Abstract
Murine double minute 2 (MDM2) and 4 (MDM4) are known as the main negative regulators of p53, a tumor suppressor. They are able to form heterodimers that are much more effective in the downregulation of p53. Therefore, the MDM2–MDM4 complex could be a target for promising therapeutic restoration of p53 function. To this aim, a deeper understanding of the molecular mechanisms underlining the heterodimerization is needed. The kinetic and thermodynamic characterization of the MDM2–MDM4 complex was performed with two complementary approaches: atomic force spectroscopy and surface plasmon resonance. Both techniques revealed an equilibrium dissociation constant (KD) in the micromolar range for the MDM2–MDM4 heterodimer, similar to related complexes involved in the p53 network. Furthermore, the MDM2–MDM4 complex is characterized by a relatively high free energy, through a single energy barrier, and by a lifetime in the order of tens of seconds. New insights into the MDM2–MDM4 interaction could be highly important for developing innovative anticancer drugs focused on p53 reactivation.
Keywords: MDM2, MDM4, atomic force spectroscopy, surface plasmon resonance
Background
p53 is a tumor suppressor protein, defined as the “guardian of the genome”, that controls response to a broad range of cellular stresses and is an important target for cancer treatment.1–3 p53 is regulated by a complex network within which Murine double minute 2 (MDM2) and 4 (MDM4 or MDMX) are the main negative regulators.4 Both MDM2 and MDM4 are able to inhibit the transcriptional activity of p53 by physically binding with its N-terminal transactivation domain.5 Moreover, MDM2 displays E3 ubiquitin ligase activity by targeting p53 for proteasomal degradation; such a function involves its C-terminal Really Interesting New Gene (RING) domain.6,7 On the other hand, the RING domain enables the formation of homodimers (MDM2–MDM2; MDM4–MDM4) or heterodimers (MDM2–MDM4).8–10 Although MDM2 and MDM4 share a similar structure, the latter does not show any significant E3 ubiquitin ligase activity toward p53 but represents an essential activator of MDM2 for p53 polyubiquitination.11 Remarkably, it is the MDM2–MDM4 heterodimer that plays a pivotal role in the p53 regulation network; primarily by controlling p53 abundance through proteasomal degradation, and also because of its involvement both in the regulation of p53 transcriptional activity and p53-induced apoptosis.12–18 Therefore, the interaction between MDM2 and MDM4 could be an appropriate target to design a highly effective cancer therapy.19–21 Indeed, designing specific antagonists for the heterodimeric complex could prevent the formation of the MDM2–MDM4 complex and thus abolish its inhibitory activity, with restoration of the p53 oncosuppressive function. In this context, the efficacy of such a strategy would benefit considerably from the study of the MDM2–MDM4 molecular interaction properties. For this purpose, we characterized MDM2–MDM4 complex by using atomic force spectroscopy (AFS) and surface plasmon resonance (SPR), which are two innovative and complementary techniques operating under nearly native conditions. AFS is a nanotechnology-based approach for studying the interactions between one protein firmly bound to the atomic force microscope (AFM) tip and the other to the substrate, under the application of an external force, and allows to measure, at the single-molecule level and with a piconewton sensitivity, the unbinding force of interaction, which is connected with the binding affinity and energy landscape of the two biomolecules.22 On the other hand, SPR is a flexible and powerful tool for studying the kinetic and equilibrium characterization of binding processes occurring between a immobilized ligand on a sensor chip and its partner free in solution.23 Both techniques witness the occurrence of a specific interaction between MDM2 and MDM4 with an equilibrium dissociation constant (KD) in the micromolar range. Moreover, AFS reveals that the formation of the MDM2–MDM4 complex overcomes a single energy barrier and is driven by a relatively high free energy. These results are discussed in connection with the thermodynamic parameters of the MDM2–p53 complex.
Materials and methods
Protein expression and purification
Full length untagged human MDM2 (hereafter MDM2) (57.8 kDa) was purchased from Sigma-Aldrich Co. (St Louis, MO, USA) and used without further purification. Full length N-terminal glutathione S-transferase (GST)-tagged human MDM4 (hereafter MDM4) (81 kDa) was expressed and purified from bacteria. Briefly, Escherichia coli BL21 cells were transformed with pGEX–MDM4–GST vector and grown at 37°C in 500 mL of Luria–Bertani broth until the optical density reached 0.5 at 600 nm. Subsequently, 0.25 mM isopropyl β-D-1-thiogalactopyranoside was added and after 2 hours, the cells were pelleted and resuspended in 25 mL NETN buffer (20 mM Tris, pH 8, 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 0.5% NP-40) containing 1 mg/mL lysozime, 1 mM dithiothreitol, and protease inhibitors (Hoffman-La Roche Ltd., Basel, Switzerland). The cells were lysed on ice by sonication and then were subjected to centrifugation at 10,000× g for 10 minutes at 4°C. The pellet containing the insoluble fraction was resuspended in 1 mL NETN–Sarkosyl buffer (NETN, 2% Sarkosyl detergent) and then recentrifuged. The supernatant was mixed with 500 µL of 50% glutathione agarose beads. After adsorption for 2 hours at 4°C, the beads were washed twice with NETN buffer and resuspended in 20 mM reduced glutathione in 400 µL of tris sodium triton (TST) buffer pH 8 (50 mM Tris, pH 8, 150 mM NaCl, 0.1% Triton ×100). After 1 and 16 hours, the eluted protein was collected by centrifugation, filtered through Amicon Ultra-15 50 kDa cutoff filter (EMD Millipore, Billerica, MA, USA), and then resuspended in phosphate-buffered saline (PBS; 50 mM K3PO4, 150 mM NaCl, pH 7.5). The protein was then checked and quantified using sodium dodecyl sulfate polyacrylamide gel electrophoresis and Coomassie Brilliant Blue R-250, while a known amount of bovine serum albumin was used as standard.
Atomic force spectroscopy experiments
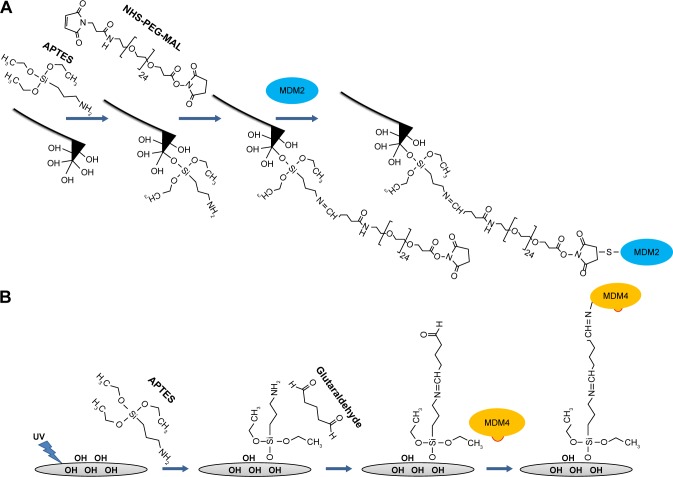
Silicon nitride AFM tips (Bruker Corporation, Billerica, MA, USA) and glass substrates (ø12 mm) were functionalized to covalently link MDM2 and MDM4, respectively, using the procedure previously reported and schematically shown in Figure 1.24 In particular, we used 10 µL of a 6 µM solution of MDM2 for tip functionalization and 30 µL of a 6 µM MDM4 solution for substrate preparation. Force measurements were performed at room temperature with a commercial AFM (Nanoscope IIIa/Multimode AFM, Veeco Instruments, Plainview, NY, USA) in 50 mM PBS buffer of pH 7.5 in a force calibration mode. Force curves were acquired using rectangular-shaped Si3N4 cantilevers (Bruker probes MSNL-10) with a nominal spring constant, knom, of 0.02 N/m functionalized as sketched in Figure 1A. A schematic representation of the approach–retraction cycle is shown in Figure 2. At the beginning, the MDM2–functionalized tip was moved toward the MDM4–functionalized substrate with a ramp size of 150 nm and the cantilever deflection 0 (point 1). The biomolecules jumped-to-contact at point 2. With further pressure of the tip onto the substrate, there was an electronic repulsion due to overlapping of molecular orbitals, producing an upward deflection. Once the preset maximum contact force value of 0.5 nN was reached, the approaching phase (dotted line) of the cantilever was stopped (point 3). After 100 ms encounter time, the cantilever was retracted from the substrate. During this retraction phase (continuous line), the adhesion forces and/or bonds formed during the contact phase caused the tip to bend downward, adhering to the substrate up to some distance beyond the initial contact point (point 4). As retraction continued, the spring force overcame the interacting force and the cantilever jumped off, sharply returning to a noncontact position (point 5). Force curves were collected by approaching the functionalized tip at different points of the substrate surface at the constant velocity of 50 nm/s, while the retraction velocity was varied from 50 to 4,200 nm/s, according to the selected nominal loading rates, defined as the product of the nominal cantilever spring constant (knom) by the tip pulling velocity (ν), and set in the range of 1 to 84 nN/s. The effective loading rates were then calculated from ksyst × ν, where the spring constant of the entire system, ksyst, was obtained, at various loading rates, from the slope of the retraction curve immediately prior to the unbinding event (in this way, taken into account was the effect on the knom of molecules, proteins, and/or linkers, bound to the AFM tip).25 To obtain a reliable quantitative information with statistical significance from the experiments, thousands of force curves were acquired at each loading rate. Finally, to check the specificity of the interactions, control experiments were performed by incubating the MDM2 functionalized tip with 10 µL of a 6 µM solution of MDM4 for 16 hours at 4°C and changes in unbinding frequency were monitored at 4 nN/s loading rate. The exerted force, which was able to break the complex, called the unbinding force, F, could be calculated by multiplying the cantilever deflection at the jump-off (d in Figure 2) by its effective spring constant (keff), which was, in turn, determined by the nondestructive thermal noise method.26 The force curves registered during the measurements showed different shapes. Curves corresponding to acceptable unbinding events were characterized, in the retraction phase, by sharp peaks, starting and ending points at zero deflection line, and by a nonlinear curved shape before the jump-off (Figure 2), which was related to the stretching features of the polyethylene glycol linker.27 Additionally, somewhat ambiguous deflection jumps were determined by using the 1/f noise approach.28,29
Figure 1.
Sketch of the immobilization strategies of MDM2 and MDM4 on the AFM tip and the glass substrate, respectively.
Notes: (A) MDM2 is anchored to the AFM tip through the SH group of cysteine residues exposed on the protein surface after the tip functionalization with APTES and NHS–PEG–maleimide crosslinker. (B) MDM4 is immobilized on a glass slide via its lysine residues through a chemical platform involving sequentially linked APTES and glutaraldehyde.
Abbreviations: AFM, atomic force microscope; APTES, (3-aminopropyl)triethoxysilane; aminopropyl triethoxysilane; MDM2, murine double minute 2; MDM4, murine double minute 4; NHS, N-hydroxysuccinimide; PEG, polyethylene glycol; SH, sulfhydryl; UV, ultraviolet.
Figure 2.
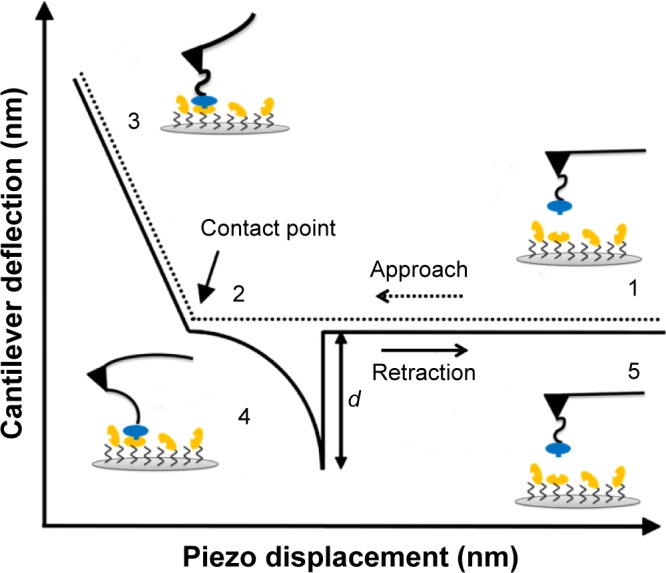
Schematic representation of the approach–retraction cycle showing a specific unbinding event.
Note: The variation of the cantilever deflection at the jump-off, d, is indicated.
Surface plasmon resonance experiments
SPR analyses were performed at 25°C with a Biacore X100 instrument (GE Healthcare BioSciences AB, Uppsala, Sweden). Full length MDM4–GST was immobilized onto the CM5 sensor chip surface using the GST Capture Kit (GE Healthcare), following the procedure recommended by the producer. This strategy involved a capturing molecule covalently immobilized on the surface in order to attach to the ligand by high affinity binding. To this aim, an anti-GST antibody (GE Healthcare) was immobilized using standard amine coupling chemistry.30 Briefly, the carboxymethylated dextran surface of the CM5 sensor chip (GE Healthcare) was first activated by a 7-minute injection of a 1:1 mixture of 0.4 M N-ethyl-N-(3-diethylaminopropyl) carbodiimide and 0.1 M N-hydroxysuccinimide at 10 µL/min to give reactive succinimide esters. Then a solution of the anti-GST antibody (30 µg/µL) in immobilization buffer (10 mM sodium acetate, pH 5.0, GE Healthcare) was fluxed on the reactive matrix using a flow rate of 10 µL/min. In such a way, the N-hydroxysuccinimide esters reacted spontaneously with the ligand amines to form covalent links (Figure 3A). We immobilized approximately 7,500 resonance units (RU) of the anti-GST antibody in flow cells 1 (Fc1) and 2 (Fc2). Unreacted sites were blocked by a 7-minute injection of 1 M ethanolamine HCl, pH 8.5, (GE Healthcare) with a flow rate of 10 µL/min. Moreover, the blocking of high-affinity sites was performed by a 3-minute injection of recombinant GST (5 µg/mL) in 50 mM PBS buffer, pH 7.5, followed by a 2-minute injection of regeneration solution (10 mM glycine–HCl pH 2.1, GE Healthcare). This procedure blocked the sites that were difficult to regenerate and could interfere with the subsequent analysis. The two flow cells were immobilized using identical conditions; therefore, Fc2 was used for ligand capture while the Fc1 was used as a reference. In the Fc2, after the baseline was stabilized by fluxing the running buffer (50 mM PBS buffer, pH 7.5, 0.005% surfactant P20 from GE Healthcare) over the surface, the ligand, MDM4–GST (20 nM), was injected at 10 µL/min flow rate until reaching an immobilized ligand level (R) of 70 RU with a theoretical analyte binding capacity (Rmax) of 50 RU calculated by using Equation 1:
| (1) |
where analyte MW is the molecular weight of MDM2 and ligand MW is the molecular weight of MDM4–GST. To prevent nonspecific binding of the analyte, MDM2, with the anti-GST antibody, we injected recombinant GST (20 µg/mL) (GE Healthcare), blocking the anti-GST antibody binding sites that did not react with the ligand in the Fc2 and saturated all the anti-GST binding sites in the Fc1. A schematic representation of the immobilization procedures of Fc1 and Fc2 is shown in Figure 3B and C, respectively. SPR analyses were performed by using a single-cycle kinetics approach that consists of sequential injections of increasing concentrations of the analyte over the functionalized sensor chip surface, without regeneration steps between each sample injection.31 Using a flow rate of 30 µL/min, five sequential increasing concentrations, ranging from 0.2 to 2 µM, of MDM2 solution in the running buffer, were fluxed on the sensor chip surface for 180 seconds, followed by a dissociation of 180 seconds with running buffer and a final dissociation of 500 seconds with the same buffer, without intermediate regeneration. Finally, the substrate was regenerated by using a 1-minute pulse of regeneration solution (GE Healthcare) at 10 µL/min. Analytical cycles were programmed by means of a wizard template, and the entire analysis was completely automated. A sketch of the MDM2 interaction over the MDM4 functionalized substrate is shown in Figure 3D. BiaEvaluation software 2.1 (GE Healthcare) was used to extract kinetic parameters from SPR data. The reference surface, Fc1, was used to correct systematic noise and instrument drift. The binding assay also included three start-up cycles using buffer to equilibrate the surface, as well as a zero concentration cycle of analyte in order to have a blank response usable for double reference subtraction.32 Sensorgrams were then globally fitted to a 1:1 interaction model including the correction for mass transfer rate. Goodness of the fit was evaluated by χ2 value and residual plots.
Figure 3.
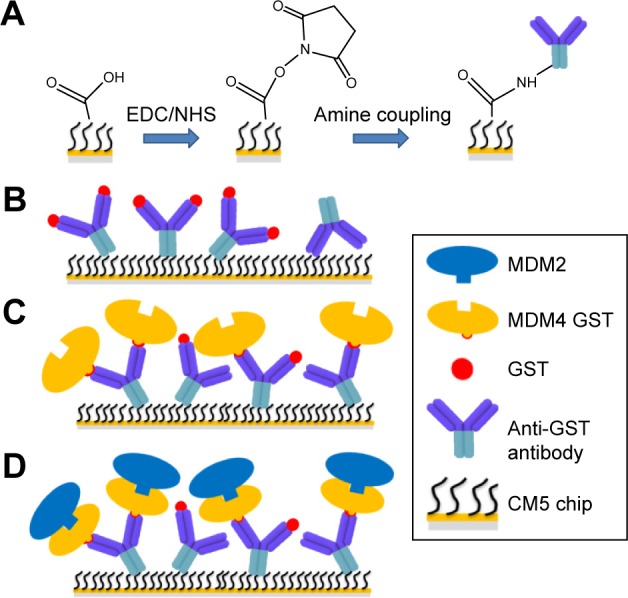
Sketch of the SPR experiment.
Notes: (A) The CM5 matrix was activated by injecting a mixture of EDC and NHS, then the amine coupling was performed; by fluxing the anti-GST antibody over the active surface, its amino groups spontaneously reacted with the NHS esters of the substrate to form covalent links. (B) In the reference Fc1, anti-GST antibody antigen sites were saturated with GST. (C) In Fc2, the MDM4–GST was captured by the anti-GST antibody, then the antigen sites were saturated with GST. (D) During binding experiments, MDM2 specifically interacted with MDM4 in the Fc2.
Abbreviations: CM5, carboxymethylated dextran; EDC, N-ethyl-N-(3-diethylaminopropyl) carbodiimide; Fc1, flow cell 1; Fc2, flow cell 2; GST, glutathione S-transferase; MDM2, murine double minute 2; MDM4, murine double minute 4; NHS, N-hydroxysuccinimide; SPR, surface plasmon resonance.
Results
AFS unbinding results
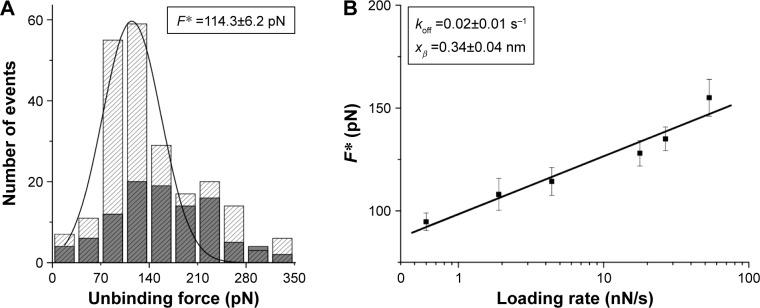
The interaction between MDM2 and MDM4 was investigated at the single-molecule level by using AFS. Approach–retraction cycles (Figure 2) were performed at five increasing loading rates by using a MDM2 functionalized tip and a MDM4 conjugated substrate (Figure 1) and specific force curves were collected. The unbinding forces were evaluated and cast into a histogram for each loading rate; in all the cases, a single mode distribution was obtained and the most probable unbinding force (F*) was extracted from the maximum of the peak of the corresponding histogram; a representative histogram corresponding to 4 nN/s loading rate is shown in Figure 4A. The recorded F* increased with the loading rate with values varying between 90 and 150 pN, which were in the range usually reported for specific biological interactions.33 The unbinding frequency, calculated as the ratio of the number of events corresponding to specific unbinding processes over the total recorded events, was approximately 17%, being consistent with values previously reported for other protein–protein interactions.24,34–36 Blocking experiments on the MDM2–MDM4 complex demonstrated that the unbinding events observed arose from a specific recognition process.37 Indeed, after incubation of the MDM2–functionalized tip with free MDM4, we noted lowering of the number of events over the whole histogram with a 60% reduction of the unbinding frequency (Figure 4A), thus confirming that the formation of the MDM2–MDM4 complex was significantly specific. As the molecular dissociation measured by AFS takes place under the application of an external force, the system is far from the thermodynamic equilibrium with an alteration of the energy profile.22 Therefore, to extract the kinetic and energy landscape parameters at the equilibrium, the use of suitable theoretical models is required.38–41 Most of them take into account the unbinding process in terms of a crossing over a single, sharp barrier through the application of a time-dependent force. However, the most widely used is the model developed by Bell and Evans, which predicts a linear dependence of the F* on the natural logarithm of the loading rate, r, as given by Equation 2:
| (2) |
where kB is Boltzmann’s constant, T is the absolute temperature, koff is the dissociation rate constant, and xβ is the width of the energy barrier along the direction of the applied force.38,39 By plotting F* versus the logarithm of the effective loading rate r (Figure 4B), we observed a single regime indicative of a single energy barrier and unique transition state of the reaction. Moreover, by fitting these data with Equation 2, we found a xβ of (0.34±0.04) nm and a koff of (0.02±0.01) s−1 (Figure 4B), with these values being typical of specific biological complexes.37 To further investigate the energy landscape of MDM2–MDM4 interaction, an estimation of the free energy ΔG was done under the assumption of a small number of involved bonds, and then by neglecting the contribution of the entropic term. In particular, the free energy of the unbinding process can be obtained by the Eyring model, through the following expression:
| (3) |
where h is the Planck’s constant. Accordingly, from koff we obtained ΔGcomplex ≅ (19.3±0.3) kcal/mol.36 To complete the kinetic profile of the interaction, we also estimated the association rate constant (kon) for the MDM2–MDM4 complex according to the expression kon = NA × Veff/t0.5, where NA is the Avogadro’s number, Veff is the effective volume of a half-sphere with a radius reff around the tip, and t0.5 is the time for the half-maximal binding probability, given by t0.5=2 reff/ν, where ν is the approach speed of the cantilever.42,43 Accordingly, a kon of approximately 104 M−1s−1 was obtained. The assessment of both the dissociation and association rate constants allowed us to determine an equilibrium dissociation constant KD, calculated as KD = koff/kon, of approximately 10−6 M for the MDM2–MDM4 complex.
Figure 4.
Analysis of AFS results for the MDM2–MDM4 complex.
Notes: (A) Histograms of the unbinding forces before (gray strips) and after (dark gray) blocking, at a loading rate of 4 nN/s. The most probable unbinding force value (F*) was determined from the maximum of the main peak of the histogram of unbinding forces before blocking by fitting with Gaussian function (black curve). (B) Plot of the most probable unbinding forces, F*, versus the logarithm of the loading rates for the MDM2–MDM4 interaction. The line is obtained by fitting the experimental data by the Bell–Evans model. The resulting koff and the xβ of the potential barrier along the direction of the applied force are shown in the insert.
Abbreviations: AFS, atomic force spectroscopy; F*, force value; koff, dissociation rate constant; MDM2, murine double minute 2; MDM4, murine double minute 4; xβ, width of the energy barrier.
SPR kinetic results
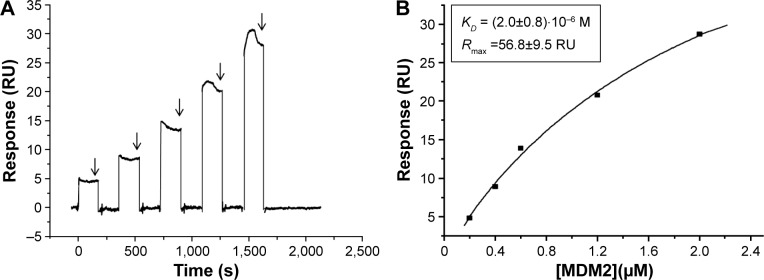
The interaction kinetics of the MDM2–MDM4 complex was studied in bulk condition by SPR using a single-cycle kinetics approach in which the analyte, MDM2, and the buffer were alternately injected into the cell where the ligand, MDM4, was previously immobilized.31 The sensorgram (Figure 5A) shows the SPR signal (RU) as a function of time obtained from the successive injection of MDM2 at five progressively higher concentrations. During the first injection with a 0.2 µM MDM2 solution, the signal increased and reached a steady state before the end of the injection. Subsequently, the buffer flowed over the ligand and the MDM2 rapidly and completely dissociated as the signal strength decreased, close to zero. The same trend was also observed for the successive injections of increasing concentration of MDM2. As far as higher MDM2 concentrations were used, progressively higher RU values at the steady state were obtained; this being indicative of increasing levels of MDM2 binding to the surface-immobilized MDM4. To extract information on the affinity between MDM2 and MDM4, the SPR data were analyzed in the framework of the Langmuir 1:1 binding model, which assumes a simple reversible bimolecular reaction between the ligand and the analyte, by using the SPR evaluation software package and considering the response at the steady state (arrows in Figure 5A).44,45 The RU values at the steady state (Req) for every sample injection were plotted against the MDM2 concentration [MDM2] (Figure 5B) and were fitted by using Equation 4 (continuous line in Figure 5B):
| (4) |
where Rmax is the analyte binding capacity of the ligand-functionalized substrate (Figure 5B) and RI, defined as the bulk refractive index, is the offset on the RU axis and is assumed to be the same for all samples. A KD of (2.0±0.8) ×10−6 M (reduced χ2=0.68 RU2) was obtained for the MDM2–MDM4 complex.
Figure 5.
SPR single-cycle kinetic of MDM2–MDM4 interaction.
Notes: (A) Sensorgram of the response (RU) versus time of the single-cycle kinetics assay performed by injecting five increasing concentrations (0.2, 0.4, 0.6, 1.2, 2.0 µM) of MDM2 on the MDM4 substrate, without any regeneration. Arrows indicate the steady state for each sample injection. (B) Plot of SPR response (RU) at the steady state versus the MDM2 concentration used for the binding assay. By fitting data with the steady state affinity model (Biacore X100 Evaluation software) (black curve), the equilibrium dissociation constant (KD) and the analyte binding capacity (Rmax) values reported in the insert were obtained.
Abbreviations: MDM2, murine double minute 2; MDM4, murine double minute 4; RU, resonance units; SPR, surface plasmon resonance.
Discussion
The kinetic and thermodynamic properties of the MDM2–MDM4 complex, investigated in vitro at the single-molecule level by using AFS, indicate the formation of a specific heterodimeric complex. The specificity of the MDM2–MDM4 interaction is clearly revealed both by the F* values and the unbinding frequency, obtained by force measurements, and by the xβ and the koff values, obtained by fitting with the Bell–Evans model, which are in the range usually reported for specific biological interactions.24,33,35,36,43 Moreover, the lifetime τ (τ =1/koff) of the MDM2–MDM4 interaction is >70 times longer than τ of the MDM2–p53 association; such a value suggests that the heterodimer is available for several cycles of association and dissociation with p53 before the displacement of the MDM2–MDM4 complex occurs.34 The relatively long lifetime of the complex with respect to that of MDM2-p53 might be consistent with the efficacy of the heterodimer in the p53 downregulation. Furthermore, the stability of the MDM2–MDM4 complex is confirmed by the high unbinding free energy obtained by AFS; being similar to that reported for some antibody–antigen pairs and higher than that reported for the unbinding of the MDM2–p53 complex.36,46 Interestingly, the micromolar KD determined by AFS at the single-molecule level is almost identical to that estimated in bulk by SPR, with this value being similar to KD values reported for related complexes involved in the p53 pathway such as MDM2–p53, MDM4–p53, and MDM4 homodimers.47,48 Although MDM2–p53 and MDM2–MDM4 complexes share a similar affinity, they display a different lifetime and unbinding free energy values. Such a difference among the affinity and the thermodynamic parameters has already been reported and attributed to the fact that some receptors might form rapidly fairly transient bonds, while others with similar affinity might require higher amounts of time to form durable bonds.49
In summary, these new insights into kinetics and energy landscape of the MDM2–MDM4 complex may contribute to further investigation on the ternary complex formed by the MDM2–MDM4 heterodimer and p53 and, more importantly, could be of significant help in designing specific antagonists that could prevent the formation of the MDM2–MDM4 complex, with subsequent restoration of the p53 oncosuppressive function.
Acknowledgments
This work was supported by the Italian Association for Cancer Research (AIRC): Grant IG15866 (SC) and Grant IG12767 (FM).
Ethical approval was not required by the institutional review boards of Università della Tuscia, Università Cattolica di Roma, or Consiglio Nazionale delle Ricerche (CNR) because the study used safe purified proteins for in vitro interaction studies.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358(6381):15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 2.Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9(9):702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 3.Duffy MJ, Synnott NC, McGowan PM, Crown J, O’Connor D, Gallagher WM. p53 as a target for the treatment of cancer. Canc Treat Rev. 2014;40(10):1153–1160. doi: 10.1016/j.ctrv.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Marine JC, Francoz S, Maetens M, Wahl G, Toledo F, Lozano G. Keeping p53 in check: essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 2006;13(6):927–934. doi: 10.1038/sj.cdd.4401912. [DOI] [PubMed] [Google Scholar]
- 5.Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010;20(5):299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420(1):25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 7.Boddy MN, Freemont PS, Borden KL. The p53-associated protein MDM2 contains a newly characterized zinc binding domain called the RING finger. Trends Biochem Sci. 1994;19:198–199. doi: 10.1016/0968-0004(94)90020-5. [DOI] [PubMed] [Google Scholar]
- 8.Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 1999;447(1):5–9. doi: 10.1016/s0014-5793(99)00254-9. [DOI] [PubMed] [Google Scholar]
- 9.Kostic M, Matt T, Martinez-Yamout MA, Dyson HJ, Wright PE. Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53. J Mol Biol. 2006;363(2):433–450. doi: 10.1016/j.jmb.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 10.Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Diff. 2008;15(5):841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Wang J, Jiang X. MdmX protein is essential for Mdm2 protein-mediated p53 polyubiquitination. J Biol Chem. 2011;286(27):23725–23734. doi: 10.1074/jbc.M110.213868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawai H, Lopez-Pajares V, Kim MM, Wiederschain D, Yuan ZM. RING domain–mediated interaction is a requirement for MDM2’s E3 ligase activity. Cancer Res. 2007;67(13):6026–6030. doi: 10.1158/0008-5472.CAN-07-1313. [DOI] [PubMed] [Google Scholar]
- 13.Pant V, Xiong S, Iwakuma T, Quintás-Cardama A, Lozano G. Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proc Natl Acad Sci. 2011;108(29):11995–12000. doi: 10.1073/pnas.1102241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang L, Yan Z, Liao X, et al. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc Natl Acad Sci. 2011;108(29):12001–12006. doi: 10.1073/pnas.1102309108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tollini LA, Jin A, Park J, Zhang Y. Regulation of p53 by Mdm2 E3 ligase function is dispensable in embryogenesis and development, but essential in response to DNA damage. Cancer Cell. 2014;26(2):235–247. doi: 10.1016/j.ccr.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Y, Regunath K, Jacq X, Prives C. Cisplatin causes cell death via TAB1 regulation of p53/MDM2/MDMX circuitry. Genes Dev. 2013;27(16):1739–1751. doi: 10.1101/gad.212258.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Conza G, Mancini F, Buttarelli M, Pontecorvi A, Trimarchi F, Moretti F. MDM4 enhances p53 stability by promoting an active conformation of the protein upon DNA damage. Cell Cycle. 2012;11(4):749–760. doi: 10.4161/cc.11.4.19208. [DOI] [PubMed] [Google Scholar]
- 18.Mancini F, Pieroni L, Monteleone V, et al. MDM4/HIPK2/p53 cytoplasmic assembly uncovers coordinated repression of molecules with anti-apoptotic activity during early DNA damage response. Oncogene. 2016;35(2):228–240. doi: 10.1038/onc.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13(2):83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu W, Xu C, Ling X, et al. Targeting RING domains of Mdm2–MdmX E3 complex activates apoptotic arm of the p53 pathway in leukemia/lymphoma cells. Cell Death Dis. 2015;6:e2035. doi: 10.1038/cddis.2015.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pellegrino M, Mancini F, Lucà R, et al. Targeting the MDM2/MDM4 interaction interface as a promising approach for p53 reactivation therapy. Cancer Res. 2015;75(21):4560–4572. doi: 10.1158/0008-5472.CAN-15-0439. [DOI] [PubMed] [Google Scholar]
- 22.Bizzarri AR, Cannistraro S. Dynamic Force Spectroscopy and Biomolecular Recognition. Boca Raton: CRC Press; 2012. pp. 193–236. [Google Scholar]
- 23.Cooper MA. Optical biosensors in drug discovery. Nat Rev Drug Discov. 2002;1(7):515–528. doi: 10.1038/nrd838. [DOI] [PubMed] [Google Scholar]
- 24.Coppari E, Yamada T, Bizzarri AR, Beattie CW, Cannistraro S. A nanotechnological, molecular-modeling, and immunological approach to study the interaction of the anti-tumorigenic peptide p28 with the p53 family of proteins. Int J Nanomedicine. 2014;9:1799–1813. doi: 10.2147/IJN.S58465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedsam C, Wehle AK, Kühner F, Gaub HE. Dynamic single-molecule force spectroscopy: bond rupture analysis with variable spacer length. J Phys Condens Matter. 2003;15:S1709–S1723. [Google Scholar]
- 26.Hutter JL, Bechhoefer J. Calibration of atomic-force microscope tips. Rev Sci Instrum. 1993;64:1868–1873. [Google Scholar]
- 27.Kienberger F, Ebner A, Gruber HJ, Hinterdorfer P. Molecular recognition imaging and force spectroscopy of single biomolecules. Acc Chem Res. 2006;39(1):29–36. doi: 10.1021/ar050084m. [DOI] [PubMed] [Google Scholar]
- 28.Bizzarri AR, Cannistraro S. 1/f α noise in the dynamic force spectroscopy curves signals the occurrence of biorecognition. Phys Rev Lett. 2013;110(4):048104. doi: 10.1103/PhysRevLett.110.048104. [DOI] [PubMed] [Google Scholar]
- 29.Bizzarri AR, Cannistraro S. Antigen–antibody biorecognition events as discriminated by noise analysis of force spectroscopy curves. Nanotechnology. 2014;25(33):335102. doi: 10.1088/0957-4484/25/33/335102. [DOI] [PubMed] [Google Scholar]
- 30.Johnsson B, Löfås S, Lindquist G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal Biochem. 1991;198(2):268–277. doi: 10.1016/0003-2697(91)90424-r. [DOI] [PubMed] [Google Scholar]
- 31.Karlsson R, Katsamba PS, Nordin H, Pol E, Myszka DG. Analyzing a kinetic titration series using affinity biosensors. Anal Biochem. 2006;349(1):136–147. doi: 10.1016/j.ab.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 32.Myszka DG. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Curr Opin Biotechnol. 1997;8(1):50–57. doi: 10.1016/s0958-1669(97)80157-7. [DOI] [PubMed] [Google Scholar]
- 33.Morfill J, Blank K, Zahnd C, et al. Affinity-matured recombinant antibody fragments analyzed by single-molecule force spectroscopy. Biophys J. 2007;93(10):3583–3590. doi: 10.1529/biophysj.107.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Funari G, Domenici F, Nardinocchi L, et al. Interaction of p53 with Mdm2 and azurin as studied by atomic force spectroscopy. J Mol Recognit. 2010;23(4):343–351. doi: 10.1002/jmr.999. [DOI] [PubMed] [Google Scholar]
- 35.Santini S, Di Agostino S, Coppari E, Bizzarri AR, Blandino G, Cannistraro S. Interaction of mutant p53 with p73: a surface plasmon resonance and atomic force spectroscopy study. Biochim Biophys Acta. 2014;1840(6):1958–1964. doi: 10.1016/j.bbagen.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 36.Coppari E, Santini S, Bizzarri AR, Cannistraro S. Kinetics and binding geometries of the complex between β2-microglobulin and its antibody: an AFM and SPR study. Biophys Chem. 2016;211:19–27. doi: 10.1016/j.bpc.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Bizzarri AR, Cannistraro S. The application of atomic force spectroscopy to the study of biological complexes undergoing a biorecognition process. Chem Soc Rev. 2010;39(2):734–749. doi: 10.1039/b811426a. [DOI] [PubMed] [Google Scholar]
- 38.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200(4342):618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 39.Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys J. 1997;72(4):1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dudko O, Hummer G, Szabo A. Theory, analysis and interpretation of single molecule force spectroscopy experiments. Proc Natl Acad Sci U S A. 2008;105(41):15755–15760. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friddle RW, Noy A, De Yoreo JJ. Interpreting the widespread nonlinear force spectra of intermolecular bonds. Proc Natl Acad Sci. 2012;109(34):13573–13578. doi: 10.1073/pnas.1202946109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taranta M, Bizzarri AR, Cannistraro S. Probing the interaction between p53 and the bacterial protein azurin by single molecule force spectroscopy. J Mol Recognit. 2008;21(1):63–70. doi: 10.1002/jmr.869. [DOI] [PubMed] [Google Scholar]
- 43.Bizzarri AR, Santini S, Coppari E, et al. Interaction of an anticancer peptide fragment of azurin with p53 and its isolated domains studied by atomic force spectroscopy. Int J Nanomedicine. 2011;6:3011–3019. doi: 10.2147/IJN.S26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Björquist P, Boström S. Determination of the kinetic constants of tissue factor/factor VII/factor VIIA and antithrombin/heparin using surface plasmon resonance. Thromb Res. 1997;85(3):225–236. doi: 10.1016/s0049-3848(97)00007-8. [DOI] [PubMed] [Google Scholar]
- 45.O’Shannessy DJ, Brigham-Burke M, Soneson KK, Hensley P, Brooks I. Determination of rate and equilibrium binding constants for macromolecular interactions using surface plasmon resonance: use of nonlinear least squares analysis methods. Anal Biochem. 1993;212(2):457–468. doi: 10.1006/abio.1993.1355. [DOI] [PubMed] [Google Scholar]
- 46.Bizzarri AR, Cannistraro S. Free energy evaluation of the p53-Mdm2 complex from unbinding work measured by dynamic force spectroscopy. Phys Chem Chem Phys. 2011;13(7):2738–2743. doi: 10.1039/c0cp01474e. [DOI] [PubMed] [Google Scholar]
- 47.Domenici F, Frasconi M, Mazzei F, D’Orazi G, Bizzarri AR, Cannistraro S. Azurin modulates the association of Mdm2 with p53: SPR evidence from interaction of the full-length proteins. J Mol Recognit. 2011;24(4):707–714. doi: 10.1002/jmr.1105. [DOI] [PubMed] [Google Scholar]
- 48.Bista M, Petrovich M, Fersht AR. MDMX contains an autoinhibitory sequence element. Proc Natl Acad Sci U S A. 2013;110(44):17814–17819. doi: 10.1073/pnas.1317398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robert P, Benoliel AM, Pierres A, Bongrand P. What is the biological relevance of the specific bond properties revealed by single-molecule studies? J Mol Recognit. 2007;20(6):432–447. doi: 10.1002/jmr.827. [DOI] [PubMed] [Google Scholar]