Abstract
Small interfering RNA (siRNA) is effective in silencing critical molecular pathways in cancer. The use of this tool as a treatment modality is limited by lack of an intelligent carrier system to enhance the preferential delivery of this molecule to specific targets in vivo. In the present study, the in vivo behavior of novel anti-NTSR1-mAb-functionalized antimutant K-ras siRNA-loaded hybrid nanoparticles, delivered by i.p. injection to non-small-cell lung cancer in mice models, was investigated and compared to that of a naked siRNA formulation. The siRNA in anti-NTSR1-mAb-functionalized hybrid nanoparticles was preferentially accumulated in tumor-bearing lungs and metastasized tumor for at least 48 hours while the naked siRNA formulation showed lack of preferential accumulation in all of the organs monitored. The plasma terminal half-life of nanoparticle-delivered siRNA was 11 times higher (17–1.5 hours) than that of the naked siRNA formulation. The mean residence time and AUClast were 3.4 and 33 times higher than the corresponding naked siRNA formulation, respectively. High-performance liquid chromatography analysis showed that the hybrid nanoparticle carrier system protected the encapsulated siRNA against degradation in vivo. Our novel anti-NTSR1-mAb-functionalized hybrid nanoparticles provide a useful platform for in vivo targeting of siRNA for both experimental and clinical purposes.
Keywords: biodistribution, lung cancer, lung tumor, nanoparticles, pharmacokinetics, siRNA, targeted delivery
Introduction
Small interfering RNAs (siRNAs) have emerged as molecules with high potential to silence critical molecular pathways involved in tumor progression. However, to harness the full potential of this treatment modality in the clinical arena, intelligent nanoparticle delivery systems need to be developed in tandem. This is crucial since siRNAs have been shown to lack the capacity to be transfected into cells on their own due to their relatively high polarity and molecular size.1 To deliver siRNA to an intracellular target, a nanoparticle delivery system is often used to “mask” the negative charges on these molecules. The small size of these nanoparticles facilitates endocytosis, enabling the internalization of siRNAs while preventing degradation within the lysosomes.2 Naked (unencapsulated) siRNA are also degraded by serum endonucleases and efficiently cleared from systemic circulation by glomerular filtration, resulting in a very short plasma half-life.3,4 Various carriers have been studied as potential delivery vehicles for siRNA therapeutic. Such carriers include cell-penetrating peptides, lipid-based carriers, and polymeric and dendrimeric carriers.1,5,6 However, these delivery systems are still limited by major challenges.4 Therefore, there is an unmet need for a smart nanoparticle delivery system for efficient and stable siRNA transfection.
Recently, our group reported on a novel hybrid nanoparticle delivery system for siRNA delivery. These nanoparticles, composed of human IgG and poloxamer-188, are designed to prevent immunogenic/inflammatory response in vivo and to reduce clearance by macrophages. These nanoparticles were able to successfully deliver siRNA to the cytosol of A549 lung cancer cell lines without eliciting immunogenic/inflammatory response in murine macrophages, RAW 264.7.4,7 Using these nanoparticles to deliver siRNA against mutant K-ras, a significant downregulation of both protein and mRNA of K-ras was achieved.4 Further, the enclosed siRNA was protected from nuclease in 50% serum for up to 48 hours.
The objective of the current study is to investigate the potential of these nanoparticles to selectively deliver the encapsulated siRNA to lung tumor in metastatic orthotopic murine models of non-small-cell lung cancer (NSCLC), following the functionalization of the nanoparticles with antineurotensin receptor 1 monoclonal antibody (anti-NTSR1-mAb). We also aim to determine the pharmacokinetic parameters of the encapsulated siRNA (antimutant K-ras siRNA) in comparison to the naked siRNA. NTSR1 is being used as a target for delivery of these nanoparticles to lung tumor because NTSR1 has been shown by immunohistochemistry to be overexpressed in ~60% of lung adenocarcinoma.8 In early stages of NSCLC, NTSR1 was one of the first 50 genes upregulated and associated with disease-free survival.8 We hypothesize that this overexpression in NSCLC can be used to achieve an optimized targeting of siRNA to NSCLC by conjugating anti-NTSR1-mAb to the surface of our hybrid nanoparticles.
Results
Nanoparticle preparation, characterization, and in vitro assessment
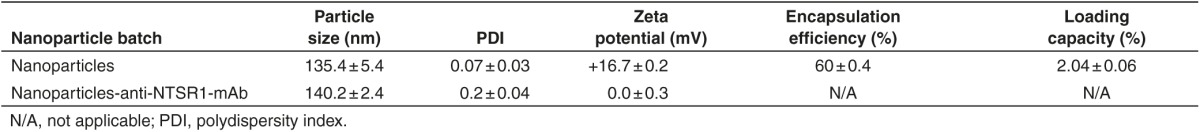
Table 1 shows that the nanoparticles were ~135 nm in diameter and the functionalization of these nanoparticles with anti-NTSR1-mAb slightly increased the size of the nanoparticles to 140 nm. Prior to the functionalization of the nanoparticles, a zeta potential of +16.7 mV was recorded. However, the nanoparticles showed no charges following the functionalization step. The encapsulation efficiency and loading capacity of antimutant K-ras siRNA in these nanoparticles were 60 and 2%, respectively.
Table 1. Particle size, PDI, and zeta potential analysis of nanoparticles measured by photon correlation spectroscopy.
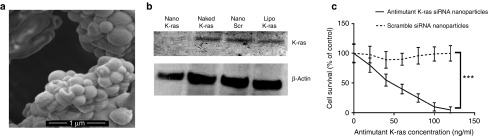
Scanning electron micrograph of the nanoparticles in Figure 1a confirms that the nanoparticles were mostly spherical in shape. The particles were fairly homogenous in morphology.
Figure 1.
Nanoparticle characterization and knockdown efficiency. (a) Scanning electron micrograph shows the spherical morphology of the nanoparticles. (b) Western blot analysis showing in vitro knockdown efficiency of siRNA nanoparticles against mutant G12S K-ras in A549 cells. Nano K-ras represents antimutant K-ras siRNA-loaded hybrid nanoparticles, naked K-ras represents unencapsulated antimutant K-ras siRNA, Nano Scr represents scramble siRNA-loaded hybrid nanoparticles, and lipo K-ras represents antimutant K-ras siRNA transfected using lipofectamine 2000. (c) MTT assay showing in vitro antiproliferative activity of antimutant K-ras siRNA nanoparticles in A549 cells, n = 5, ***P ≤ 0.001.
Figure 1b demonstrates the knockdown efficiency of these antimutant K-ras siRNA-loaded hybrid nanoparticles against G12S mutant K-ras normally expressed in A549 cells. These nanoparticles produced superior downregulation of G12S mutant K-ras when compared to lipofectamine 2000 as a transfection reagent.
Antiproliferative effect of these antimutant K-ras siRNA-loaded nanoparticles in A549 cells is further demonstrated in Figure 1c. In contrast, scramble siRNA-loaded hybrid nanoparticles did not demonstrate any antiproliferative effect in A549 cells (Figure 1c).
Metastatic orthotopic murine model of NSCLC
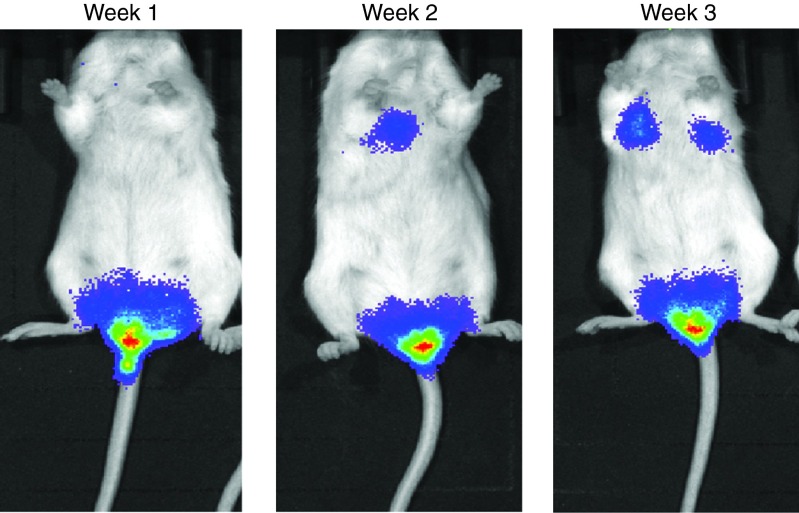
Mouse models used for these were developed over a 3-week period. Following the injection of A549-luciferase cells into the tail vein of the mice, tumor burden was monitored using Xenogen IVIS bioluminescence imaging system. Tumor was initially established in the colorectal region after the first week. This tumor quickly metastasizes to the two lungs of the mice after 3 weeks of injection as seen in Figure 2.
Figure 2.
SCID beige mice showing tumor burden in the lungs and colorectal region over a 3-week period.
Plasma pharmacokinetics
Antimutant K-ras siRNA was isolated from mice plasma using the analytical procedure described in Figure 6. Ion-pair high-performance liquid chromatography (HPLC) analysis showed a linear standard curve ranging from 50 to 50,000 ng/ml (r = 0.9998). The lowest detection limit was 50 ng/ml. siRNA recovery from plasma following isolation with Clarity OTX isolation and ion-pair HPLC analysis was consistently above 90%.
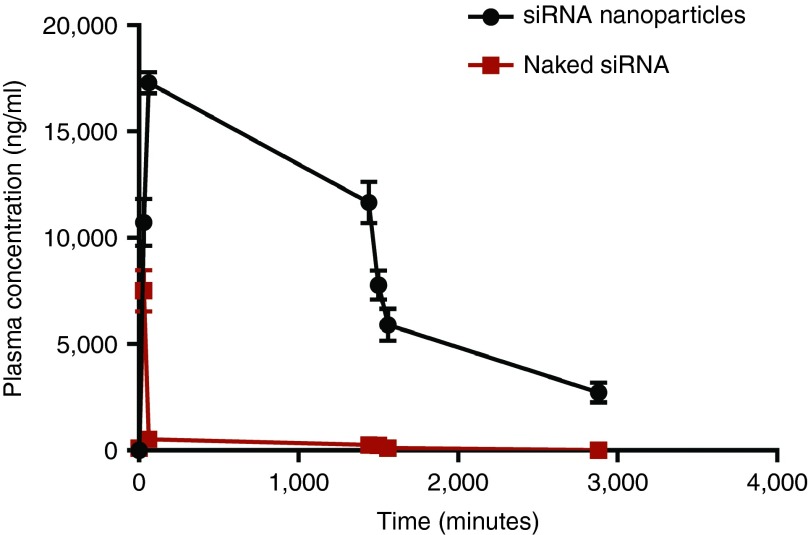
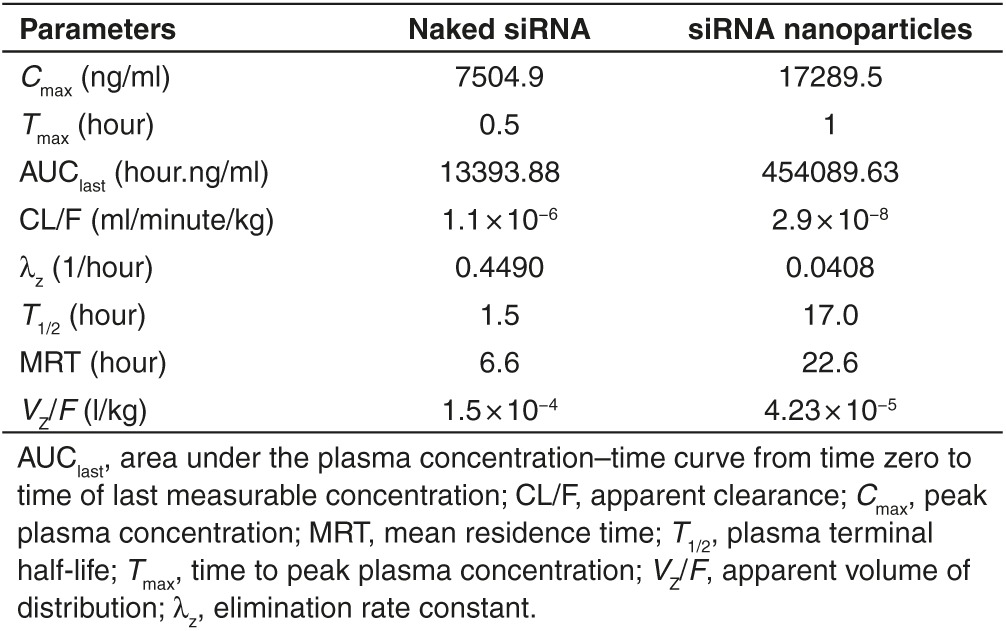
Figure 3 demonstrates the plasma concentration versus time curves when equivalent of 0.9 mg/kg of siRNA either naked or loaded into anti-NTSR1-mAb-functionalized hybrid nanoparticles. The maximum concentration (Cmax) of siRNA delivered by anti-NTSR1-mAb-functionalized hybrid nanoparticles was 17289.5 ng/ml compared to 7504.9 ng/ml observed for the naked siRNA formulation. These maximum concentrations were observed at 30 and 60 minutes (Tmax) for naked siRNA and siRNA in anti-NTSR1-mAb-functionalized hybrid nanoparticles, respectively. Figure 4 demonstrates the superiority of using novel anti-NTSR1-mAb-functionalized hybrid nanoparticles for siRNA delivery in murine models of lung cancer in comparison to the naked siRNA formulation.
Figure 3.
siRNA plasma concentration over time. Tumor-bearing mice were given a single dose of 0.9 mg/kg of antimutant K-ras siRNA either in anti-NTSR1-mAb-functionalized nanoparticles or as naked siRNA. n = 3.
Figure 4.
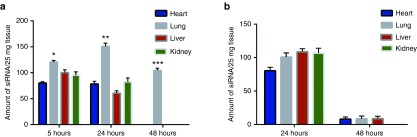
In vivo biodistribution studies. Biodistribution of antimutant K-ras siRNA following i.p. injection of siRNA in (a) functionalized hybrid nanoparticles and (b) naked siRNA. Lung data statistically compared to the next highest accumulation in tissues of the same time point. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. n = 3.
Key pharmacokinetic parameters presented in Table 2 further demonstrate the superiority of using these novel anti-NTSR1-mAb-functionalized hybrid nanoparticles for siRNA delivery in murine models of lung cancer in comparison to the naked siRNA formulation.
Table 2. Key pharmacokinetic parameters of naked siRNA and siRNA-loaded anti-NTSR1-mAb-functionalized hybrid nanoparticles in SCID models of metastatic orthotopic non-small-cell lung cancer.
Tissue distribution of siRNA in NSCLC murine models
Figure 4 demonstrates the discriminatory delivery of siRNA to tumor-bearing lungs by anti-NTSR1-mAb-functionalized hybrid nanoparticles over a 3-day period. In contrast, naked siRNA was not discriminatory in tissue delivery. After 5 hours, a significant accumulation of siRNA delivered by anti-NTSR1-mAb-functionalized hybrid nanoparticles was observed in tumor-bearing lungs in comparison to the liver (P ≤ 0.05). On the second day after siRNA administration, an even more significant accumulation in the lung tumor was observed when compared to the kidneys (P ≤ 0.01). On the third day after siRNA administration, total clearance was observed in the liver, heart, and kidney, leaving the lung tissue as the only tissue with siRNA accumulation.
In vivo stability of siRNA
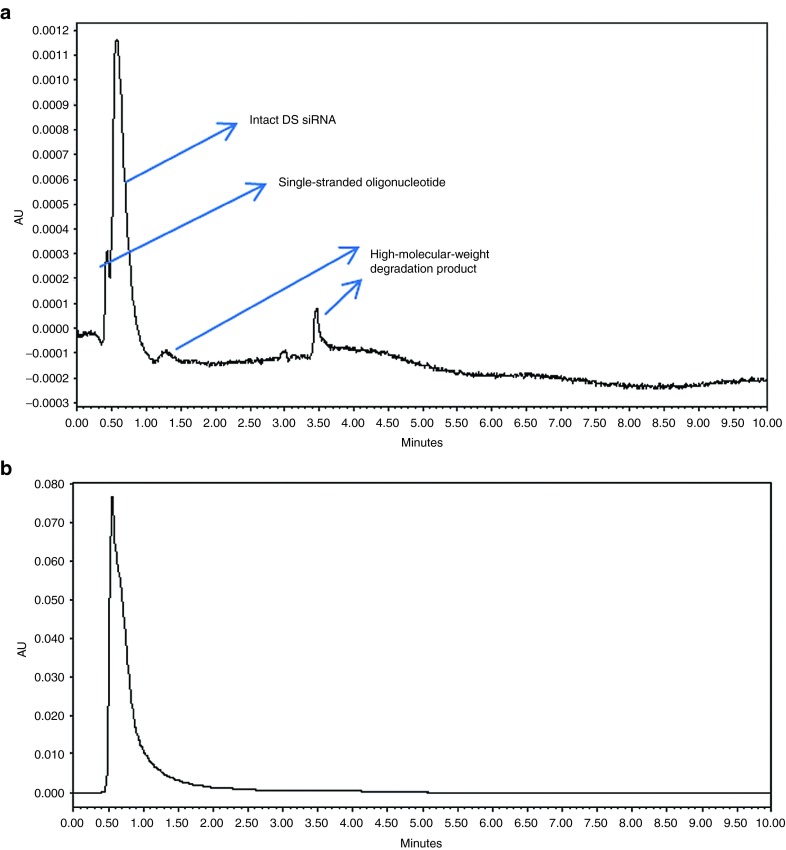
In vivo stability of siRNA delivered by anti-NTSR1-mAb-functionalized hybrid nanoparticles was compared to that of the naked siRNA using ion-pair HPLC-UV following siRNA isolation using Clarity OTX, an oligonucleotide extraction cartridge (Phenomenex, Torrance, CA). Blood samples collected by orbital puncture at 0-, 30-, and 90-minute and 24-, 25-, 26-, and 48-hour time points were analyzed for signs of degradation. The presence of single-stranded oligonucleotides in the naked siRNA samples was observed immediately after 30 minutes of administration. However, degradation products were not observed in the corresponding sample for hybrid nanoparticle-delivered siRNA. Figure 5a demonstrates the presence of single-stranded oligonucleotides as well as the presence of higher molecular weight degradation products in the naked siRNA samples, 25 hours after administration. However, such degradation/metabolic products were not observed in the corresponding hybrid nanoparticle-delivered siRNA samples. The first sign of degradation in the hybrid nanoparticle-delivered siRNA samples was not observed until 48 hours postadministration. This confirms the ability of the hybrid nanoparticles to protect the encapsulated siRNA in vivo similar to what was observed in vitro.4
Figure 5.
In vivo stability studies. High-performance liquid chromatogram of (a) naked siRNA and (b) hybrid nanoparticle-delivered siRNA 25 hours after i.p. administration to tumor-bearing mice. DS siRNA, double-stranded siRNA.
Discussion
This study presents the biodistribution and pharmacokinetics properties of siRNA encapsulated in an anti-NTSR1-mAb-functionalized hybrid nanoparticles in comparison to naked siRNA. These hybrid nanoparticles combine the benefits derived from human immunoglobulin G (human IgG) and poloxamer-188 (polyoxyethylenepolyoxypropylene block copolymer), for stable and efficient siRNA delivery. Human IgG is the main antibody isotype found in blood, it is the main immunoglobulin that protects the body against infection. Since human IgG is part of the body's natural defends mechanism, we hypothesized that a nanoparticle composing IgG would reduce the well-documented immunogenic reaction experienced with most nanoparticle formulations. This hypothesis has been tested and proved to be correct.4 Poloxamer-188, a nonionic triblock copolymer, has been previously used as a stealth polymer to prevent macrophage uptake of nanoparticles hence circumventing the reticuloendothelial system during systemic circulation. It was proposed that it reduces opsonization by serum proteins hence reducing macrophageal uptake.9,10 The combination of these two components in a single nanoparticle delivery system makes these nanoparticles quite unique. In this study, hybrid nanoparticles were functionalized with anti-NTSR1-mAb to convert them into an actively targeted nanoparticle delivery system for delivering siRNAs to NSCLC cells using NTSR1 as a delivery target. NTSR1 has been shown by immunohistochemistry to be overexpressed in ~60% of lung adenocarcinoma.7 In early stages of NSCLC, NTSR1 was one of the first 50 genes upregulated and associated with disease-free survival.8 We aimed to take advantage of this overexpression in NSCLC to achieve an optimized targeting of siRNA to NSCLC by conjugating anti-NTSR1-mAb to the surface of our hybrid nanoparticles. Anti-NTSR1-mAb was attached to the nanoparticles to the preformed nanoparticles by thiolating anti-NTSR1-mAb using 2-iminothiolane (Traut's reagent) as reported previously.11,12 N-[p-maleimidophenyl] isocyanate was used as a heterobifunctional, sulfhydryl and hydroxyl-reactive linker and was conjugated to the hydroxyl groups present in poloxamer-188 on the surface of the hybrid nanoparticles.13 The isocyanate end of N-[p-maleimidophenyl] isocyanate reacted with the hydroxyl group on the nanoparticles to form carbamate linkage while the maleimide end reacted with the sulfhydryl groups in the thiolated anti-NTSR1.14 The particle size of anti-NTSR1-mAb-functionalized nanoparticles was slightly higher than the corresponding nonfunctionalized nanoparticles as shown in Table 1. Particle size, zeta potential, loading efficiency, and encapsulation efficiency of hybrid nanoparticles presented in Table 1 have been previously reported by our group.7 The increase in size may be attributed to the conjugation of mAb to the surface of the nanoparticles. Further, the fact that several centrifugation steps and lyophilization were involved in the conjugation process may also contribute to the increase in particle size.12,13
Metastatic model of lung cancer developed in Figure 2 shows that tumor was initially implanted in the colorectal region of the mice before finally metastasizing to the lung rather than the commonly observed trend of initial implantation in the lungs before metastasis to other areas of the body. This is because of the relatively lesser number (5 × 105) of A549-luciferase cells injected into the tail vain of the mice. Observations in our laboratory showed that when higher number of cells such as 5 × 106 was injected into the tail vain, initial implantation of tumor in the lung was observed.
One of the main limitations to the application of siRNA in translational medicine is the fact that they are very vulnerable to endonuclease enzymes found in body fluids.4 The rapid clearance of naked siRNA from the blood of the mice could be attributed to degradation by endonucleases in the blood. However, the hybrid nanoparticle delivery system offered protective armor to the siRNA when delivered using the anti-NTSR1-mAb-functionalized hybrid nanoparticles, hence the lower clearance rate. The protection of the siRNA by the nanoparticle delivery system could also be responsible for the longer plasma terminal half-life and the higher mean residence time in comparison to the naked siRNA formulation.
Figure 4 demonstrates that siRNA delivered using anti-NTSR1-mAb-functionalized hybrid nanoparticles were mostly accumulated in the lung after 5 hours. However, the liver and the kidney also had substantial amount of siRNA delivered at this time. The liver among other organs such as spleen belong to the reticuloendothelial system organs.15 Liver is also a highly perfused organ; hence, nanoparticle carriers are rapidly distributed to this organ.15,16 Further, the microvessels of liver have relatively large fenestrations which allow entry of particles as high as ~200 nm.17 In view of this, systemically delivered nanoparticles usually accumulate in the liver and spleen.17 The kidney also demonstrated a high amount of siRNA probably because it is highly perfused and it is the main organ for elimination. However, after 24 hours, a more discriminatory delivery of siRNA to tumor-bearing lungs began to manifest due to the effect of the presence of anti-NTSR1-mAb on the hybrid nanoparticles coupled with the fact that tumors are naturally more permeable to the penetration of particles between 100 and 800 nm due to enhanced permeation and retention effect.1,15 After 48 hours, siRNA from anti-NTSR1-mAb-functionalized hybrid nanoparticles could only be seen in the tumor-bearing lung and was completely cleared from other organs probably due to the interaction between anti-NTSR1-mAb on the surface of the nanoparticles and the NTSR1 receptors present in the tumor-bearing lungs. Out of curiosity, we wanted to know if anti-NTSR1-mAb-functionalized nanoparticles were able to deliver siRNA to metastasized tumor in the colorectal region. Tumor in this area was harvested in some mice and analyzed for siRNA delivery. A significant accumulation of siRNA was detected in these metastasized tumors. In contrast, naked siRNA formulation showed a conspicuous lack of selectivity in the delivery of siRNA to tumor-bearing lungs and other organs after 24 hours. Further, naked siRNA was also significantly cleared from the tumor-bearing lungs after 48 hours.
In conclusion, Anti-NTSR1-mAb-functionalized hybrid nanoparticles were able to drive the pharmacokinetics and biodistribution of siRNA in tumor-bearing mice. These nanocarriers were able to significantly reduce the clearance and increase the circulation time of siRNA in plasma when compared to that of naked siRNA formulation. siRNA delivered to anti-NTSR1-mAb-functionalized hybrid nanoparticles were selectively delivered in tumor-bearing lungs over a period of 48 hours. Hybrid nanoparticles also protected encapsulated siRNA from degradation in systemic circulation. Thus, these novel hybrid nanoparticles can serve as an effective nonviral vector for siRNA delivery.
Materials and methods
Chemicals and reagents. Human IgG was purchased from Equitech Bio (TX). Poloxamer-188, RNase-free water, and fetal bovine serum were obtained from Fisher Scientific. siRNA against mutated K-ras G12S was designed and purchased from Thermo Scientific (Rockford, IL; formerly Dharmacon). siG12S sense and antisense sequences are GUUGGAGCUAGUGGCGUAGdTdT and CUACGCCACUAGCUCCAACdTdT, respectively. Clarity OTX solid-phase extraction was obtained from Phenomenex. Erlotinib hydrochloride and HPLC grade solvents were purchased from Fisher Scientific (Pittsburgh, PA). XenoLight RediJect D-Luciferin Bioluminescent Substrate was obtained from Perkin Elmer (Waltham, MA). Mouse antineurotensin receptor 1 monoclonal antibody was purchased from Santa Cruz Biotechnology (Dallas, TX). Monoclonal antibodies specific for mutant G12S K-ras proteins was obtained from NewEast Biosciences, King of Prussia, PA.
Cell culture. Human adenocarcinoma cell line A549-luciferase, expressing K-ras mutation at G12S, was obtained from Perkin Elmer. They were maintained in F12K medium supplemented with 10% fetal bovine serum and 1% antibiotics. They were kept in a humidified air atmosphere with 5% carbon dioxide.
Western blot analysis. Cells were washed twice with ice-cold phosphate-buffered saline. Cell lysates were prepared in Pierce RIPA buffer (Thermo Scientific) with addition of Pierce Protease Inhibitor Mini Tablets (Thermo Scientific). The protein concentration was determined with the Coomassie Plus (Bradford) Assay Kit (Thermo Scientific). About 80 µg of total proteins was separated on NuPAGE 4–12% Bis-Tris gels (Life Technologies, Carlsbad, CA) with NuPAGE MES SDS running buffer (Life Technologies) and subsequently transferred onto nitrocellulose membrane of 0.45 µm pore size (Life Technologies). The membrane with proteins was blocked according to manufacturer's instructions (Invitrogen, Carlsbad, CA) for 1 hour at room temperature and probed with primary antibodies—K-ras G12S monoclonal antibody (1:500; New East Biosciences, Malvern, PA) and β-actin (1:5,000; Sigma-Aldrich, St. Louis, MO) overnight at 4 °C. The membranes were washed three times for 5 minutes in wash buffer according to manufacturer's instructions (Invitrogen) and incubated with secondary goat anti-mouse secondary antibodies, conjugated with horseradish peroxidase (Molecular Probes, Eugene, OR) at a dilution 1:1,000. Immune complexes were detected with chemiluminescent substrate, Pierce ECL Western Blotting Substrate (Thermo Scientific) in a dark room on tabletop processor SRX-101A (Konica Minolta, Japan). Images were quantified in the ImageJ software.
Cytotoxicity assay. MTT assay was used to determine the effect of siG12S-loaded nanoparticles on the proliferation of A549 cells. Cells (1 × 104)/well were seeded in 96-well plates and incubated at 37 °C in a humidified atmosphere with 5% carbon dioxide for 24 hours. The cells were then treated with different concentrations of siG12S-loaded hybrid nanoparticles and scramble-siRNA-loaded hybrid nanoparticles. Cells were incubated for 72 hours. About 10 µl of 12 mmol/l MTT reagent was then added to each well. This was then incubated at 37 °C for 4 hours. The medium was aspirated and 50 µl of sterile dimethyl sulfoxide was added to each well and mixed thoroughly with pipet. The cells were then incubated at 37 °C for 10 minutes. The plate was read at 540 and 650 nm.
Animals. Female severe combined immunodeficient (SCID) beige mice, 7–8 weeks old and weighing ~25 g were supplied by Taconic and maintained in the American Association for the Accreditation of Laboratory Animal Care (AAALAC)-accredited facility at Thomas Jefferson University. All animal studies were approved by the Institutional Animal Care and Use Committee of Thomas Jefferson University.
Preparation of siRNA-loaded hybrid nanoparticles. Hybrid nanoparticles were prepared based on our previously reported protocol.4 About 50 mg of excipient-free human IgG was dissolved in 0.01 N HCl containing 20 mg of poloxamer-188 and 187 µg of siRNA to make a 10 ml total solution in a 50 ml beaker. The final concentration of human IgG in each solution amounted to 5 mg/ml. This solution was then slowly titrated with 0.01 N NaOH to bring the pH of the mixture to 7, which is the isoelectric point (pI) of human IgG as determined in our laboratory using isoelectric focusing. The nanoparticles were continuously mixed on a magnetic stirrer for additional 10 minutes. At the pI, siRNA-loaded nanoparticles were spontaneously precipitated. The colloidal suspension was then centrifuged with a micro centrifuge (Eppendorf centrifuge 5418) at 2,000 rpm for 5 minutes. The nanoparticles were rinsed with double distilled deionized water before being redispersed in water and snap-frozen using liquid nitrogen. This was then loaded into a freeze-dryer (Labconco FreezeZone 4.6), and lyophilization was performed for 48 hours.
Thiolation of anti-NTSR1-mAb. Thiolation of anti-NTSR1-mAb was performed based on a method adopted from a previously used method.18 Briefly, 1.3 × 10−4 mol/l of Traut's reagent (2-Iminothiolane HCl) was prepared in phosphate-buffered saline (pH 7.4). Further, 500 µl of this solution was then added to 1 ml of 1.3 × 10−6 mol/l (0.2 mg/ml) of anti-NTSR1-mAb solution. The reaction was stirred for 2 hours at 25 °C. The mixture then centrifuged at 4,000 rpm and 10 °C for 15 minutes using 30 kDa cutoff centrifugal ultrafilters (Millipore Corp.) to exclude unreacted Traut's reagent.
Activation of hybrid nanoparticles with heterobifunctional cross-linker. About 5 mg/ml of hybrid nanoparticles were dispersed in 0.1 mol/l phosphate buffer (pH 7.2). This was then added to 2 mg/ml of N-[p-maleimidophenyl] isocyanate, a heterobifunctional cross-linker that links sulfhydryl to hydroxyl groups.10 N-[p-maleimidophenyl] isocyanate was dissolved in 50 mmol/l phosphate buffer (pH 8).The reaction was performed for 3 hours at 25 °C, after which the activated nanoparticles were centrifuged at 4,000 rpm at 10 °C for 15 minutes using 30 kDa ultrafilters (Millipore Corp.).
Preparation of anti-NTSR1-mAb-functionalized hybrid nanoparticles. The activated nanoparticles were finally functionalized with the thiolated anti-NTSR1-mAb by adding 500 µl of 2 mg/ml thiolated anti-NTSR1-mAb to 4 ml of activated nanoparticle suspension (5 mg/ml) and then incubated for 3 hours at 25 °C. The functionalized nanoparticles were then centrifuged for 30 minutes at 4,000 rpm and 10 °C. Unconjugated anti-NTSR1-mAb in the supernatant was quantified using Total Protein Kit (Micro Lowry, Sigma) based on the supplier's instructions.
Nanoparticle characterization. Photon correlation spectroscopy using ZetaSizer Nano ZS (Malvern Instruments, UK) was used to measure the particle size and zeta potential of the nanoparticles. Pellets were redispersed in deionized water and sonicated for ~5 minutes. Intensity autocorrelation was measured at a scattering angle (θ) of 173°. The Z-average and polydispersity index were recorded in triplicate. For zeta potential, samples were taken in a universal dip cell (Malvern Instruments) and the zeta potential recorded in triplicate.
The morphology of nanoparticles was obtained using scanning electron microscopy. Powder was layered on a scanning electron microscopy stub and spread gently using a spatula. Samples were then coated with a thin layer of palladium. Coated samples were imaged using a Zeiss Supra 50 V system (Zeiss, Jena, Germany).
Murine models of metastatic NSCLC. Metastatic models of NSCLC were created by injecting 5 × 105 A549-luciferase cells suspended in sterile phosphate-buffered saline into the tail veins of SCID beige mice. Tumor burden was monitored using Xenogen IVIS bioluminescence imaging system. About 100 µl of 30 mg/ml Xenolight Rediject D-Luciferin was administered by i.p. injection into the mice ~10 minutes before animals were imaged.
Animal treatment and blood sampling. Tumor-bearing mice were divided into two groups. The first group was treated with a single dose of 0.9 mg/ml siRNA loaded into anti-NTSR1-mAb-functionalized hybrid nanoparticles while the second group was treated with 0.9 mg/ml naked siRNA. Both nanoparticles and naked siRNA were dispersed in sterile phosphate-buffered saline. Blood samples were collected from three mice per group per time point at 0, 30, and 60 minutes and 24, 25, 26, and 48 hours post-dose by retro-orbital puncture into ethylenediaminetetraacetic acid tubes. Animals were also sacrificed on days 1, 2, and 3 to harvest the lungs, kidneys, heart, and livers for analysis. Blood samples were mixed with equal amount of lysis-loading buffer of Clarity OTX kit (Phenomenex, CA) and stored at −80 °C until ready to be used. Tissues were snap-frozen in the liquid nitrogen stored at −80 °C before analysis.
siRNA isolation and ion-pair HPLC. siRNA in blood and tissue was isolated for HPLC using Clarity OTX kit (Phenomenex) with Thermo Scientific HyperSep Vacuum Manifold (Thermo Sci National Scientific, Rockwood, TN) according to manufacturer's instructions. Briefly, equal aliquots of Clarity OTX loading buffer was mixed with plasma samples prior to loading on the solid-phase extraction cartridge. Solid-phase extraction isolation cartridge (Clarity OTX 100 mg/3 ml: Phenomenex) was first wetted with methanol before equilibrating with Clarity equilibration buffer (10 mmol/l phosphate, pH 5.5) before sample loading. For tissue samples, they were first homogenized in 0.1 mol/l Tris buffer, pH 8.0 and then mixed with equal amount of lysis-loading buffer. After sample loading, equilibration buffer was added to the cartridge to rinse twice before rinses with Clarity OTX wash buffer (10 mmol/l phosphate, pH 5.5/50% acetonitrile). Elution buffer (100 mmol/l ammonium bicarbonate, pH 8.0/40% acetonitrile/10% tetrahydrofuran) was then added to elute the siRNA from the cartridge. Samples were then speed vacuum evaporated to 100 µl before liquid chromatography analysis.
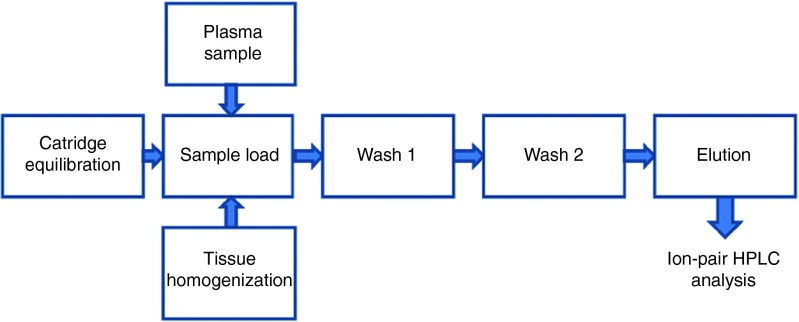
Figure 6 shows the schematic of the siRNA isolation protocol. SiRNA was analyzed using a well-validated ion-pair HPLC method.4,19,20 Briefly, analysis was performed using an Alliance HPLC system, Waters 2695 Separation Module combined with a Water 2998 photodiode array detector (Waters, Milford, MA). A Clarity 3 µm Oligo-RP column (Phenomenex) with column dimension 50 × 2.0 mm was used. One microliter of siRNA sample was injected using 20 mmol/l triethylamine-acetic acid (pH 7) and 5–12% acetonitrile, gradient elusion as mobile phase. Analysis was performed at a flow rate of 0.2 ml/minute. UV detection was performed at 269 nm and chromatogram was recorded using Empower Prosoftware.
Figure 6.
Schematic of the siRNA isolation protocol using Clarity OTX. HPLC, high-performance liquid chromatography.
Pharmacokinetic data analysis. Pharmacokinetic parameters were estimated using noncompartment analysis of the composite data with WinNonlin software version 6.0 (Pharsight, Mountain View, CA).
Statistical analysis. Results are expressed as mean ± SD, unless otherwise indicated. Statistically significant difference between two groups was determined by two-tailed Student's t-test. A P value of 0.005 was taken as statistically significant.
Acknowledgments
We acknowledge the support of the Science Center and Thomas Jefferson University towards this project through the award of QED grant number: S1402.
References
- Wang, J, Lu, Z, Wientjes, MG and Au, JL (2010). Delivery of siRNA therapeutics: barriers and carriers. AAPS J 12: 492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng, YC, Mozumdar, S and Huang, L (2009). Lipid-based systemic delivery of siRNA. Adv Drug Deliv Rev 61: 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Water, FM, Boeman, OC, Wouterse, AC, Peters, JG, Russel, FG and Masereeuw, R (2006). Intravenously administered short interfering RNA accumulates in the kidney and selectively suppresses gene function in renal proximal tubes. Drug Metab Dispos 34: 1393–1397. [DOI] [PubMed] [Google Scholar]
- Lakshmikuttyamma, A, Sun, Y, Lu, B, Undieh, AS and Shoyele, SA (2014). Stable and efficient transfection of siRNA for mutated KRAS silencing using novel hybrid nanoparticles. Mol Pharm 11: 4415–4424. [DOI] [PubMed] [Google Scholar]
- Meade, BR and Dowdy, SF (2007). Exogenous siRNA delivery using peptide transduction domains/cell penetrating peptides. Adv Drug Deliv Rev 59: 134–140. [DOI] [PubMed] [Google Scholar]
- de Martimprey, H, Vauthier, C, Malvy, C and Couvreur, P (2009). Polymer nanocarriers for the delivery of small fragments of nucleic acids: oligonucleotides and siRNA. Eur J Pharm Biopharm 71: 490–504. [DOI] [PubMed] [Google Scholar]
- Dim, N, Perepelyuk, M, Gomes, O, Thangavel, C, Liu, Y, Den, R et al. (2015). Novel targeted siRNA-loaded hybrid nanoparticles: preparation, characterization and in vitro evaluation. J Nanobiotechnology 13: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alifano, M, Souazé, F, Dupouy, S, Camilleri-Broët, S, Younes, M, Ahmed-Zaïd, SM et al. (2010). Neurotensin receptor 1 determines the outcome of non-small cell lung cancer. Clin Cancer Res 16: 4401–4410. [DOI] [PubMed] [Google Scholar]
- Jain, D, Athawale, R, Bajaj, A, Shrikhande, S, Goel, PN and Gude, RP (2013). Studies on stabilization mechanism and stealth effect of poloxamer 188 onto PLGA nanoparticles. Colloids Surf B Biointerfaces 109: 59–67. [DOI] [PubMed] [Google Scholar]
- Zhang, WL, Liv, JP and Chen, ZQ (2009). Stealth tanshinone IIA-loaded solid lipid nanoparticles in rats. Acta Pharm Sin 44: 1422–1428. [PubMed] [Google Scholar]
- Akinc, A, Goldberg, M, Qin, J, Dorkin, JR, Gamba-Vitalo, C, Maier, M et al. (2009). Development of lipidoid-siRNA formulations for systemic delivery to the liver. Mol Ther 17: 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, AR and Shoyele, SA (2014). Influence of surface modification and the pH on the release mechanisms and kinetics of Erlotinib from antibody-functionalized chitosan nanoparticles. Ind Eng Chem Res 53: 2987–2993. [Google Scholar]
- Ishida, O, Maruyama, K, Sasaki, K and Iwatsuru, M (1999). Size-dependent extravasation and interstitial localization of polyethyleneglycol liposomes in solid tumor-bearing mice. Int J Pharm 190: 49–56. [DOI] [PubMed] [Google Scholar]
- PMPI-Instruction Manual. ThermoScientific. (2012). <www.thermoscientific.com/pierce>. Accessed 18 February 2015.
- Christensen, J, Litherland, K, Faller, T, van de Kerkhof, E, Natt, F, Hunziker, J et al. (2014). Biodistribution and metabolism studies of lipid nanoparticle-formulated internally [3H]-labeled siRNA in mice. Drug Metab Dispos 42: 431–440. [DOI] [PubMed] [Google Scholar]
- Juliano, R, Bauman, J, Kang, H and Ming, X (2009). Biological barriers to therapy with antisense and siRNA oligonucleotides. Mol Pharm 6: 686–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L, Sullenger, B and Juliano, R (2010). The role of carrier size in the pharmacodynamics of antisense and siRNA oligonucleotides. J Drug Target 18: 567–574. [DOI] [PubMed] [Google Scholar]
- Yousefpour, P, Atyabi, F, Vasheghani-Farahani, E, Movahedi, AA and Dinarvand, R (2011). Targeted delivery of doxorubicin-utilizing chitosan nanoparticles surface-functionalized with anti-Her2 trastuzumab. Int J Nanomedicine 6: 1977–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge, J, Scott, G, Hail, M and McGinley, M (2011). Preparation and LC/MS analysis of oligonucleotides therapeutics from biological matrices. Chromatography Today <https://www.chromatographytoday.com/article_read/983/> (March 2011). Accessed 4 March 2015.
- Scott, G, Gause, H, Rivera, B and McGinley, M (2011). Rapid extraction of therapeutic oligonucleotides from primary tissues for LC/MS analysis using Clarity® OTX™, an oligonucleotides extraction cartridge <www.phenomenex.com>. Accessed 4 March 2015.