal obtained after round 7 (SELMNKRd7) and round 10 (SELMNKRd10), relative to the initial RND40 population described in the Materials and Methods section (Supplementary Figure S1a). Because of these results, we proceeded with the cloning of the SELMNKRd10 aptamer population to isolate and characterize individual aptamers, as described in the Materials and Methods section. We obtained 28 aptamers and analyzed their sequences to identify four groups of sequences (apMNK1, apMNK2, apMNK3, and apMNK4). The sequence corresponding to apMNK1 was repeated 17 times, although 3 of them had mutations from the addition of one or more nucleotides. The sequence corresponding to apMNK2 was found once, apMNK3 appeared eight times, one was a substitution, and the sequence apMNK4 was found repeated twice (Supplementary Figure S1b). Although the starting population contained molecules of 76 nucleotides in length, we observed sequences of 66–67 nucleotides in all samples, except for apMNK2 which had 75 nucleotides (Supplementary Table S1).
The two chains produced from the polymerase chain reaction (PCR) were used during the selection process, and it was necessary to analyze both strands of each clones, named chain F and chain R. ELONA was used to study the binding capacity of different aptamers to their targets, and each chain was labeled with digoxigenin or phosphate through PCR and treated with exonuclease to remove the complementary strand as described in the Materials and Methods section. The results showed that the aptamers had a high binding capacity to the target, reaching values at least twice the blank value for apMNK2F, apMNK3F, apMNK3R, and apMNK4F (Figure 1a).
Figure 1.
Analysis of the affinity of the aptamers. (a) His-MNK1b (5 pmol) protein was incubated with 8 pmol/well (40 nmol/l) of R- or F-digoxigenin-labeled aptamers. The dotted line indicates the double value of the target. (b) The study of individual aptamers by recovery of the aptamer–target complexes and quantification by qPCR as described in the Materials and Methods section. Ct values for each aptamer in the presence (black bars) or absence (white bars) of the protein are shown. The bars show the mean of a representative experiment.
To use an alternative method for studying the binding ability of aptamers to the target, quantitative PCR (qPCR) assays were performed using template aptamers that would bind to the resin–MNK1b complex, as described in the Materials and Methods section. These assays allow us to check if the selected aptamers are specific against MNK1b and not the Ni-NTA resin. We considered aptamers positive if there was a difference between the presence and absence of the protein of at least 3 Cts. The Cts of the aptamers apMNK1F, apMNK2F, apMNK3R, and apMNK4R were threefold to fivefold lower in the presence of MNK1b compared to its absence (Figure 1b).
In summary, these results (ELONA and analysis of the aptamer–target complex by qPCR) indicated that the apMNK2F and apMNK3R aptamers should be selected for further characterization.
Bioinformatics analysis of the secondary structures
To predict the most stable secondary structures of apMNK2F and apMNK3R aptamers, we performed a bioinformatic analysis of their sequences using the mFold software. In Figure 2a, we showed the most probable secondary structures of these aptamers, taking into account the lower free energy (ΔG). Because many of the aptamers described in the literature contain G-quadruplex structures,22,23,24 which strongly stabilize the tertiary structure, the capacity of apMNK2F and apMNK3R to form G-quadruplex structures was studied using the QGRS Mapper program. The analysis results indicated that the aptamer apMNK2F could form two of these structures, one with three and one with two flat planes, while apMNK3R could form a two plane G-quadruplex structure (Figure 2a). These structures provide greater stability for the molecules; therefore, the theoretically apMNK2F and apMNK3R aptamers are very stable aptamers despite their relative high free energies.
Figure 2.
MNK1b aptamers are very stable molecules. (a) Secondary structures and G-quadruplex were obtained by bioinformatics sequence analysis using the mFold programs and QGRS Mapper. (b) Biostability of the DNA aptamers analyzed by incubation in the presence of DNAse I. Representative agarose gels are shown.
The nucleotide composition of the aptamer is also important for its stability. In fact, a higher G+C content stabilizes the aptamer structure. An analysis of the G+C content for the selected aptamers showed that both of them have a G+C content higher than 50% (66.7% apMNK2F and 60.6% apMNK3R).
The high susceptibility of aptamers to nucleases limits their potential clinical application. In this study, we developed ssDNA aptamers that are significantly more stable than RNA aptamers. To analyze the stability, we performed a time-course analysis of gel electrophoresis for apMNK2F and apMNK3R by incubating them with the endonuclease DNAse I, leading to complete degradation after only 5 minutes of treatment (Figure 2b, left panel). Therefore, we performed the same experiment with several dilutions of the enzyme for a 5-minute incubation. The results showed that apMNK3R was more sensitive to the enzymatic activity. In fact, the treatment with 0.1 units of DNAse I produced 70% of apMNK3R degradation and only 30% of apMNK2F degradation (Figure 2b, right panel).
Characterization of the interaction of the aptamers with MNK1 by ELONA
To study the affinity of the aptamers apMNK2F and apMNK3R for the MNK1b protein, we used ELONA assays where the GST-MNK1b protein was incubated with increasing concentrations of biotin-labeled aptamers. The data obtained were analyzed using a nonlinear regression showing a response to a hyperbola whose equation is y = (x × Bmax)/(x + Kd), where Bmax is the maximal binding and Kd (dissociation constant) is the concentration of ligand required to reach half-maximal binding. The aptamers apMNK2F and apMNK3R were capable of detecting the MNK1b protein in a concentration-dependent manner with a Kd of 1.79 ± 0.48 and 6.36 ± 1.72 nmol/l, respectively (Figure 3a).
Figure 3.
MNK1b aptamers recognize a region between amino acids common to both isoforms and the specific MNK1b region, MNK1bSR. (a) Analysis of the binding of aptamers with MNK1b was performed by ELONA. GST-MNK1b was plated at 0.2 µg/well (3 pmol/well) and then incubated with Bio-apMNK2F and Bio-apMNK3R at 0–40 nmol/l concentrations. The bars represent the mean ± SEM of 4–6 independent experiments. (b) Proteins MNK1a, MNK1b, MNK1aΔ77, and MNK1ΔCt were plated at 3 pmol/well and then incubated with aptamers Bio-apMNK2F, Bio-apMNK3R, and Bio-38x(AG) at 20 nmol/l. Bars represent the mean ± SEM of 4–6 independent experiments (**P < 0.01 compared to apMNK2F; aP < 0.001 compared to blank). (c) A scheme displaying MNK1a, MNK1b, MNK1aΔ77, and MNK1ΔCt possible sites of interaction with the aptamers and the extent of binding for aptamers to the different forms of the kinases.
Once the aptamer's affinity for MNK1b was analyzed, we performed ELONA experiments to study the ability of aptamers to differentially recognize MNK1a and MNK1b and analyze the region of MNK recognized by the aptamers. In these experiments, we used the targets MNK1a, MNK1b, and MNK1aΔ77, which was as short as MNK1b but differs in the 12 amino acid residues of the specific MNK1b region (MNK1bSR), and MNK1ΔCt, which has the sequence of MNK1a but lacks the 89 amino acids of the C-terminus.25 A ssDNA with a 38x(AG) sequence that does not acquire any type of secondary structure was used as a negative control. The results indicated that the aptamers apMNK2F and apMNK3R were significantly able to bind to different proteins (Figure 3b). Furthermore, although these aptamers bind MNK1b with higher affinity than to other proteins, this difference is significant only with MNK1ΔCt. Interestingly, aptamer apMNK2F showed higher affinity for MNK1a compared to apMNK3R (P < 0.01).
MNK1b aptamers detect MNK1a/b in cytochemistry
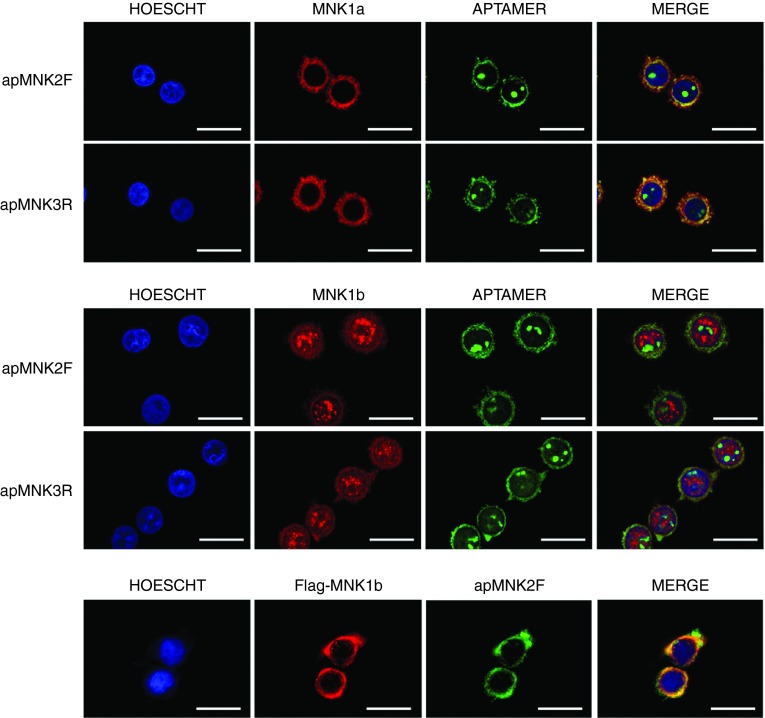
To check whether the selected aptamers were able to specifically recognize MNK1a or MNK1b, we performed cytochemical assays using Alexa 488-labeled aptamers as described in the Materials and Methods section. Therefore, using the stably transfected MDA-MB-231 cells described in the Materials and Methods section (Supplementary Figure S2), we incubated the stable cell lines with the aptamers apMNK2F and apMNK3R labeled with Alexa-488, and the antibodies recognized MNK1a (MNK1 (C-20)) or MNK1b (MNK1 (M-20)) as described in the Materials and Methods section. The results obtained by confocal microscopy using the MDA-MB-231 cells overexpressing MNK1a indicated that both apMNK2F and apMNK3R aptamers showed a cytoplasmic pattern and some accumulation in the cell nucleus (Figure 4a). However, we have also observed co-localization between aptamers and the MNK1a antibody in the cytoplasm for both cases.
Figure 4.
Aptamers detect MNK1a/b in cells. Aptacytochemistry in MDA-MB-231 cells stably overexpressing (a) MNK1a, (b) MNK1b, or (c) transiently overexpressing MNK1b. Experiments were performed using Alexa-488-conjugated aptamers apMNK2F and apMNK3R (2 pmol/coverslip) and (a) MNK1 (C-20), (b) MNK1 (M-20), or (c) anti-Flag antibodies as described in the Materials and Methods section. Confocal microscopy images corresponding to the staining of nuclei with Hoechst (blue), antibodies (red), and aptamers (green). The overlay images (merge) are also shown. Bar = 20 µm.
We performed the same type of experiment with MDA-MB-231 cells stably expressing MNK1b, noting that the subcellular distribution of the aptamers was similar to that shown in cells overexpressing MNK1a (Figure 4b). However, in these experiments, we did not observe co-localization of the aptamers with the MNK1b antibody in the nucleus. In addition, although no co-localization was observed in the cytoplasm, this may be due to the intensity of the labeled antibody is significantly lower than that observed for MNK1a. Therefore, new experiments were used to analyze co-localization of the aptamer apMNK2F and anti-Flag antibody in cells transiently transfected with the pcDNA3-Flag-MNK1b plasmid. The results of these assays showed that the aptamer and antibody co-localized in the cytoplasm of cells (Figure 4c).
MNK1b aptamers inhibit translation in vitro
One of the main objectives of selecting aptamers against MNK1 is to regulate its kinase activity. To assess whether the selected aptamers affect activating or inhibiting MNK1 activity, we performed in vitro kinase activity assays. In these experiments, we analyze the effect of the aptamers apMNK2F and apMNK3R on MNK1a and MNK1b activity using the peptide RRRLSSLRA as a substrate and rMNK1a (BPS Bioscience, San Diego, CA) or Myc-MNK1b immunoprecipitated from cells overexpressing the protein as kinases. We used the aptamer:protein ratio of 2:1 to ensure the presence of enough aptamers to inhibit kinase activity. In parallel, we used the same concentration of the MNK inhibitor, CGP57380, as a positive control. Only the aptamer apMNK3R significantly reduced MNK1a activity (26.5%), which was roughly half of the MNK1 inhibitor (41.6%) at the same concentration (Figure 5a). However, none of the aptamers produced a significant effect on activation or inhibition compared to the control on MNK1b activity under the conditions of these assays (Supplementary Figure S3).
Figure 5.
Effects of aptamers on kinase activity and translation in vitro. (a) The kinase activity assay was performed using the peptide RRRLSSLRA as a substrate and the recombinant activated MNK1a (GST-MNK1a T385D) as a kinase in the absence (control) or the presence of 6 pmol for each aptamer. In parallel, the same concentration of CGP57380 was used as a positive control. Percentages were calculated relative to the control. Bars represent the mean ± SEM of 2–4 independent experiments (*P < 0.05 compared to the control). (b) In vitro translation assay performed with a Rabbit Reticulocyte Lysate System in the absence (control) or presence of 4 µmol/l aptamers. Luciferase activity was measured after different incubation times. The data are expressed as arbitrary units (A.U.) and represent the mean ± SEM of 2–4 independent experiments.
We assayed the ability of the aptamers to affect translation using a rabbit reticulocyte translation system. In these experiments, exogenous luciferase mRNA was translated in the absent or presence of 4 µmol/l of each aptamers or 38x(AG), and luciferase levels were measured at different incubation times (Figure 5b). Results showed that the aptamers apMNK2F and apMNK3R inhibited protein synthesis significantly (98.9 and 52.4%, respectively). Aptamer 38x(AG) did not produce any change relative to the control.
MNK1b aptamers inhibit tumor cell proliferation and migration, and colony formation
One of the objectives of this work was to identify specific MNK1b aptamers that cause inhibition of tumor cell viability. We first investigated the effect of novel aptamers on the growth, proliferation, migration, and colony formation capacities of MDA-MB-231 breast cancer cells.
Therefore, MDA-MB-231 cells were transfected with the aptamers at different concentrations, and cell viability was measured after 72 hours by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction. Results revealed that the aptamers apMNK2F and apMNK3R potently inhibited the growth of MDA-MB-231 cells with IC50 values of 15 and 20.5 nmol/l, respectively. The unspecific 38x(AG) aptamer did not significantly affect cell viability (Figure 6a). We performed the same experiment with a 25 nmol/l aptamer concentration using HEK293T and MCF7 cells, and the results showed a similar decrease in MTT activity (Supplementary Figure S4). Next, we evaluated the effects of aptamers on the migratory potential of MDA-MB-231 cells using wound-healing assays (Figure 6b). We found that 24 hours after the well monolayer was wounded, control cells almost completely filled the scratched area. Our results showed that apMNK2F potentially inhibited cell migration (43%) while apMNK3R and the unspecific 38x(AG) control were significantly less effective than apMNK2F (20 and 7%, respectively). These data suggest the involvement of MNK1 in MDA-MB-231 cell migration. Finally, to evaluate the effect of aptamers on the clonogenic capacity of the MDA-MB-231 cells, we carried out colony formation experiments as described in the Materials and Methods section. The results demonstrated that apMNK2F and apMNK3R significantly reduced MDA-MB-231 colony number (57.0 and 39.7%, respectively), while the unspecific 38x(AG) aptamer did not produce any effects (Figure 6c).
Figure 6.
MNK1b aptamers inhibit proliferation, migration, and colony formation in vitro. (a) MDA-MB-231 cells were plated at a density of 6 × 103 cells/well in p96 plates. After 16–24 hours, aptamers were transfected at 5–50 nmol/l concentrations for 72 hours, and MTT activity assays were performed. The graph represents the mean ± SEM of 2–3 independent experiments. (*P < 0.05, **P < 0.01, ***P < 0.001 compared to the control; aP < 0.001, bP < 0.01, cP < 0.05 compared to 38x(AG)). (b) Effects of aptamers on MDA-MB-231 cell migration with a wound-healing assay for 24 hours. Transfected cells were allowed to form a confluent monolayer and were treated with mitomycin C. Representative photomicrographs of initial and final wounds are shown. Bar = 1,000 µm (upper and middle panels). The lower panel shows a higher magnification (bar = 200 µm) of the wounds after 24 hours. Cells transfected in the absence of aptamers were used as a control. (c) Effect of the aptamers on colony formation. Transfected MDA-MB-231 cells were seed at 1 × 103 cells/well in six-well plates. After 8–9 days, the colonies were fixed, stained, and counted. Bars represent the mean ± SEM of four independent experiments. *P < 0.05. Cells transfected in the absence of aptamers were used as a control. (d) MDA-MB-231 cells stably transfected with Myc-pcDNA3 (control), Myc-MNK1a (MNK1a), and Myc-MNK1b (MNK1b) were plated at a density of 6 × 103 cells/well in p96 plates. After 16–24 hours, aptamers were transfected at 12.5 nmol/l concentration for 72 hours and MTT activity assays were performed. The graph represents the mean ± SEM of four independent experiments. bP < 0.01, cP < 0.05 compared to the control.
To compare the effects of the aptamers with those produced by MNK inhibitors (CGP57380, SB203580, and U0126), we performed similar cell proliferation and colony formation assays. We observed that CGP57380 and U0126 displayed similar effects compared to the aptamers; however, SB203580 only affected colony formation (Supplementary Figure S5).
In addition, we confirmed that the effect of the aptamers on cell viability was specific for MNK1a/b when additional experiments were performed using the stable cells overexpressing both kinases (Figure 6d). Therefore, MNK1a/b overexpression, significantly MNK1a, partially blocked the antiproliferative effect of both apMNK2F and apMNK3R aptamers.
MNK1b aptamers activity is independent of eIF4E phosphorylation
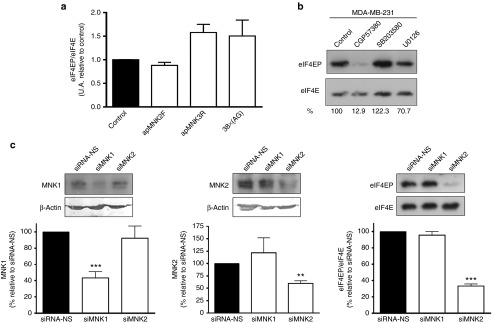
To gain further insight into the mechanism of aptamers inducing cell growth inhibition, we studied the effects of apMNK2F and apMNK3R on the phosphorylation levels of eIF4E, which is the substrate for both isoforms of MNK1. For this analysis, aptamer-transfected MDA-MB-231 cells were lysed after 24 hours of transfection, followed by western blots analysis of eIF4EP levels as described in the Materials and Methods section. We found that none of the aptamers inhibited eIF4E phosphorylation. In fact, apMNK3R and 38x(AG) induced eIF4EP levels, although this was not statistically significant (Figure 7a).
Figure 7.
MNK1b aptamer activity does not affect eIF4E phosphorylation. (a) MDA-MB-231 cells transfected with aptamers for 24 hours were lysed, and eIF4EP levels were analyzed with a western blot using specific antibodies as described in the Materials and Methods section. eIF4EP levels, with respect to eIF4E, were calculated and expressed as a percentage relative to the control. Bars represent the mean ± SEM of 2–3 independent experiments. (b) MDA-MB-231 cells were incubated in the presence of CGP57380, SB203580, and U0126, and eIF4E phosphorylation was analyzed with a western blot as in a. A representative blot is shown. Numbers at the bottom are the percentage of eIF4EP relative to the control (cell treated with the vehicle). (c) Effects of suppressing expression of MNK1/2 on eIF4E phosphorylation. MDA-MB-231 cells were seed on p6 plates and transfected with 75 pmol/well of siRNA (control), siMNK1, and siMNK2. After 72 hours, cell lysates were analyzed with a western blot using specific antibodies. MNK1 and MNK2 levels were normalized relative to β-actin, a loading control, and eIF4EP levels were calculated as shown above. Bars represent the mean ± SEM of 4–5 independent experiments. **P < 0.01, ***P < 0.001.
It is interesting to note that diverse MNK inhibitors affected eIF4E phosphorylation in a different way. In an attempt to study the aptamer mechanisms, we performed experiments where MDA-MB-231 cells were incubated in the presence of CGP57380, SB203580, and U0126, followed by eIF4E phosphorylation analyzed with western blots. The results showed that CGP57380 produces a strong decrease on eIF4EP levels, SB203580 increases eIF4E phosphorylation, and U0126 partially blocks the phosphorylation of the factor (Figure 7b).
These results demonstrated that aptamers against MNK1 do not produce changes in the phosphorylation of eIF4E, and because phosphorylation of the factor may be produced by both MNK1 and MNK2 activity, we decided to transfect MDA-MB-231 cells with siRNAs against MNK1 and MNK2 to analyze the role of both kinases in the phosphorylation of eIF4E. Results showed that both siRNAs (siMNK1 and siMNK2) produced a specific decrease in the levels of their targets (56.4 and 40%, respectively), without affecting the expression of the other kinase (Figure 7c, left and central panels). When we analyzed eIF4E phosphorylation levels, we observed that they do not change in the cells transfected with siMNK1 relative to the control, whereas eIF4EP levels decreased by 66.4% in cells transfected with siMNK2 (Figure 7c, right panel). These results clearly demonstrated that eIF4E phosphorylation is mainly mediated by MNK2, not MNK1, in MDA-MB-231 cells.
Discussion
Breast cancer is a complex and heterogeneous disease in regards to histology, cellular origin, mutations, capacity to metastasize, disease progression, and treatment response. Most breast cancer-related deaths are due to a poor diagnosis and resistance to therapies. Much effort has been made to identify new diagnostic markers and/or therapeutic targets. In this study, we aimed to select and characterize aptamers against MNK1b. The selected aptamers were highly specific against MNK1, showing higher affinity to MNK1b than to MNK1a. Interestingly, both aptamers were able to produce significant translation inhibition and prevent tumor cell proliferation and migration and colony formation in breast cancer cells.
As indicated in the Results section, the aptamers obtained from selection round 10 mostly showed a size of 66–67 nucleotides, which was shorter than the starting population. To further explore this surprising result, high-throughput DNA sequencing from round 7 was performed (data not shown). Most of the aptamers from this round were also 66–67 nucleotides long, and the most frequently represented aptamers were identical to those obtained in round 10. In addition, the heterogeneity of the sequences obtained in these rounds was very low, and the sequence apMNK1 was strongly represented. This discrepancy in the size of the aptamers, perhaps due to the introduction of changes during PCR, was not considered relevant because regardless of the sequence length, structure and functionality are more important for aptamer activity.
After analyzing the results of ELONA and qPCR, the apMNK2F and apMNK3R aptamers were identified as the most promising. Analysis of these sequences revealed that these two aptamers showed a G+C abundance higher than 50%, suggesting high structural stability. Furthermore, bioinformatic analysis of the sequences provided the more stable secondary structures and the free energy values, and the possibility of forming G-quadruplex structures. In our laboratory, we observed that aptamers having higher values of free energy, and consequently thermodynamically unstable, are those that can potentially form G-quadruplex structures, allowing them to increase their stability. This occurs for the selected aptamers apMNK2F and apMNK3R, which exhibited relatively high free energy and were able to form G-quadruplexes. It is reasonable to think that the aptamers must have a high stability, which is important for subsequent applications in biological systems because it is necessary to enter cells and reach their target proteins without being degraded. In fact, stability studies showed that the aptamer apMNK2F was more resistant to nuclease treatment than apMNK3R, which correlates with the fact that apMNK2F can potentially form two G-quadruplex structures.
The studies determining the specificity of aptamers showed that both recognized the isoforms of the protein, although they bind with higher affinity to MNK1b. In addition, we demonstrated that aptamers recognized the MNK1aΔ77 mutant, which is the same size as MNK1b. However, the MNK1ΔCt mutant, which lacks the 89 amino acids at the C-terminus, has a significantly lower recognition. This indicated that the sequence bound by the aptamers is located in the region between the amino acids common to both isoforms and the specific MNK1b region, MNK1bSR. Although the selection process was conducted against MNK1b, it is not surprising to find aptamers that also recognize MNK1a due to the similarities between the two protein isoforms.
ApMNK2F and apMNK3R inhibit translation and decrease MTT activity, migration, and colony formation in MDA-MB-231 cells, which suggested an antiproliferative effect. However, we do not know the exact biological mechanism that is mediating this effect. Recently, it has been described that Mnk-I1, an effective MNK inhibitor that potently blocks eIF4E phosphorylation, also blocks the migration of fibroblasts and cancer cells.26 Although both aptamers are able to recognize and/or bind MNK1, we were not able to determine a clear effect on the kinase activity of the protein. The results showed that only apMNK3R produces significant inhibition of kinase activity in vitro. However, experiments where eIF4E phosphorylation was evaluated in cells transfected with aptamers demonstrated that apMNK2F and apMNK3R do not affect the levels of eIF4E phosphorylation. This result could be explained, as demonstrated in experiments using siRNAs, because eIF4E phosphorylation in MDA-MB-231 cells is mediated primarily by MNK2 and not MNK1. It would be interesting to conduct this study in other breast cell lines to determine if the aptamers do not recognize MNK2. In addition, we cannot rule out the effect of aptamers specific for MNK1 by regulating the phosphorylation of other substrates of MNKs, such as hnRNP A1, PSF, and Sprouty 2, which are also implicated in tumorigenesis. Several studies indicated that hnRNP A1 is overexpressed in hepatocellular carcinoma cell lines and promotes tumor invasion27 and that the expression of hnRNP A1 is altered in renal cancers affecting alternative splicing of genes related to tumor growth.28 The PSF protein is involved in the survival and growth of colon cancer cells,29 and Sprouty 2 regulates prostate cancer30 and is a prognostic factor in breast cancer, especially for patients treated with trastuzumab.31
MNK inhibitors that block kinase activity and reduce phosphorylation of eIF4E, proliferation, and cell survival could treat different tumor processes in which these proteins are involved. There are several inhibitors of these kinases, such as CGP57380 and cercosporamide. Although CGP57380 is an inhibitor of MNK,32 studies have shown that this compound also inhibits other kinases, so it cannot be considered a specific inhibitor.33 Cercosporamide is also an inhibitor of MNKs, particularly MNK2, and has inhibitory effects on other kinases, albeit with lower potency.13 Furthermore, prolonged treatment with mTOR inhibitors, such as rapamycin in tumor cells, produces high PI3K activity promoting cell survival.34 Recently, it was reported that retinamides, a type of blocking agents of retinoic acid metabolism, are capable of degrading MNK1, blocking the phosphorylation of eIF4E, inhibiting cell proliferation, decreasing colony formation, inducing apoptosis, and preventing tumor invasion in breast cancer cells.35,36 The MTT activity assays performed in these different cell lines with retinamides showed IC50 values in the µmol/l range, whereas apMNK2F and apMNK3R aptamers exhibit IC50 values one order of magnitude lower (nmol/l).
In summary, we presented two ssDNA aptamers that bind with high affinity and specificity to MNK1. Interestingly, both aptamers are able to produce significant translation inhibition and prevent tumor cell proliferation and migration in vitro, and colony formation in breast cancer cells (Table 1). These effects on cancer cells open the possibility to study the functionality of the aptamers as antitumor therapeutics.37
Table 1. Main characteristics of the aptamers.

Materials and methods
Reagents. All the chemical reagents unless otherwise stated were obtained from Sigma-Aldrich (Hamburg, Germany).
Cell culture and generation of stable cell lines. Human embryonic kidney (HEK293T) and breast adenocarcinoma (MDA-MB-231 and MCF7) cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (PAA, Pasching, Austria) or DMEM/Ham F-12 medium (PAA) (mixed 1:1) with 10% fetal calf serum (Gibco, Grand island, NY) and 100 U/ml penicillin, 100 µg/ml streptomycin, and 25 µg/ml amphotericin (Sigma-Aldrich) in a humidified 5% CO2/95% air incubator at 37 °C. For stable transfection, MDA-MB-231 cells were seeded at 2 × 106 cells/plate in P100 plate and after 16–24 hours transfected with 10 µg/plate of Myc-pcDNA3, Myc-pcDNA3-MNK1a, or Myc-pcDNA3-MNK1b using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to manufacturer's instructions. After 24 hours, medium was changed with new medium containing 1 mg/ml of Geneticin (Invitrogen). Untransfected MDA-MB-231 cells were treated in parallel to check geneticin-induced cell death. After several passages, the presence of MNK1a or MNK1b were checked by immunocytochemistry, western blot, and mRNA quantitation, and the cells were frozen in 10% dimethyl sulfoxide /fetal bovine serum in liquid nitrogen until use (Supplementary Figure S2).
Protein extraction, dodecyl sulphate-polyacrylamide gel electrophoresis, and immunoblotting. To obtain cell lysates, cells were mechanically harvested and washed once with cold buffer A (20 mmol/l Tris–HCl pH 7.6, 1 mmol/l dithiothreitol (DTT), 1 mmol/l ethylenediaminetetraacetic acid, 1 mmol/l phenylmethylsulfonyl fluoride, 1 mmol/l benzamidine, 10 mmol/l sodium molybdate, 10 mmol/l sodium β-glycerophosphate, 1 mmol/l sodium orthovanadate, 120 mmol/l potassium chloride (KCl), 10 µg/ml antipain, 1 µg/ml pepstatin A, and leupeptin). Next, we lysed the cells in the same buffer containing 1% Triton X-100 (volume ratio 1:2) and centrifuged at 12,000g for 10 minutes. Afterwards, we determined the protein concentration by the method of Bradford,38 and the supernatant was aliquoted and stored at −80 °C until use.
Proteins were resolved by 12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis, at the conditions indicated in figure legends, and transferred onto polyvinylidene difluoride membranes. Membranes were incubated with monoclonal antibodies for 2 hours at room temperature and with polyclonal antibodies overnight at 4 °C. After washed, membranes were incubated with the corresponding peroxidase-conjugated secondary antibody for 1 hour at room temperature, developed with enhanced chemiluminescence's kits (GE Healthcare, Barcelona, Spain). Full Range Rainbow molecular weight markers (GE Healthcare) were used in all the experiments. The blots were probed with anti-eIF4E (ser209P; Cell Signaling, Danvers, MA), anti-eIF4E (BD Biosciences, Franklin Lake, NJ), anti-MNK1 (C-20), MNK2 and c-Myc (Santa Cruz Biotechnology, Santa Cruz, CA), and β-actin (Sigma-Aldrich) antibodies.
Expression and purification of recombinant proteins. Recombinant HIS-MNK1b (rMNK1b) was cloned in the pDest expression vector, and the recombinant protein was purified by affinity chromatography on Ni-NTA resin columns as described.39 Briefly, the BL21 cells expressing rMNK1b were harvested and suspended in sonication buffer (20 mmol/l Tris–HCl pH 7.8, 0.5 mol/l NaCl, 20 mmol/l imidazole, 2 mmol/l MgCl2, 1 mg/ml lysozime) and incubated in ice for 30 minutes. Afterwards bacteria were subjected to sonication for four cycles of 15 seconds at maximum amplitude and centrifuged at 10000g for 30 minutes at 4 °C. rMNK1b present in the soluble fraction was purified using the Histrap FF column (GE Healthcare) with an AKTA prime system plus (GE Healthcare) by elution at 0.5 mol/l imidazole. Purified HIS-MNK1b was dialyzed against phosphate-buffered saline (PBS), concentrated using Amicon columns (Millipore, Darmstadt, Germany) aliquoted and frozen at −80 °C. Protein concentration was determined as above.
Human GST-tagged MNK1a, MNK1b, and the truncated forms MNK1aΔ77 and MNK1ΔCt were subcloned into the BamHI and NotI sites of pGEX-4T3 and expressed in Escherichia coli Rosetta cells. The proteins were purified with glutathione–Sepharose (GenScript, Piscataway, NJ) according to the manufacturer's instructions. Briefly, expression was induced with 1 mmol/l isopropyl β-D-1-thiogalactopyranoside for 2 hours at 25 °C. The cells were suspended in buffer containing 5 mmol/l sodium phosphate, 150 mmol/l NaCl, 1 mmol/l ethylenediaminetetraacetic acid, pH 7.4, and 1 mg/ml lysozyme and incubated in ice for 30 minutes. Afterwards, 0.5% Triton X-100 was added, and bacteria were subjected to sonication. After removal of cell debris by centrifugation, the supernatant was incubated with glutathione–Sepharose equilibrated in the same buffer by rocking for 2 hours at 4 °C. After extensive washes with the same buffer, the proteins were eluted with 10 mmol/l glutathione in 50 mmol/l Tris/HCl at pH 8.
In vitro selection. Selection of DNA aptamers for recombinant rMNK1b was performed as described previously by Ramos et al.40 Briefly, synthetic random ssDNA (IBA Life Sciences, Goettingen, Germany), containing a central randomized region of 40 nucleotides flanked by two conserved 18-nucleotides regions in each end (RND40, 5′-GCGGATGAAGACTGGTCT-40N-GTTGCTCGTATTTAGGGC-3′), was denatured at 90 °C for 10 minutes and then cooled on ice for 10 minutes. For the initial SELEX round, 50 µg (2 nmol) of RND40 were mixed with 4 μg (100 pmol) of rMNK1b in 200 μl of selection buffer (20 mmol/l Tris–HCl pH 7.4, 1 mmol/l MgCl2, 150 mmol/l NaCl, and 5 mmol/l KCl) and incubated at 37 °C for 1 hour. The aptamer–rMNK1b complexes were purified by adding 20 μl of Ni-NTA superflow (Qiagen, Madrid, Spain) for 1 hour at 4 °C. After washing three times with 1 ml of selection buffer, the aptamer–rMNK1b complexes were suspended in 20 μl of distilled H2O and amplified by PCR using the primers named F3 (5′ GCGGATGAAGACTGGTGT 3′) and R3 (5′ GTTGCTCGTATTTAGGGC 3′) (IBA Life Sciences) under the conditions of 0.8 μmol/l/primer, 200 μmol/l dNTPs, 2 mmol/l MgCl2, and 2 U Taq polymerase (Biotools, Madrid, Spain) in a final volume of 50 μl for 15 cycles (95 °C for 30 seconds, 56 °C for 30 seconds, and 72 °C for 30 seconds), and PCR product was ethanol-precipitated. In the next rounds of selection, 25 µg (1 nmol) of previously selected population were denatured at 90 °C for 10 minutes and then cooled on ice for 10 minutes and used as above. In addition, after round 5, the incubation time was reduced to 30 minutes. Contraselection against Ni-NTA resin was performed after rounds 4, 7, and 10.
Enzyme-linked oligonucleotide assay. To assess the enrichment of the selected population and the affinity of the individual aptamers for the target, we performed ELONA assays in which aptamers were labeled by PCR using 5′ digoxigenin-labeled F3/5′ phosphate-labeled or 5′ phosphate-labeled F3/5′ digoxigenin-labeled R3 primers (IBA Life Sciences) and removing the phosphate-labeled strand with 1 U λ-exonuclease (New England Biolabs, Ipswich, MA) during 30 minutes at 37 °C. Alternatively, we use digoxigenin or biotin-labeled ssDNA aptamers provided by IBA Life Sciences. Recombinant proteins were diluted to 1 µg/ml in selection buffer, and 200 μl of the solution were incubated in a 96-well microtiter plate (NUNC, Rochester, NY) overnight at 4 °C and, then, washed four times in selection buffer. Afterwards, digoxigenin-labeled aptamers or digoxigenin-labeled RND40 library were diluted in selection buffer at concentrations indicated in the figures, denatured for 10 minutes at 95 °C and cooled for 10 minutes on ice. Next, 200 µl of the solution were added to each well, the plate incubated at 37 °C for 1 hour, and then washed four times with selection buffer to remove unbound ssDNA. Afterwards, 200 μl of a 1/1,000 dilution of anti-digoxigenin antibody (Roche, Basel, Switzerland) or streptavidin (GE Healthcare) conjugated with horseradish peroxidase were added to the individual wells. Following 1-hour incubation at 37 °C on a shaking platform, the plates were washed four times and developed using ABTS solution (Roche) according to the manufacturer's instruction. OD405nm values were determined using a SpectraFluor microplate reader (TECAN, Barcelona, Spain).
Analysis of aptamer-MNK1 complexes by real-time PCR. An alternative method to determine the affinity of aptamers for MNK1b is quantifying aptamers capable to bind to resin–MNK1b complex by qPCR. The complexes were obtained by incubating His-MNK1b with Ni-NTA agarose resin for 1 hour at 4 °C on a shaker. The individual aptamers (2 µg; 80 pmol) were incubated with 10 µl resin–MNK1b complexes (200 ng/tube, 5 pmol/tube) for 30 minutes at 37 °C with stirring. In parallel, the same amount of each aptamer was incubated with 10 µl of Ni-NTA resin without MNK1b. After centrifugation at 12,000g for 10 minutes, complexes were washed three times with 250 µl of selection buffer, and finally, the resin was suspended in 20 µl of H2O and incubated at 90 °C for 10 minutes. Quantitative analysis was performed by qPCR using SYBR Premix Ex kit TaqTM (Takara Bio, Shiga, Japan) and F3 and R3 oligonucleotides following the manufacturer's instructions in a iQ5 equipment (Bio-Rad, Barcelona, Spain). The reaction mixture consisted of 1× SYBR Premix Ex Taq, 0.2 µmol/l oligonucleotide, and 1 µl of template in a 20 µl/tube final volume.
Aptamer cloning and sequencing and secondary structure prediction. The dsDNA products with “A”-overhangs from SEL7MNK1b or SEL10MNK1b were cloned onto pGEM-T Easy-cloning vector (Invitrogen) following manufacturer's instructions. Individual clones were sequenced using T7 (5′-TAATACGACTCACTATAGGG-3′) and Sp6 primers (5′- ATTTAGGTGACACTATAGAA-3′) (IBA Life Sciences). Selected ssDNA molecules were subjected to secondary structure prediction using the mFold software (http://mfold.rna.albany.edu/?q=mfold/DNA-Folding-Form)41 at 37 °C in 150 mmol/l [Na+] and 1 mmol/l [Mg+2] and QGRS Mapper, a web-based server for predicting G-quadruplexes in nucleotide sequence.42
Aptamer stability assays. Three-hundred nanograms of aptamer were incubated with 0.1 U DNAse I for 2 hours, and samples were collected at the times indicated in the figures using phenol-chloroform extraction. The digested aptamer was ran on a 3% concentration MS-8 Agarose gel (Conda, Madrid, Spain) in 1× TAE buffer and visualized with GelRed (Biotium, Hayward, CA).
In vitro kinase assay. In vitro MNK1a activity was assayed as follows: 0.25 μg recombinant activated MNK1a (GST-MNK1a T385D) (BPS Bioscience, EEUU) was preincubated with 0.2 μmol/l of each aptamers (6 pmol/tube) or the same concentration of the MNK1 inhibitor (CGP57380) for 5 minutes at 30 °C, and the kinase reaction was performed using as substrate 200 μmol/l of the peptide substrate (RRRLSSLRA) in 20 mmol/l Tris–HCl, pH 7.5; 50 mmol/l KCl, 10 mmol/l MgCl2, 100 μmol/l ATP, and 1 μCi [γ -32P] ATP (Hartmann Analytic, Germany) for 30 minutes at 30 °C. Reactions were stopped onto Whatman P81 filters (GE Healthcare), filters washed three times in 1% phosphoric acid for 10 minutes, and the radioactivity read on a scintillation counter.
For in vitro MNK1b activity assay, Myc-MNK1b were purified from transfected HEK293T cell lysates (1.5–2 mg of protein) by immunoprecipitation. Briefly, Myc antibodies were added for 2 hours at 4 °C, and complexes were collected on protein G-agarose for 1 hour at 4 °C. The beads were washed three times with 1 ml of buffer A each, once with 0.5 mol/l LiCl and twice with kinase buffer (20 mmol/l Tris–HCl, pH 7.5; 50 mmol/l KCl, 10 mmol/l MgCl2). From the last washing, beads (1/10 volume) were transferred at new tubes, and kinase reactions were performed as above.
Translation assays. The assay was performed with the Rabbit Reticulocyte Lysate System, Nuclease Treated (Promega, Madrid, Spain). The aptamers were heated at 95 °C for 10 minutes in selection buffer and cooled on ice prior to addition. The reaction (25 µl) was performed following kit instructions, with 0.25 µl of luciferase RNA control (provided in the kit) and the aptamers at a final concentration of 4 µmol/l. The reaction was stopped at different incubation times (with emetine), and 2 µl of sample were used for measurement of luciferase activity with luciferase assay reagent (Promega) in a luminometer (Berthold, Bad Wildbad, Germany).
Aptacytochemistry. Aptacytochemistry is a technique that allows the microscopic localization of proteins present in the cells using aptamers. For this assay, stable MDA-MB-231 cells expressing Myc-MNK1a or Myc-MNK1b (4 × 104 cells/well) were seed on glass coverslips pretreated with poly-l-lysine (Sigma-Aldrich). After 16–24 hours, the cells were fixed with cold methanol for 20 minutes at −20 °C, washed three times with PBS, and blocked with 10% fetal bovine serum diluted in PBS (blocking buffer) for 1 hour at room temperature. Next, the cells were incubated with 2 pmol of 5′ Alexa 488-conjugated aptamer (IBA Life Sciences) in selection buffer with 0.2% bovine serum albumin for 1 hour at room temperature. Subsequently, cells were washed three times with PBS and incubated with anti-MNK1 (C-20) (1/50 dilution) or anti-MNK1 (M-20) (1/25 dilution) antibodies in blocking buffer overnight at 4 °C. After incubation, cells were washed with PBS and incubated with rhodamine-conjugated goat antibody (Jackson ImmunoResearch Laboratories, Suffolk, UK; 1/200 dilution) in blocking buffer for 1 hour at room temperature. In other assay, MDA-MB-231 cells transiently transfected with pcDNA3-Flag-MNK1b were incubated with mouse anti-Flag antibody (Sigma-Aldrich; 1/3,000 dilution) and mouse Alexa 568-conjugated IgG (Invitrogen; 1/300 dilution) as secondary antibody. Finally, the cells were mounted on glass slides using glycerol-buffer containing p-phenyl-enediamine and 30 µmol/l bis-benzamide (Hoechst 33342) for nuclear staining. Controls were made by omitting the primary antibody. Co-localization was assessed by confocal microcopy using a Nikon ECLIPSE Ti-e inverted fluorescence microscope equipped with a Nikon C1 laser scanning confocal microscope system (Nikon, Tokyo, Japan) and a 60× oil immersion objective.
Cell viability (MTT) assays. HEK293T cells (2 × 104 cells/well), MDA-MB-231 cells, or MCF7 cells (6 × 103 cells/well) were plated in p96. After 16–24 hours, the aptamers or the 38x(AG) unstructured ssDNA were transfected at concentrations indicated in the figure legends using Lipofectamine 2000 (Invitrogen) following to manufacturer's instructions for siRNA transfection. After 72 hours, medium was removed and 100 μl of MTT (1 mg/ml in culture medium) was added to each well, and plates were incubated at 37 °C for 4 hour. Next, 100 µl/well of lysis buffer (10% sodium dodecyl sulphate and 10 mmol/l HCl) were added and, after 24 hours of incubation, absorbance was read at 540 nm on a SpectraFluor microplate reader (TECAN). Percent inhibition was calculated relative to the cells transfected in the absence of aptamers (control).
Scratch wound-healing assays. MDA-MB-231 cells were plated in six-well plates at 5 × 105 cells/well in 2 ml growth medium. After 24 hours, cells were transfected with the aptamers or the 38x(AG) unstructured ssDNA at 5 nmol/l concentration as indicated above and allowed to form a confluent monolayer for 24–48 hours. Cells were treated with 0.5 µmol/l mitomycin C for 2 hours to ensure that wounds are filled due to cell migration and not by cell proliferation.43 Subsequently, the monolayer was scratched with a P-10 pipette tip, washed with media to remove floating cells, and photographed with an Olympus IX70 (time 0). Plates were then incubated at 37 °C, and images were taken after 24 hours. Cells that have migrated into the wounded area were counted, and the percent inhibition of cell migration calculated relative to the control.
Colony-forming assays. For colony formation assays, MDA-MB-231 cells were plated (3 × 104 cells/well) in 24-well plates. After 16–24 hours, the different aptamers or the 38x(AG) unstructured ssDNA were transfected at 20 nmol/l as above. After 16–24 hours, alive cells were counted by Trypan blue exclusion assay (Sigma-Aldrich) using the counter TC10 (Bio-Rad) and seed at 1 × 103 cells/well in six-well plates. Approximately 8–9 days later, the colonies were fixed, stained for 30 minutes with Giemsa 0.02% (Sigma-Aldrich), and counted with a eCount Colony Counter Pen (Heathrow Scientific, Vernon Hills, IL) and a magnifying glass (×1.75) (Bel-Art Scienceware, Wayne, NJ). Percent inhibition was calculated relative to the control.
Statistical analysis. Data are presented as an average value ± SEM from three to six independent measurements in separate experiments and analyzed using Graphpad Prism 6 (San Diego, CA). The statistical significance was performed by analysis of variance followed by Tukey's test or one-sample t-test against a control value. Significance was assumed at P < 0.05.
SUPPLEMENTARY MATERIAL Figure S1. Evolution of selection and relative abundance of each aptamer family. Figure S2. Characterization of MDA-MB-231 stable lines. Figure S3. Effects of aptamers on MNK1b kinase activity in vitro. Figure S4. Efect of aptamers on HEK293T and MCF7 cell viability. Figure S5. Viability and colony formation activity of MDA-MB-231 cells treated with the MNK1 inhibitors. Table S1. Sequence and size of the selected aptamers.
Acknowledgments
The authors thank Christopher G. Proud (South Australian Health and Medical Research Institute, Australia) for GST-plasmids coding for MNKs and Ruben Martinez-Buey (Universidad de Salamanca, Spain) for E. coli Rosetta cells and the expression vector pGEX-4T3. Support of Aptus Biotech SL is also acknowledged. E.M.G-R. and C.P-D. were supported by grants IPT-2011-1057-010000 and RTC-2014-1986-1 from Ministerio de Economía y Competitividad M.E.M. and V.M.G. are researchers from FIBio-HRC. This work was supported by grants SAF2010/21663 (Plan Nacional de I+D+I 2008–2013) from Ministerio de Economía y Competitividad (Spain) and FMM2013/0067 from Fundación Mutua Madrileña (Spain).
M.G-H. and G.F. are employed by Aptus Biotech SL. The funder provided support in the form of salaries but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The other authors declare no conflicts of interest. There are no further patents, products in development, or marketed products to declare.
Supplementary Material
Evolution of selection and relative abundance of each aptamer family.
Characterization of MDA-MB-231 stable lines.
Effects of aptamers on MNK1b kinase activity in vitro.
Efect of aptamers on HEK293T and MCF7 cell viability.
Viability and colony formation activity of MDA-MB-231 cells treated with the MNK1 inhibitors.
Sequence and size of the selected aptamers.
References
- Fukunaga, R and Hunter, T (1997). MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J 16: 1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Loghlen, A, González, VM, Piñeiro, D, Pérez-Morgado, MI, Salinas, M and Martín, ME (2004). Identification and molecular characterization of Mnk1b, a splice variant of human MAP kinase-interacting kinase Mnk1. Exp Cell Res 299: 343–355. [DOI] [PubMed] [Google Scholar]
- Parra-Palau, JL, Scheper, GC, Wilson, ML and Proud, CG (2003). Features in the N and C termini of the MAPK-interacting kinase Mnk1 mediate its nucleocytoplasmic shuttling. J Biol Chem 278: 44197–44204. [DOI] [PubMed] [Google Scholar]
- Morley, SJ and McKendrick, L (1997). Involvement of stress-activated protein kinase and p38/RK mitogen-activated protein kinase signaling pathways in the enhanced phosphorylation of initiation factor 4E in NIH 3T3 cells. J Biol Chem 272: 17887–17893. [DOI] [PubMed] [Google Scholar]
- Flynn, A and Proud, CG (1995). Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. J Biol Chem 270: 21684–21688. [DOI] [PubMed] [Google Scholar]
- Joshi, B, Cai, AL, Keiper, BD, Minich, WB, Mendez, R, Beach, CM et al. (1995). Phosphorylation of eukaryotic protein synthesis initiation factor 4E at Ser-209. J Biol Chem 270: 14597–14603. [DOI] [PubMed] [Google Scholar]
- Makkinje, A, Xiong, H, Li, M and Damuni, Z (1995). Phosphorylation of eukaryotic protein synthesis initiation factor 4E by insulin-stimulated protamine kinase. J Biol Chem 270: 14824–14828. [DOI] [PubMed] [Google Scholar]
- Topisirovic, I, Ruiz-Gutierrez, M and Borden, KL (2004). Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer Res 64: 8639–8642. [DOI] [PubMed] [Google Scholar]
- Phillips, A and Blaydes, JP (2008). MNK1 and EIF4E are downstream effectors of MEKs in the regulation of the nuclear export of HDM2 mRNA. Oncogene 27: 1645–1649. [DOI] [PubMed] [Google Scholar]
- Proud, CG (2015). Mnks, eIF4E phosphorylation and cancer. Biochim Biophys Acta 1849: 766–773. [DOI] [PubMed] [Google Scholar]
- Chrestensen, CA, Shuman, JK, Eschenroeder, A, Worthington, M, Gram, H and Sturgill, TW (2007). MNK1 and MNK2 regulation in HER2-overexpressing breast cancer lines. J Biol Chem 282: 4243–4252. [DOI] [PubMed] [Google Scholar]
- Wheater, MJ, Johnson, PW and Blaydes, JP (2010). The role of MNK proteins and eIF4E phosphorylation in breast cancer cell proliferation and survival. Cancer Biol Ther 10: 728–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konicek, BW, Stephens, JR, McNulty, AM, Robichaud, N, Peery, RB, Dumstorf, CA et al. (2011). Therapeutic inhibition of MAP kinase interacting kinase blocks eukaryotic initiation factor 4E phosphorylation and suppresses outgrowth of experimental lung metastases. Cancer Res 71: 1849–1857. [DOI] [PubMed] [Google Scholar]
- Furic, L, Rong, L, Larsson, O, Koumakpayi, IH, Yoshida, K, Brueschke, A et al. (2010). eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci USA 107: 14134–14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, HG, Silva, RL, Malina, A, Mills, JR, Zhu, H, Ueda, T et al. (2007). Dissecting eIF4E action in tumorigenesis. Genes Dev 21: 3232–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, T, Watanabe-Fukunaga, R, Fukuyama, H, Nagata, S and Fukunaga, R (2004). Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol Cell Biol 24: 6539–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington, AD and Szostak, JW (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822. [DOI] [PubMed] [Google Scholar]
- Ellington, AD and Szostak, JW (1992). Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 355: 850–852. [DOI] [PubMed] [Google Scholar]
- Tuerk, C and Gold, L (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510. [DOI] [PubMed] [Google Scholar]
- White, RR, Sullenger, BA and Rusconi, CP (2000). Developing aptamers into therapeutics. J Clin Invest 106: 929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel, KW and Giangrande, PH (2009). Therapeutic applications of DNA and RNA aptamers. Oligonucleotides 19: 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J, Wu, J, Li, C, Zhu, L, Zhang, WY, Kong, G et al. (2011). A G-quadruplex aptamer inhibits the phosphatase activity of oncogenic protein Shp2 in vitro. Chembiochem 12: 424–430. [DOI] [PubMed] [Google Scholar]
- Nagatoishi, S, Isono, N, Tsumoto, K and Sugimoto, N (2011). Loop residues of thrombin-binding DNA aptamer impact G-quadruplex stability and thrombin binding. Biochimie 93: 1231–1238. [DOI] [PubMed] [Google Scholar]
- Fujita, H, Imaizumi, Y, Kasahara, Y, Kitadume, S, Ozaki, H, Kuwahara, M et al. (2013). Structural and affinity analyses of g-quadruplex DNA aptamers for camptothecin derivatives. Pharmaceuticals (Basel) 6: 1082–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Loghlen, A, González, VM, Jurado, T, Salinas, M and Martín, ME (2007). Characterization of the activity of human MAP kinase-interacting kinase Mnk1b. Biochim Biophys Acta 1773: 1416–1427. [DOI] [PubMed] [Google Scholar]
- Beggs, JE, Tian, S, Jones, GG, Xie, J, Iadevaia, V, Jenei, V et al. (2015). The MAP kinase-interacting kinases regulate cell migration, vimentin expression and eIF4E/CYFIP1 binding. Biochem J 467: 63–76. [DOI] [PubMed] [Google Scholar]
- Zhou, ZJ, Dai, Z, Zhou, SL, Fu, XT, Zhao, YM, Shi, YH et al. (2013). Overexpression of HnRNP A1 promotes tumor invasion through regulating CD44v6 and indicates poor prognosis for hepatocellular carcinoma. Int J Cancer 132: 1080–1089. [DOI] [PubMed] [Google Scholar]
- Piekielko-Witkowska, A, Wiszomirska, H, Wojcicka, A, Poplawski, P, Boguslawska, J, Tanski, Z et al. (2010). Disturbed expression of splicing factors in renal cancer affects alternative splicing of apoptosis regulators, oncogenes, and tumor suppressors. PLoS One 5: e13690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara, T, Haniu, H and Matsuda, Y (2013). PTB-associated splicing factor (PSF) is a PPARγ-binding protein and growth regulator of colon cancer cells. PLoS One 8: e58749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, R, Gao, M, Ahmad, I, Fleming, J, Singh, LB, Rai, TS et al. (2013). Sprouty2, PTEN, and PP2A interact to regulate prostate cancer progression. J Clin Invest 123: 1157–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faratian, D, Sims, AH, Mullen, P, Kay, C, Um, I, Langdon, SP et al. (2011). Sprouty 2 is an independent prognostic factor in breast cancer and may be useful in stratifying patients for trastuzumab therapy. PLoS One 6: e23772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauf, U, Tschopp, C and Gram, H (2001). Negative regulation of protein translation by mitogen-activated protein kinase-interacting kinases 1 and 2. Mol Cell Biol 21: 5500–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain, J, Plater, L, Elliott, M, Shpiro, N, Hastie, CJ, McLauchlan, H et al. (2007). The selectivity of protein kinase inhibitors: a further update. Biochem J 408: 297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X, Yue, P, Chan, CB, Ye, K, Ueda, T, Watanabe-Fukunaga, R et al. (2007). Inhibition of mammalian target of rapamycin induces phosphatidylinositol 3-kinase-dependent and Mnk-mediated eukaryotic translation initiation factor 4E phosphorylation. Mol Cell Biol 27: 7405–7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbatia, HW, Ramalingam, S, Ramamurthy, VP, Martin, MS, Kwegyir-Afful, AK and Njar, VC (2015). Novel C-4 heteroaryl 13-cis-retinamide Mnk/AR degrading agents inhibit cell proliferation and migration and induce apoptosis in human breast and prostate cancer cells and suppress growth of MDA-MB-231 human breast and CWR22Rv1 human prostate tumor xenografts in mice. J Med Chem 58: 1900–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam, S, Gediya, L, Kwegyir-Afful, AK, Ramamurthy, VP, Purushottamachar, P, Mbatia, H et al. (2014). First MNKs degrading agents block phosphorylation of eIF4E, induce apoptosis, inhibit cell growth, migration and invasion in triple negative and Her2-overexpressing breast cancer cell lines. Oncotarget 5: 530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarpia, L, Lippman, SM and El-Naggar, AK (2012). Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin Ther Targets 16: 103–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, MM (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Iborra, S, Soto, M, Carrión, J, Alonso, C and Requena, JM (2004). Vaccination with a plasmid DNA cocktail encoding the nucleosomal histones of Leishmania confers protection against murine cutaneous leishmaniosis. Vaccine 22: 3865–3876. [DOI] [PubMed] [Google Scholar]
- Ramos, E, Piñeiro, D, Soto, M, Abanades, DR, Martín, ME, Salinas, M et al. (2007). A DNA aptamer population specifically detects Leishmania infantum H2A antigen. Lab Invest 87: 409–416. [DOI] [PubMed] [Google Scholar]
- Zuker, M (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikin, O, D'Antonio, L and Bagga, PS (2006). QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res 34(Web Server issue): W676–W682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa, S, Fujiwara, T, Matsuzaki, S, Shingaki, K, Taniguchi, M, Miyata, S et al. (2010). bFGF regulates PI3-kinase-Rac1-JNK pathway and promotes fibroblast migration in wound healing. PLoS One 5: e12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Evolution of selection and relative abundance of each aptamer family.
Characterization of MDA-MB-231 stable lines.
Effects of aptamers on MNK1b kinase activity in vitro.
Efect of aptamers on HEK293T and MCF7 cell viability.
Viability and colony formation activity of MDA-MB-231 cells treated with the MNK1 inhibitors.
Sequence and size of the selected aptamers.