Abstract
Context
Renal cancer is a common urologic malignancy, and therapeutic options for metastatic disease are limited. Most clear cell renal cell carcinomas (ccRCC) are associated with loss of von Hippel-Lindau tumor suppressor (pVHL) function and deregulation of hypoxia pathways.
Objective
This review summarizes recent evidence from genetic and biological studies showing that hypoxia and hypoxia-related pathways play critical roles in the development and progress of renal cancer.
Evidence acquisition
We used a systematic search for articles using the keywords hypoxia, HIF, renal cancer, and VHL.
Evidence synthesis
Identification of the tumor suppressor pVHL has allowed the characterization of important ccRCC-associated pathways. pVHL targets α-subunits of hypoxia-inducible transcription factors (HIF) for proteasomal degradation. The two main HIF-α isoforms have opposing effects on RCC biology, possibly through distinct interactions with additional oncogenes. Furthermore, HIF-1α activity is commonly diminished by chromosomal deletion in ccRCCs, and increased HIF-1 activity reduces tumor burden in xenograft tumor models. Conversely, polymorphisms at the HIF-2α gene locus predispose to the development of ccRCCs, and HIF-2α promotes tumor growth. Genetic studies have revealed a prominent role for chromatin-modifying enzyme genes in ccRCC, and these may further modulate specific aspects of the HIF response. This suggests that, rather than global activation of HIF, specific components of the response are important in promoting kidney cancer. Some of these processes are already targets for current therapeutic strategies, and further dissection of this pathway might yield novel methods of treating RCC.
Conclusions
In contrast to many tumor types, HIF-1α and HIF-2α have opposing effects in ccRCC biology, with HIF-1α acting as a tumor suppressor and HIF-2α acting as an oncogene. The overall effect of VHL inactivation will depend on fine-tuning of the HIF response.
Patient summary
High levels of hypoxia-inducible transcription factors (HIF) are particularly important in the clear cell type of kidney cancer, in which they are no longer properly regulated by the von Hippel-Lindau protein. The two HIF-α proteins have opposing effects on tumor evolution.
Keywords: Hypoxia, Hypoxia-inducible transcription factors, Clear cell renal cell carcinoma, von Hippel-Lindau
1. Introduction
Renal cell carcinoma (RCC) is the 14th most common malignancy and the third most common urologic cancer [1,2]. It has an age-standardized population incidence rate (per 100 000) of 15.8 in men and 7.1 in women in the European Union and mortality rates of 6.5 and 2.7, respectively [3]. Worldwide kidney cancer causes >100 000 deaths per year. A number of environmental risk factors for the development of renal cancer have been identified including smoking, obesity, hypertension, and diabetes [3]. Histopathologic classifications distinguish three major subtypes: clear cell RCC (ccRCC; 70–75%), papillary RCC (pRCC; 10–16%), and chromophobe RCC (chRCC; 5%) [4]. Each subtype is associated with a separate hereditary syndrome, and together they account for 2–3% of all RCC cases. ccRCC is associated with von Hippel-Lindau (VHL) disease, which also features development of hemangioblastomas and pheochromocytomas. Patients with familial mutations in the c-Met proto-oncogene have a high risk of developing type 1 pRCC, whereas patients with germline mutations in fumarate hydratase (FH) develop cutaneous and uterine leiomyomas and type 2 pRCC (hereditary leiomyomatosis and RCC [HLRCC]). chRCCs and other tumors of the kidney are seen in patients with the Birt-Hogg-Dubé syndrome, which is caused by mutations in the folliculin tumor suppressor gene. In addition, other genes such as tuberous sclerosis 1 or succinate dehydrogenase B are also associated with syndromes that predispose to the development of RCC [5]. In accordance with the Knudson hypothesis, persons with each syndrome have a hypomorphic germline mutation in one allele of the relevant tumor suppressor gene. Somatic inactivation of the remaining wild-type allele within the cancer cells then “exposes” this dysfunctional gene product.
The majority of sporadic ccRCCs have somatic inactivation of both VHL alleles with loss of function of the VHL tumor suppressor protein (pVHL). About 60–80% of ccRCC cases display loss-of-function coding mutations in the VHL gene, chromosomal aberrations on chromosome 3p25 that affect the VHL locus, or hypermethylation of the VHL promoter [6–8]. Re-expression of pVHL in VHL-defective RCC xenografts reduces tumor growth, confirming that VHL is a bona fide tumor suppressor gene [9].
The best-understood molecular function of pVHL is as the recognition component of an E3 ubiquitin ligase complex that targets proteins for proteasomal degradation by tagging them with ubiquitin [10]. Recently, ccRCC-associated mutations in TCEB1, which encodes for the elongin C component of the VHL E3 ligase complex, have also been described in ccRCC, suggesting that this function is important in its role as a tumor suppressor [11]. To date, the best-characterized targets of pVHL are the α-subunits of the hypoxia-inducible transcription factors (HIFs; HIF-1α and HIF-2α, also known as EPAS1). Oxygen-dependent hydroxylation of HIFs at specific proline residues by prolyl hydroxylase (PHD) enzymes triggers binding of pVHL, ubiquitination, and subsequent proteasomal degradation [12–15]. Consequently, when oxygen is abundant, HIF is rapidly degraded. In a hypoxic environment, hydroxylation is suppressed, and HIF-α escapes degradation to form dimers with the constitutively expressed HIF-1β isoform, also called the aryl hydrocarbon receptor nuclear translocator (ARNT) protein. This complex is imported into the nucleus and binds DNA at the hypoxia response elements to activate the transcription of a wide variety of genes [16,17]. Similarly, loss of function of pVHL also leads to stabilization of HIF-α and subsequent “pseudohypoxic” transcriptional responses; therefore, loss of pVHL in RCC is tightly associated with the activation of HIF and its transcriptional consequences.
Interestingly, activation of HIFs also has been described in the context of the other major RCC subtypes [18–20]. The loss of FH or succinate dehydrogenase B, for example, leads to increased levels of the Krebs cycle intermediates fumarate or succinate, respectively, which in turn inhibit PHD-mediated hydroxylation of HIF-α by competing with the cosubstrate 2-oxoglutarate [18,20,21]. In addition, many types of tumors frequently outgrow their blood supply, generating hypoxic regions that can activate HIF in both tumor and stromal cells, despite a functional degradation apparatus [22].
This review summarizes current knowledge of the effects of hypoxia and HIF in the context of RCC biology.
2. Evidence acquisition
A systematic literature search in PubMed was conducted using the keywords or phrases hypoxia, HIF, VHL, renal cancer, and kidney cancer. Articles were selected by relevance and the novelty of their findings. In addition, key publications in the field of hypoxia research were added.
3. Evidence synthesis
3.1. VHL and hypoxia-inducible transcription factors
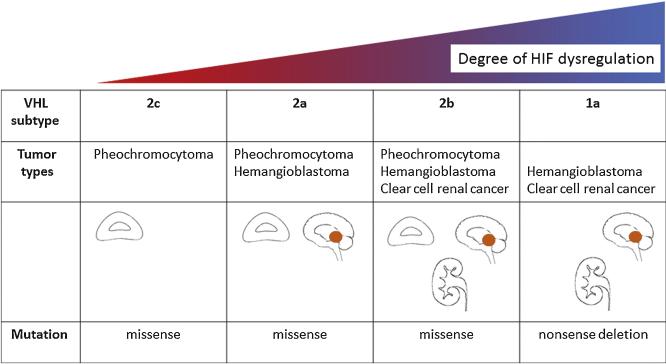
Loss of function of VHL due to gene deletions, inactivating mutations, or epigenetic silencing is observed in the vast majority of both familial and sporadic ccRCCs [6,8,23]; however, the inherited RCC syndrome VHL disease may be subclassified according to the relative risk of different types of tumor (Fig. 1). Type 1 VHL syndrome is associated with ccRCC and hemangioblastoma, whereas patients with type 2 VHL syndrome develop pheochromocytomas. Type 2 VHL syndrome is subdivided into type 2a (pheochromocytomas and hemangioblastomas but low risk of ccRCC), type 2b (pheochromocytomas, hemangioblastomas and ccRCC), and type 2c (pheochromocytomas only).
Fig. 1.
Subtypes of von Hippel-Lindau disease. Phenotypic characteristics correlate with the degree of hypoxia-inducible transcription factor dysregulation.
HIF = hypoxia-inducible transcription factors; VHL = von Hippel-Lindau protein.
These phenotypes were found to have distinct VHL genotypes that had disparate effects on the HIF pathway [24–26]. Specifically, all ccRCC-associated mutations caused complete HIF dysregulation, whereas those associated with pheochromocytoma alone regulated HIF normally (although pheochromocytoma and paraganglioma have since been associated with somatic activating mutations at the EPAS1 locus). Although this correlation between ccRCC risk and HIF dysregulation is not absolute (some HIF dysregulation was seen with type 2a mutations at low risk of ccRCC), it further emphasizes the importance of the HIF pathway in the pathogenesis of ccRCC and suggests that both quantitative and qualitative windows of HIF activation are necessary for oncogenesis.
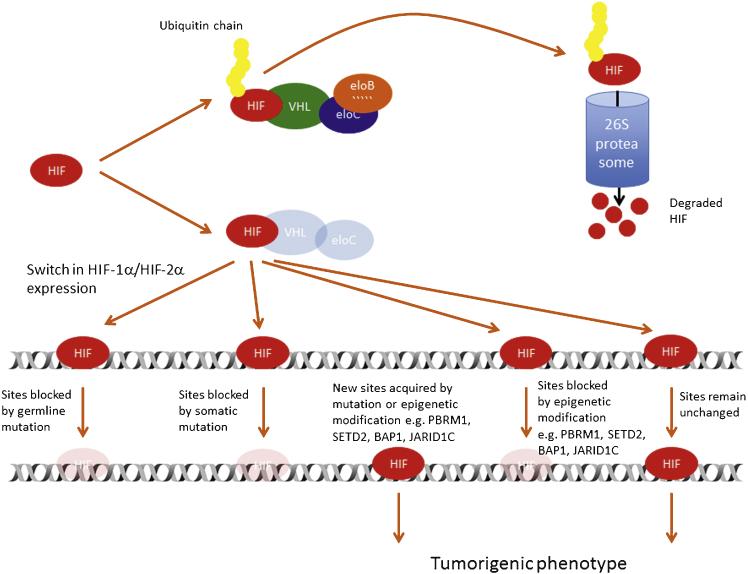
ccRCCs display a highly angiogenic and glycolytic phenotype, and several associated genes such as VEGFA or GLUT1 appear to be consistently overexpressed [27,28]. These genes have previously been shown to be inducible by hypoxia [29–31], but in ccRCC cells, this regulation is lost [15]. Importantly, reintroduction of functional pVHL in ccRCC cells restores their ability to downregulate hypoxia-inducible genes [15,32]. Moreover, pVHL binds directly to the master transcriptional regulators of hypoxic gene expression, HIF-1α and HIF-2α [14,15]. In complex with other components of the ubiquitin E3 ligase complex, elongin B, elongin C, cullin 2, and Rbx1 [10,33,34], pVHL catalyzes the ubiquitination of HIFs, which are then degraded by the proteasome [10,33,34]. This binding of pVHL to HIF-α molecules (and hence the degradation of HIF-α) is dependent on the availability of molecular oxygen as well as iron and ascorbate. Enzymatic modification, leading to hydroxylation of HIF-α subunits at specific proline residues, allows pVHL to recognize and bind HIF-α [12,13]. Importantly, deletions or mutations in the VHL gene that lead to RCC development (types 1 and 2b) preferentially affect pVHL binding to HIF and are associated with high levels of HIF expression [24–26]. Furthermore, recent whole-genome and/or whole-exome sequencing in combination with RNA sequencing has identified mutations in TCEB1, the gene that encodes elongin C in ccRCC (Fig. 2) [11]. All TCEB1 mutant tumors were VHL wild type, suggesting that the common role of these proteins in the E3 ubiquitin ligase complex is important for the development of ccRCC.
Fig. 2.
The hypoxia-inducible transcription factor (HIF) pathway in clear cell renal cancer. In normal epithelial cells, ubiquitination of HIF-α by the E3 ubiquitin ligase leads to degradation via the 26 proteasome. In precancerous cells, defective tumor suppressor pVHL or elongin C results in stabilization of both HIF-α isoforms. Subsequent deletions of HIF-1α and/or chromatin-modifying enzymes as well as somatic mutations in HIF DNA-binding sites can affect the transcriptional program of HIF and eventually lead to a tumorigenic phenotype.
eloB = elongin B; eloC = elongin C; HIF = hypoxia-inducible transcription factors; VHL = von Hippel-Lindau protein.
Functions of pVHL outside of the hypoxia-signaling pathway have also been described. The pVHL E3 ubiquitin ligase complex also targets protein kinase C and Rpb1 (a subunit of RNA polymerase 2) proteins for ubiquitination and proteasomal degradation [35,36]. Moreover, interactions of pVHL that do not result in polyubiquitylation have been described with fibronectin and collagen IV α2 [37,38]. VHL is also important for maintaining cell morphology and polarity by interacting with microtubules and promoting correct cilia formation, an important feature of epithelial cell integrity [39]; however, to date, the best-described function for pVHL and the most clearly associated with the pathogenesis of ccRCC is the clearance of proline-hydroxylated HIF-α subunits from normoxic cells.
3.2. Hypoxia-inducible transcription factors
HIF was discovered in the course of investigating hypoxic erythropoietin (EPO) regulation [40,41]. Analysis of the EPO gene uncovered hypoxia-dependent binding of a transcription factor to a 3′ enhancer region that was responsible for hypoxic EPO messenger RNA (mRNA) induction. This factor was termed hypoxia-inducible factor 1 and is a heterodimer of two basic helix-loop-helix Per/ARNT/SIM (PAS)-domain proteins [42], namely, HIF-1α and ARNT (or HIF-1β).
Subsequent analyses revealed that two additional paralogues (HIF-2α and HIF-3α) are also present in human cells. HIF-2α is regulated similarly to HIF-1α by an oxygen-dependent mechanism [43,44]. This involves hydroxylation of the both HIF-1α and HIF-2α subunits at specific prolyl residues in the oxygen-dependent degradation domains by PHD1–3 [12,13,45]. The hydroxylated residues are specifically recognized by pVHL, which facilitates binding of an E3 ubiquitin ligase complex. The enzymatic reaction depends on the availability of α-ketoglutarate (2-oxoglutarate), ascorbate, and iron as well as oxygen. Thus, in hypoxia, hydroxylation is impaired, and HIF-α subunits escape degradation. Hydroxylation of a specific asparagyl residue in the C-terminal transactivation domain (CTAD) of HIF-α by factor inhibiting HIF-1 (FIH1) reduces transcriptional activity and adds another layer of hypoxia-dependent gene regulation [46,47]. FIH1 has a different affinity for oxygen and is active at lower pO2 levels compared with the PHDs, potentially expanding the dynamic range of HIF activation [48]. Both HIF-1α and HIF-2α display different susceptibility to FIH1 and PHD-mediated prolyl hydroxylation. The CTAD of HIF-1α appears to be preferentially hydroxylated by FIH1 [48,49].
HIF-1α is widely expressed in human tissues, whereas the expression of HIF-2α is more restricted to specific cell types in some organs [50]. In the kidney, for example, HIF-1α expression can be detected predominantly in tubular cells. In contrast, renal interstitial fibroblasts, endothelial cells, and some glomerular cells show HIF-2α positivity [51,52]. Besides the regulation of EPO, analyses of other cis-acting regulatory sequences has revealed that hypoxic gene regulation by HIF is a common and widespread mechanism that includes the regulation of important genes for maintaining oxygen and metabolic homeostasis. The repertoire of HIF-dependent transcriptional targets includes genes that regulate angiogenesis, glycolysis, chromatin remodeling, cell cycle, and even the oxygen-sensing pathway itself. Indeed, genomewide analysis of the transcriptomic response to hypoxia coupled with HIF binding revealed that at least 500–1000 genes are under the control of HIF [17,53–57] in any particular cell type. Specifically, HIF is an activator (rather than repressor) of gene transcription, and its binding to chromatin can transactivate gene expression by interacting with promoters over long genomic distances (up to several hundred kilobases) [17,53,54]. Furthermore, the repertoire of HIF targets is highly cell-type dependent, with only a small number of HIF-regulated genes conserved across all cell types [58]. In the setting of pVHL-defective ccRCC cells, however, microarray studies have shown extensive overlap between VHL and hypoxia-regulated genes [59–61]. Subsequently, integration of HIF DNA-binding sites in the pVHL-defective 786-O RCC cell line with these transcriptomic profiles has revealed that a number of HIF targets are consistently overexpressed across a variety of ccRCC cell lines and tumors and thus are likely to be important for RCC biology [62,63].
3.3. HIF-1 versus HIF-2 in renal cancer
3.3.1. Functional evidence that hypoxia-inducible transcription factors are important for clear cell renal cancer
HIF-1α and HIF-2α subunits display distinct but overlapping target gene repertoires (Table 1) [64,65]; however, depending on tissue type and microenvironment, different expression profiles and contrasting effects on tumor biology can be observed [65,66]. This may result in part from differential patterns of DNA binding between the two isoforms. Nonetheless, a number of gene loci bind both HIF-1 and HIF-2, whereas only one isoform is able to effect transactivation. The functional relevance of the different HIF-α isoforms in the early stages of renal tumor development is not entirely clear. In the kidneys of patients with VHL disease, HIF-α isoforms are present in early precancerous lesions that lack functional pVHL [52,67,68]. This overexpression is accompanied by the induction of ubiquitous HIF target genes such as CA9 or GLUT1. Importantly, in immunohistochemical experiments, both α-isoforms could be detected in these lesions, whereas in normal tubular cells, only protein of the HIF-1α isoform was found, suggesting that the release of HIF-2α expression is an early event in pVHL-defective tubular cells [51,67,69]. The mechanisms are unclear but may involve reduced methylation of a CpG island at the EPAS1 locus following suppression of the DNA methyltransferase 3a [70]. In addition, the expression of more renal cancer–specific HIF targets such as the cell cycle oncogene cyclin D1 is also increased in these early foci [52,68].
Table 1.
Selected hypoxia-inducible transcription factor target genes in clear cell renal cell carcinoma
| Gene | Reported function | HIF-1/HIF-2 | Reference |
|---|---|---|---|
| VEGFA | Angiogenesis | HIF-1/HIF-2 | [126] |
| CXCR4 | Chemotaxis | HIF-1/HIF-2 | [118] |
| CAXII | pH regulation | HIF-1/HIF-2 | [126] |
| SLC2A1 | Glucose transport | HIF-1/HIF-2 | [126] |
| mir210 | Mitochondrial function | HIF-1/HIF-2 | [127] |
| GAS6/AXL | Metastasis | HIF-1/HIF-2 | [123] |
| CDCP1 | Metastasis | HIF-1/HIF-2 | [128] |
| JMJD1A | Histone demethylation | HIF-1/HIF-2 | [55] |
| JARID1C | Histone demethylation | HIF-1/HIF-2 | [92] |
| BNIP3 | Apoptosis | HIF-1 | [68] |
| HK2 | Glycolysis | HIF-1 | [126] |
| PFK | Glycolysis | HIF-1 | [126] |
| ALDOA | Glycolysis | HIF-1 | [126] |
| PGK1 | Glycolysis | HIF-1 | [126] |
| LDHA | Glycolysis | HIF-1 | [126] |
| FBP1 | Gluconeogenesis | HIF-1 | [129] |
| SPAG4 | Cell migration | HIF-1 | [130,131] |
| CAIX | pH regulation | HIF-1 | [68,132] |
| FYN | Metastasis | HIF-1 | [128] |
| HIG2 | Lipid storage | HIF-1 | [133] |
| Plin2 | Lipid storage | HIF-2 | [126] |
| CCND1 | Cell cycle control | HIF-2 | [68] |
| MMP1 | Matrix remodeling | HIF-2 | [134] |
| TGFα | Inflammation | HIF-2 | [68] |
| SK1 | Invasion, angiogenesis | HIF-2 | [135] |
In vitro studies using mouse embryo fibroblast cells or nonmalignant human tubular cells has shown that the immediate loss of VHL leads to senescence in these cells, indicating that additional genetic or biological events are necessary for malignant transformation of VHL-mutant tubular cells [71]. This also becomes clear when considering that in kidneys of patients with VHL syndrome, hundreds of VHL-defective lesions can be detected but only a fraction progress to full-blown cancer [67,72]. Taken together, these studies suggest that biallelic loss of VHL in tubular cells primarily leads to stabilization of HIF-1α and de novo release of HIF-2α with increased expression of their target genes and that additional HIF-dependent or independent events are necessary to overcome cell senescence and to promote tumor progression.
The importance of the HIF-1α and HIF-2α subunits in renal tumor progression has been well studied. Overexpression of the HIF-2α subunit in RCC cells leads to an increased tumor burden in a mouse ccRCC-xenograft model [68,73]. This effect was restricted to HIF-2α proteins with intact DNA-binding domains, suggesting that transcriptional activity is essential for the tumor-promoting effect [74]. Similarly, short hairpin RNA–mediated knockdown of HIF-2α reduces tumor mass in the same model [74]. Opposing effects have been described for the HIF-1α isoform. A reduced tumor size was observed when HIF-1α protein was overexpressed, whereas knockdown of HIF-1α increased cell proliferation [68,75]. Similarly, inactivation of HIF-1α, but not HIF-2α, exacerbates renal cyst development following FH inactivation in a mouse model [76]. These observations led to the conclusion that in RCCs, HIF-2α has tumorigenic activity, whereas HIF-1α is antitumorigenic. This finding is surprising because overexpression of both HIF-α subunits (HIF-1α and/or HIF-2α) is usually associated with poor outcome in many other types of cancer including breast, colorectal, and prostate cancers [77]. Similarly, both HIF-α subunits promote tumor growth in many nonccRCC xenograft or autochthonous tumor models [77].
3.3.2. Genetic evidence for divergent roles of the hypoxia-inducible transcription factor subunits in renal cell carcinoma biology
Recently, further evidence has emerged for a divergent role of the HIF-α isoforms in the context of ccRCC. Immunohistochemical analysis of VHL-defective cancers has shown that although all VHL-defective tumors express the HIF-2α isoform, many have inactivated HIF-1α [78]. These workers classified ccRCC into three groups: those with wild type VHL, those with defective pVHL and expressing both HIF-1α and HIF-2α together, or those with defective pVHL and expressing HIF-2α exclusively. In this study, cancers with exclusive HIF-2α expression were associated with a larger tumor volume, although no other clinical parameter showed significant association.
Genomic analyses of renal tumors have revealed that deletions in a region of chromosome 14 that harbors the HIF-1α gene are a common feature of RCC [75,79]. In keeping with this finding, some commonly used RCC cell lines (eg, 786-O and A498) have lost expression of full-length HIF-1α [75]. Restoring expression of wild-type HIF-1α in these cells resulted in reduced xenograft tumor growth. This re-expression of HIF-1α also resulted in the transcriptional activation of established HIF-1α target genes such as BNIP3, PGK1, HK1, and TP11 [75]. This transcriptional signature may contribute to the protective effects of HIF-1α. Alternatively, HIF-1α could outcompete HIF-2α from shared DNA-binding sites and oppose the action of HIF-2α at gene loci at which HIF-1α is inactive. Together with the findings already noted, this suggests that in the context of RCC, HIF-1α acts a tumor suppressor. The importance of HIF-2α in RCC biology became more apparent through recent genomewide screens. A genomewide association study (GWAS) revealed that DNA polymorphisms in the first intron of the EPAS1 gene (coding for HIF-2α) are associated with an increased risk of developing RCC [80,81]; however, to date, the functional consequences of these single-nucleotide polymorphisms regarding regulation of the EPAS1 gene or tumor development have not been resolved, although cross-reference with ENCODE data sets suggests that the polymorphisms are in regulatory regions of the EPAS1 gene. In addition, several other loci were associated with RCC development in these GWASs. The strongest association was found for polymorphisms in a gene-poor region on chromosome 11 that were protective for RCC development [80]. These associations were reproduced in several other studies [82–84]. Integrating transcriptomic analyses of the HIF response and genomewide HIF DNA-binding data revealed that the locus on chromosome 11 corresponded to an HIF-binding enhancer of cyclin D1 expression, a well-characterized oncogene regulated by HIF exclusively in the context of VHL-defective RCC [63]. Interestingly, although both HIF-α subunits have overlapping DNA-binding profiles and can bind to the 11q13.3 enhancer, cyclin D1 is exclusively regulated by the HIF-2α subunit, suggesting postbinding mechanisms of transcriptional regulation. Cyclin D1 was also induced in early VHL-defective lesions in kidneys from patients with VHL disease, implying a functional role of the VHL–HIF-2α–cyclin D1 axis in early RCC development [68].
Recently, genomewide exome-sequencing analyses have revealed that the chromatin-modifying enzymes lysine (K)-specific demethylase 6A (UTX), lysine (K)-specific demethylase 5C (JARID1C), SET domain containing 2 (SETD2), and polybromo 1 (PBRM1) and the nuclear deubiquitinase BRCA1 associated protein-1 (BAP1) are frequently mutated in ccRCC [85–88]. PBRM1 has the highest mutation rate, with a frequency of approximately 40% in ccRCC samples, followed by BAP1, which has a mutation frequency of approximately 14% [87,88]. Together with the finding that the mutational landscape of ccRCC displays a pronounced intratumoral and metastatic heterogeneity, the current concept of tumorigenesis involves inactivation of pVHL as the initiating step, followed by additional hits in the above-mentioned genes to further promote tumor growth (Fig. 2) [89,90]. How mutations in chromatin-modifying enzymes interact with the pseudohypoxic response in VHL-defective cells is not fully understood. Knockout of PBRM1 in the PBRM1 competent but VHL-defective RCC cell line 786-O increased cell proliferation [88]. The HIF response is dependent on open chromatin configuration, and HIF binding is sensitive to CpG methylation [17,91]. Consequently, it is conceivable that disturbances in chromatin accessibility caused by the loss of chromatin modifier activity could influence the HIF response in a VHL-defective background. In line with this hypothesis, the first studies examining the function of JARID1C and SETD2 found a significant impact of these genes on the HIF response [92,93]. Suppression of JARID1C, a histone-3 lysine-4 trimethyl (H3K4me3) demethylase, in 786-O renal cancer cells restored H3K4me3 levels at promoters and mRNA expression of HIF-target genes such as IGFBP3. Interestingly, JARID1C was also shown to be an HIF-target gene, adding more evidence to a functional link between HIF and chromatin configuration. Simon et al found that differences in chromatin accessibility are a major factor influencing ccRCC biology [93]. Moreover, integration of HIF binding sites revealed that tumor-specific open chromatin strongly associates with HIF binding. The authors found that mutations in the SETD2 gene, coding for a methyltransferase responsible for H3K36 trimethylation, leads to loss of H3K36me3 and increases chromatin accessibility, as determined by formaldehyde-assisted isolation of regulatory elements and DNA methylation assays. In a recent study, combined genetic deletion of BAP1 and VHL in the mouse resulted in increased tumor development [94]. Interestingly, BAP1 or VHL tubular-specific knockout mice developed a cystic renal phenotype and renal failure, but only the combined knockout led to the appearance of tumors. HIF target genes were induced in the VHL-defective background; however, whether HIF is relevant for tumor development or whether BAP1 loss and VHL loss cooperate in regulating HIF target genes in this model has not been addressed. These findings from pangenomic studies underpin the important role of HIF-α in ccRCC development. Besides the initial loss of VHL, tumor-acquired defects in HIF-1α are frequent events in ccRCCs, and germline polymorphisms in the EPAS1 gene are associated with the occurrence of ccRCCs. Newly identified tumor suppressor genes are mainly associated with chromatin-modifying processes, and recent studies confirm that JARID1C and SETD2 functions are linked to the HIF response in ccRCCs [92,93].
3.4. Regulators and interacting proteins of hypoxia-inducible transcription factors in clear cell renal cell carcinoma
Each HIF isoform has a different transactivation capacity, in part due to different sensitivity of the C-terminal transactivation domain to hydroxylation by FIH1 [48,49], with HIF-1α being more sensitive to inactivation by FIH1. Interestingly, knockdown of FIH1 increased HIF-1α transcriptional activity and apoptosis of RCC cells [95]. This indicates that in RCC, the presence of FIH1 can contribute to reduced HIF-1α activity and promote cancer cell survival. In contrast, and in keeping with previous findings that HIF-1α acts as a tumor suppressor in ccRCC, low expression of FIH1 in the nuclei of RCC specimens correlated with poor overall survival [96].
HIF-3α (also called inhibitory PAS domain [IPAS]) has been identified as another paralogue of HIF-1α [44,97]. Although some HIF-3α splice variants lack an oxygen-dependent degradation domain and thus are not directly regulated by pVHL, HIF-3α is itself an HIF-1α target gene and so may be indirectly regulated by pVHL [44,97]. Splice variants lacking a transactivation domain can act as a dominant negative by competing with HIF-1α and HIF-2α for binding to HIF-1β binding. This adds an additional layer of negative feedback to hypoxic gene regulation because HIF-3α is a potential HIF-dependent inhibitor of the HIF pathway [98].
Maynard et al detected reduced levels of HIF-3α mRNA in ccRCC samples compared with normal kidney [98]; however, in ccRCC sections with high levels of HIF-1α, an increased signal for HIF-3α protein was observed [99]. Overexpression of the HIF-3α isoform in RCC cells led to reduced expression of HIF target genes and delayed tumor growth in a xenograft model [100]. Consequently, HIF-3α appears to be an HIF-1α–specific target that primarily interferes with both HIF-1α– and HIF-2α–dependent transcription to reduce hypoxic gene expression. Given that its expression seems protective in the context of ccRCC biology, it seems likely that its effect in antagonizing HIF-2α outweighs its effect on HIF-1α. Furthermore, a recent report suggests that some HIF-3α splice variants retain oxygen-dependent regulation and the ability to directly bind DNA and transactivating gene expression [101]. To date, however, further evidence of a functional role of HIF-3α in the context of RCC biology and data gained from a larger RCC cohort is lacking.
In addition to directly controlling pathways relevant to tumor metabolism and survival via transcriptional activation, HIF-α subunits also interact with a number of important oncogenes, including regulation of MYC activity by HIF. The MYC gene locus is commonly amplified in ccRCCs, and the MYC protein contributes to tumor cell biology by regulating the expression of genes that control cell cycle progression and cell metabolism [7,102], thereby enhancing cell proliferation and growth. Gordan et al showed that, again, the different HIF-α subunits influence MYC activity in opposing ways [78,103,104]. HIF-1α reduces MYC transcriptional activity at a number of different levels. First, it can bind to SP1 and thus remove MYC from target gene promoters. Second, HIF-1α can disrupt binding of MYC to its binding partners MAX and MIZ1. Third, HIF-1α induces MAX interactor 1 (MXI1) expression, which inhibits expression of MAX target genes. Finally, HIF-1α enhances MYC degradation under chronic hypoxia. Consequently, HIF-1α is an antagonist of MYC-mediated processes including cell proliferation and survival. In contrast, HIF-2α enhances MYC activity in malignant cells and, in doing so, increases the expression of cell cycle regulators such as cyclin D2. The interaction of HIF-2α and MYC pathways appeared to be highly relevant for genomic integrity and resistance to replication stress in tumor cells [78].
TP53 mutations have also been observed in ccRCC [7]; however, compared with most types of cancer, they are relatively infrequent, suggesting that other mutations somehow inactivate the p53 pathway. Indeed, HIF-2α can inhibit p53 phosphorylation, thereby reducing its activity. Conversely, HIF-2α knockdown leads to increased p53 protein and activity promoting cell cycle arrest, increased cell death, and reduced colony formation [105]. In ccRCC samples, HIF-2α–positive tumors showed reduced yH2AX, pS15-p53, and p53 target gene levels. Interestingly, PBRM1 has also been identified as a gene with inactivation that rescues p53-dependent senescence [106].
In summary, genetic and functional data indicate opposing actions of HIF-1α and HIF-2α in ccRCC biology. This finding is further corroborated by differential interactions of each HIF-α isoform with other regulators of the hypoxic pathway or other crucial oncogenes.
3.5. The role of HIF on clinical outcome and therapeutic approaches in renal cancer patients
Although experimental work would suggest that HIF isoforms are important predictive factors for ccRCC development and progression, evidence from large cohort studies on HIF expression and its association with clinical outcome is sparse. Biswas et al found that high tumor expression of either isoform alone was associated with worse patient survival (although only HIF-1α reached statistical significance); paradoxically, high expression of both HIF-1α and HIF-2α together did not significantly alter patient survival [107]. Consistent with earlier data, HIF-2α expression correlated with increased tumor volume [78,107]. Interestingly, expression of HIF target genes such as CCND1 or GLUT1 did not predict outcome in this study. Similarly, in a larger cohort of 357 patients, high HIF-1α expression was correlated with a worse outcome in patients with metastatic disease [108]. In contrast, in a cohort of 92 patients, high HIF-1α expression determined by Western blotting showed favorable overall survival [109]. In neither study was HIF-2α level measured.
Common targeted therapies used to treat ccRCC include inhibitors of both the vascular endothelial growth factor (VEGF) pathway and the mammalian target of rapamycin (mTOR) pathway. Because VEGFA is a well-characterized target gene of HIF and mTOR regulates HIF translation, both therapeutic approaches interfere with the HIF signaling pathway [110]. Consequently, one might hypothesize that the level of HIF expression in tumors would predict response to these targeted therapies. Pazopanib, a multikinase inhibitor of VEGF and platelet-derived growth factor receptors and c-Kit, has been shown to prolong progression-free survival in patients with metastatic disease [111]; however, Choueiri et al found no correlation between HIF expression (HIF-1α or HIF-2α) and either the overall response rate or progression-free survival with pazopanib [112]. Similarly, in another study surveying 123 patients with progressive disease who were receiving sunitinib, sorafenib, axinitib, or bevacizumab, neither VHL inactivation nor HIF status had any effect on response rate [113]. These findings are not surprising, given that attempts to correlate VHL status with patient prognosis in the era before targeted therapies led to heterogeneous findings [114–117]. Despite the lack of correlation between perturbations in the VHL–HIF axis and clinical outcome, experimental data strongly suggest that the HIF pathway plays an essential role in ccRCC tumor progression and metastasis and that this can be further exploited for therapeutic approaches. Recently, Vanharanta and colleagues provided evidence for an epigenetic switch in PRC2-dependent H3K27 trimethylation affecting the HIF-dependent regulation of the important target genes CXCR4 and CYTIP [118,119]. These findings fit with the data from genetic studies that assigned a large proportion of tumor progression to epigenetic changes and the findings that chromatin-modifying enzymes are targets of HIF themselves [53,120,121]. How these pathways could be used for designing novel drugs active against ccRCC is currently not known. In another approach, Chan et al used a synthetic lethality screen to identify putative therapeutic targets in VHL-defective ccRCC [122]. This work identified GLUT1 as an essential component of tumor cell survival in ccRCC. Pharmacologic inhibition of this transporter led to decreased tumor growth in a xenograft model. The same group also identified HIF-induced expression of the AXL receptor tyrosine kinase as an aggressive driver of MET-dependent metastasis in RCC [123]. Inhibition of the AXL signaling pathway reduced metastatic colonization to the lung in a xenograft model. These findings further confirm the importance of the HIF pathway for tumor progression in ccRCC and its potential for novel drug development.
3.6. The role of hypoxia-inducible transcription factors in other renal cell carcinoma entities
Although HIFs have been most extensively studied in the context of VHL-defective ccRCCs, increasing evidence suggests that they play additional roles in other RCC subtypes such as pRCC or chRCC. The activation of HIF in kidneys of patients with HLRCC has been described [18]. Accumulation of the Krebs cycle intermediate fumarate in these patients leads to competitive inhibition of PHD activity and stabilization of HIF-α subunits. In a mouse model, FH deficiency leads to the development of cysts as well as activation of the HIF pathway [21]; however, subsequent analysis has revealed that cyst formation in this model is unexpectedly increased by inactivation of HIF-1α and that other pathways might be crucially involved in FH-related pRCC [76]. In human cells, the metabolic shift in FH-deficient cells seems to activate pathways that promote oncogenic cell behavior, including HIF-α [124]. HIF activation has also been described in cells and tissue from patients with the Birt-Hogg-Dubé syndrome [19,125]. These tumors mainly express HIF-2α; however, evidence is sparse with regard to the significance of HIF in tumorigenesis or tumor progression in the context of chRCC outside of potential hypoxic activation similar to other nonrenal cancer types.
4. Conclusions
Functional and genetic evidence tightly links the hypoxia-signaling pathway with ccRCCs. In experimental settings, HIF-α paralogues HIF-1α and HIF-2α have opposing effects on tumor development and progression. HIF-1α acts as a tumor suppressor, whereas HIF-2α has oncogenic potential. So far, this dichotomy is unique to ccRCC tumors. HIF stabilization is an early event in cancerous renal lesions. Multiple secondary events in RCC evolution can affect HIF function. These events include genetic loss of the protective HIF-1α isoform, interaction with other oncogenic pathways such as the cell cycle, or additional mutations in genes with tumor-suppressing or -promoting functions. These mutations predominantly affect genes associated with chromatin-modifying functions, suggesting that epigenetic integrity is commonly altered in ccRCC. Because HIF's action as a transcription factor is dependent on chromatin accessibility at its hypoxia response element motifs, it seems reasonable that epigenetic changes strongly affect the HIF pathway. The net effect of tumor-repressing and -promoting events influences whether single lesions can outgrow their environment and progress to full-blown cancer; therefore, it would be interesting to further understand which events in early lesions lead to tumor progression.
Current therapies for advanced ccRCC already target components of the HIF pathway such as HIF translation (mTOR inhibitors) or the function of important HIF target genes (VEGFA inhibitors). The specific contribution of each HIF-α isoform with respect to tumor treatment is not clear. Furthermore, these therapies are of limited efficacy and carry significant toxicity. Further research is required to precisely parse the effects of the different HIF-α isoforms on each aspect of tumor progression (eg, proliferation, invasion, angiogenesis, and metastasis) to guide novel therapeutic strategies.
Take-home message.
In contrast to many tumor types, HIF-1α and HIF-2α have opposing effects on clear cell renal cell carcinoma biology, with HIF-1α acting as a tumor suppressor and HIF-2α as an oncogene. The overall effect of VHL inactivation will depend on fine-tuning of the hypoxia-inducible transcription factor response.
Acknowledgments
Funding/Support and role of the sponsor: This study was supported by the German Research Foundation (SCHO 1598/1; J.S.), the Else Kröner-Fresenius-Stiftung (2014_EKES.11; J.S.), the Wilhelm Sander-Stiftung (2014.168.1, J.S.), Cancer Research UK (A16016, D.R.M.), the Ludwig Institute for Cancer Research, and the Higher Education Funding Council for England.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: Johannes Schödel had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Schödel, Mole.
Acquisition of data: Schödel, Grampp.
Analysis and interpretation of data: Schödel, Mole.
Drafting of the manuscript: Schödel, Mole.
Critical revision of the manuscript for important intellectual content: Maher, Moch, Russo, Ratcliffe.
Statistical analysis: None.
Obtaining funding: None.
Administrative, technical, or material support: None.
Supervision: None.
Other (specify): None.
Financial disclosures: Johannes Schödel certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/ affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Ljungberg B, Campbell SC, Choi HY, et al. The epidemiology of renal cell carcinoma. Eur Urol. 2011;60:615–21. doi: 10.1016/j.eururo.2011.06.049. [DOI] [PubMed] [Google Scholar]
- 4.Shuch B, Amin A, Armstrong AJ, et al. Understanding pathologic variants of renal cell carcinoma: distilling therapeutic opportunities from biologic complexity. Eur Urol. 2015;67:85–97. doi: 10.1016/j.eururo.2014.04.029. [DOI] [PubMed] [Google Scholar]
- 5.Verine J, Pluvinage A, Bousquet G, et al. Hereditary renal cancer syndromes: an update of a systematic review. Eur Urol. 2010;58:701–10. doi: 10.1016/j.eururo.2010.08.031. [DOI] [PubMed] [Google Scholar]
- 6.Gnarra JR, Tory K, Weng Y, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herman JG, Latif F, Weng Y, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A. 1994;91:9700–4. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iliopoulos O, Kibel A, Gray S, Kaelin WG., Jr Tumour suppression by the human von Hippel-Lindau gene product. Nat Med. 1995;1:822–6. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 10.Kibel A, Iliopoulos O, DeCaprio JA, Kaelin WG., Jr Binding of the von Hippel-Lindau tumor suppressor protein to elongin B and C. Science. 1995;269:1444–6. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 11.Sato Y, Yoshizato T, Shiraishi Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–7. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 12.Ivan M, Kondo K, Yang H, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–8. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 13.Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–72. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 14.Cockman ME, Masson N, Mole DR, et al. Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J Biol Chem. 2000;275:25733–41. doi: 10.1074/jbc.M002740200. [DOI] [PubMed] [Google Scholar]
- 15.Maxwell PH, Wiesener MS, Chang GW, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–5. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 16.Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Sci STKE. 2005;2005:re12. doi: 10.1126/stke.3062005re12. [DOI] [PubMed] [Google Scholar]
- 17.Schodel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117:e207–17. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isaacs JS, Jung YJ, Mole DR, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–53. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 19.Kim CM, Vocke C, Torres-Cabala C, et al. Expression of hypoxia inducible factor-1alpha and 2alpha in genetically distinct early renal cortical tumors. J Urol. 2006;175:1908–14. doi: 10.1016/S0022-5347(05)00890-6. [DOI] [PubMed] [Google Scholar]
- 20.Selak MA, Armour SM, MacKenzie ED, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 21.Pollard PJ, Spencer-Dene B, Shukla D, et al. Targeted inactivation of FH1 causes proliferative renal cyst development and activation of the hypoxia pathway. Cancer Cell. 2007;11:311–9. doi: 10.1016/j.ccr.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8:967–75. doi: 10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latif F, Tory K, Gnarra J, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–20. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 24.Li L, Zhang L, Zhang X, et al. Hypoxia-inducible factor linked to differential kidney cancer risk seen with type 2A and type 2B VHL mutations. Mol Cell Biol. 2007;27:5381–92. doi: 10.1128/MCB.00282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clifford SC, Cockman ME, Smallwood AC, et al. Contrasting effects on HIF-1alpha regulation by disease-causing pVHL mutations correlate with patterns of tumourigenesis in von Hippel-Lindau disease. Hum Mol Genet. 2001;10:1029–38. doi: 10.1093/hmg/10.10.1029. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman MA, Ohh M, Yang H, Klco JM, Ivan M, Kaelin WG., Jr von Hippel-Lindau protein mutants linked to type 2C VHL disease preserve the ability to downregulate HIF. Hum Mol Genet. 2001;10:1019–27. doi: 10.1093/hmg/10.10.1019. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi A, Sasaki H, Kim SJ, et al. Markedly increased amounts of messenger RNAs for vascular endothelial growth factor and placenta growth factor in renal cell carcinoma associated with angiogenesis. Cancer Res. 1994;54:4233–7. [PubMed] [Google Scholar]
- 28.Nagase Y, Takata K, Moriyama N, Aso Y, Murakami T, Hirano H. Immunohistochemical localization of glucose transporters in human renal cell carcinoma. J Urol. 1995;153:798–801. [PubMed] [Google Scholar]
- 29.Liu Y, Cox SR, Morita T, Kourembanas S. Hypoxia regulates vascular endothelial growth factor gene expression in endothelial cells. Identification of a 5′ enhancer. Circ Res. 1995;77:638–43. doi: 10.1161/01.res.77.3.638. [DOI] [PubMed] [Google Scholar]
- 30.Forsythe JA, Jiang BH, Iyer NV, et al. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–13. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bashan N, Burdett E, Hundal HS, Klip A. Regulation of glucose transport and GLUT1 glucose transporter expression by O2 in muscle cells in culture. Am J Physiol. 1992;262:C682–90. doi: 10.1152/ajpcell.1992.262.3.C682. [DOI] [PubMed] [Google Scholar]
- 32.Iliopoulos O, Levy AP, Jiang C, Kaelin WG, Jr., Goldberg MA. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc Natl Acad Sci U S A. 1996;93:10595–9. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duan DR, Pause A, Burgess WH, et al. Inhibition of transcription elongation by the VHL tumor suppressor protein. Science. 1995;269:1402–6. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 34.Kishida T, Stackhouse TM, Chen F, Lerman MI, Zbar B. Cellular proteins that bind the von Hippel-Lindau disease gene product: mapping of binding domains and the effect of missense mutations. Cancer Res. 1995;55:4544–8. [PubMed] [Google Scholar]
- 35.Okuda H, Hirai S, Takaki Y, et al. Direct interaction of the beta-domain of VHL tumor suppressor protein with the regulatory domain of atypical PKC isotypes. Biochem Biophys Res Commun. 1999;263:491–7. doi: 10.1006/bbrc.1999.1347. [DOI] [PubMed] [Google Scholar]
- 36.Kuznetsova AV, Meller J, Schnell PO, et al. von Hippel-Lindau protein binds hyperphosphorylated large subunit of RNA polymerase II through a proline hydroxylation motif and targets it for ubiquitination. Proc Natl Acad Sci U S A. 2003;100:2706–11. doi: 10.1073/pnas.0436037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohh M, Yauch RL, Lonergan KM, et al. The von Hippel-Lindau tumor suppressor protein is required for proper assembly of an extracellular fibronectin matrix. Mol Cell. 1998;1:959–68. doi: 10.1016/s1097-2765(00)80096-9. [DOI] [PubMed] [Google Scholar]
- 38.Grosfeld A, Stolze IP, Cockman ME, et al. Interaction of hydroxylated collagen IV with the von Hippel-Lindau tumor suppressor. J Biol Chem. 2007;282:13264–9. doi: 10.1074/jbc.M611648200. [DOI] [PubMed] [Google Scholar]
- 39.Schermer B, Ghenoiu C, Bartram M, et al. The von Hippel-Lindau tumor suppressor protein controls ciliogenesis by orienting microtubule growth. J Cell Biol. 2006;175:547–54. doi: 10.1083/jcb.200605092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem. 1995;270:1230–7. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- 41.Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12:5447–54. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang GL, Semenza GL. Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J Biol Chem. 1993;268:21513–8. [PubMed] [Google Scholar]
- 43.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 44.Gu YZ, Moran SM, Hogenesch JB, Wartman L, Bradfield CA. Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3alpha. Gene Expr. 1998;7:205–13. [PMC free article] [PubMed] [Google Scholar]
- 45.Epstein AC, Gleadle JM, McNeill LA, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 46.Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002;295:858–61. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 47.Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–71. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koivunen P, Hirsila M, Gunzler V, Kivirikko KI, Myllyharju J. Catalytic properties of the asparaginyl hydroxylase (FIH) in the oxygen sensing pathway are distinct from those of its prolyl 4-hydroxylases. J Biol Chem. 2004;279:9899–904. doi: 10.1074/jbc.M312254200. [DOI] [PubMed] [Google Scholar]
- 49.Bracken CP, Fedele AO, Linke S, et al. Cell-specific regulation of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha stabilization and transactivation in a graded oxygen environment. J Biol Chem. 2006;281:22575–85. doi: 10.1074/jbc.M600288200. [DOI] [PubMed] [Google Scholar]
- 50.Wiesener MS, Jurgensen JS, Rosenberger C, et al. Widespread hypoxia-inducible expression of HIF-2alpha in distinct cell populations of different organs. FASEB J. 2003;17:271–3. doi: 10.1096/fj.02-0445fje. [DOI] [PubMed] [Google Scholar]
- 51.Rosenberger C, Mandriota S, Jurgensen JS, et al. Expression of hypoxia-inducible factor-1alpha and -2alpha in hypoxic and ischemic rat kidneys. J Am Soc Nephrol. 2002;13:1721–32. doi: 10.1097/01.asn.0000017223.49823.2a. [DOI] [PubMed] [Google Scholar]
- 52.Schietke RE, Hackenbeck T, Tran M, et al. Renal tubular HIF-2alpha expression requires VHL inactivation and causes fibrosis and cysts. PLoS One. 2012;7:e31034. doi: 10.1371/journal.pone.0031034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xia X, Lemieux ME, Li W, et al. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci U S A. 2009;106:4260–5. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mole DR, Blancher C, Copley RR, et al. Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem. 2009;284:16767–75. doi: 10.1074/jbc.M901790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krieg AJ, Rankin EB, Chan D, Razorenova O, Fernandez S, Giaccia AJ. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Biol. 2010;30:344–53. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanimoto K, Tsuchihara K, Kanai A, et al. Genome-wide identification and annotation of HIF-1alpha binding sites in two cell lines using massively parallel sequencing. Hugo J. 2010;4:35–48. doi: 10.1007/s11568-011-9150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mimura I, Nangaku M, Kanki Y, et al. Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Mol Cell Biol. 2012;32:3018–32. doi: 10.1128/MCB.06643-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ortiz-Barahona A, Villar D, Pescador N, Amigo J, del Peso L. Genome-wide identification of hypoxia-inducible factor binding sites and target genes by a probabilistic model integrating transcription-profiling data and in silico binding site prediction. Nucleic Acids Res. 2010;38:2332–45. doi: 10.1093/nar/gkp1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wykoff CC, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ. Identification of novel hypoxia dependent and independent target genes of the von Hippel-Lindau (VHL) tumour suppressor by mRNA differential expression profiling. Oncogene. 2000;19:6297–305. doi: 10.1038/sj.onc.1204012. [DOI] [PubMed] [Google Scholar]
- 60.Wykoff CC, Sotiriou C, Cockman ME, et al. Gene array of VHL mutation and hypoxia shows novel hypoxia-induced genes and that cyclin D1 is a VHL target gene. Br J Cancer. 2004;90:1235–43. doi: 10.1038/sj.bjc.6601657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang Y, Zhang W, Kondo K, et al. Gene expression profiling in a renal cell carcinoma cell line: dissecting VHL and hypoxia-dependent pathways. Mol Cancer Res. 2003;1:453–62. [PubMed] [Google Scholar]
- 62.Schodel J, Mole DR, Ratcliffe PJ. Pan-genomic binding of hypoxia-inducible transcription factors. Biol Chem. 2013;394:507–17. doi: 10.1515/hsz-2012-0351. [DOI] [PubMed] [Google Scholar]
- 63.Schodel J, Bardella C, Sciesielski LK, et al. Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression. Nat Genet. 2012;44:420–5. S1–2. doi: 10.1038/ng.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elvidge GP, Glenny L, Appelhoff RJ, Ratcliffe PJ, Ragoussis J, Gleadle JM. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: the role of HIF-1alpha, HIF-2alpha, and other pathways. J Biol Chem. 2006;281:15215–26. doi: 10.1074/jbc.M511408200. [DOI] [PubMed] [Google Scholar]
- 65.Warnecke C, Zaborowska Z, Kurreck J, et al. Differentiating the functional role of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha (EPAS-1) by the use of RNA interference: erythropoietin is a HIF-2alpha target gene in Hep3B and Kelly cells. FASEB J. 2004;18:1462–4. doi: 10.1096/fj.04-1640fje. [DOI] [PubMed] [Google Scholar]
- 66.Warnecke C, Weidemann A, Volke M, et al. The specific contribution of hypoxia-inducible factor-2alpha to hypoxic gene expression in vitro is limited and modulated by cell type-specific and exogenous factors. Exp Cell Res. 2008;314:2016–27. doi: 10.1016/j.yexcr.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 67.Mandriota SJ, Turner KJ, Davies DR, et al. HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in the nephron. Cancer Cell. 2002;1:459–68. doi: 10.1016/s1535-6108(02)00071-5. [DOI] [PubMed] [Google Scholar]
- 68.Raval RR, Lau KW, Tran MG, et al. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol. 2005;25:5675–86. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hackenbeck T, Knaup KX, Schietke R, et al. HIF-1 or HIF-2 induction is sufficient to achieve cell cycle arrest in NIH3T3 mouse fibroblasts independent from hypoxia. Cell Cycle. 2009;8:1386–95. doi: 10.4161/cc.8.9.8306. [DOI] [PubMed] [Google Scholar]
- 70.Lachance G, Uniacke J, Audas TE, et al. DNMT3a epigenetic program regulates the HIF-2alpha oxygen-sensing pathway and the cellular response to hypoxia. Proc Natl Acad Sci U S A. 2014;111:7783–8. doi: 10.1073/pnas.1322909111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Young AP, Kaelin WG., Jr Senescence triggered by the loss of the VHL tumor suppressor. Cell Cycle. 2008;7:1709–12. doi: 10.4161/cc.7.12.6124. [DOI] [PubMed] [Google Scholar]
- 72.Walther MM, Lubensky IA, Venzon D, Zbar B, Linehan WM. Prevalence of microscopic lesions in grossly normal renal parenchyma from patients with von Hippel-Lindau disease, sporadic renal cell carcinoma and no renal disease: clinical implications. J Urol. 1995;154:2010–4. discussion 2014–5. [PubMed] [Google Scholar]
- 73.Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG., Jr Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–46. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 74.Kondo K, Kim WY, Lechpammer M, Kaelin WG., Jr Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 2003;1:E83. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shen C, Beroukhim R, Schumacher SE, et al. Genetic and functional studies implicate HIF1alpha as a 14q kidney cancer suppressor gene. Cancer Discov. 2011;1:222–35. doi: 10.1158/2159-8290.CD-11-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Adam J, Hatipoglu E, O'Flaherty L, et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell. 2011;20:524–37. doi: 10.1016/j.ccr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gordan JD, Lal P, Dondeti VR, et al. HIF-alpha effects on c-Myc distinguish two subtypes of sporadic VHL-deficient clear cell renal carcinoma. Cancer Cell. 2008;14:435–46. doi: 10.1016/j.ccr.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Monzon FA, Alvarez K, Peterson L, et al. Chromosome 14q loss defines a molecular subtype of clear-cell renal cell carcinoma associated with poor prognosis. Mod Pathol. 2011;24:1470–9. doi: 10.1038/modpathol.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Purdue MP, Johansson M, Zelenika D, et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat Genet. 2011;43:60–5. doi: 10.1038/ng.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han SS, Yeager M, Moore LE, et al. The chromosome 2p21 region harbors a complex genetic architecture for association with risk for renal cell carcinoma. Hum Mol Genet. 2012;21:1190–200. doi: 10.1093/hmg/ddr551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Audenet F, Cancel-Tassin G, Pierre B, et al. Germline genetic variations at 11q13 and 12p11 locus modulate age at onset for renal cell carcinoma. J Urol. 2014;191:487–92. doi: 10.1016/j.juro.2013.07.064. [DOI] [PubMed] [Google Scholar]
- 83.Su T, Han YF, Yu YW, et al. A GWAS-identified susceptibility locus on chromosome 11q13.3 and its putative molecular target for prediction of postoperative prognosis of human renal cell carcinoma. Oncol Lett. 2013;6:421–6. doi: 10.3892/ol.2013.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu X, Scelo G, Purdue MP, et al. A genome-wide association study identifies a novel susceptibility locus for renal cell carcinoma on 12p11.23. Hum Mol Genet. 2012;21:456–62. doi: 10.1093/hmg/ddr479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dalgliesh GL, Furge K, Greenman C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–3. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Haaften G, Dalgliesh GL, Davies H, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–3. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44:751–9. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–42. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gerlinger M, Horswell S, Larkin J, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–33. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wenger RH, Kvietikova I, Rolfs A, Camenisch G, Gassmann M. Oxygen-regulated erythropoietin gene expression is dependent on a CpG methylation-free hypoxia-inducible factor-1 DNA-binding site. Eur J Biochem. 1998;253:771–7. doi: 10.1046/j.1432-1327.1998.2530771.x. [DOI] [PubMed] [Google Scholar]
- 92.Niu X, Zhang T, Liao L, et al. The von Hippel-Lindau tumor suppressor protein regulates gene expression and tumor growth through histone demethylase JARID1C. Oncogene. 2012;31:776–86. doi: 10.1038/onc.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Simon JM, Hacker KE, Singh D, et al. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res. 2014;24:241–50. doi: 10.1101/gr.158253.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang SS, Gu YF, Wolff N, et al. Bap1 is essential for kidney function and cooperates with Vhl in renal tumorigenesis. Proc Natl Acad Sci U S A. 2014;111:16538–43. doi: 10.1073/pnas.1414789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Khan MN, Bhattacharyya T, Andrikopoulos P, et al. Factor inhibiting HIF (FIH-1) promotes renal cancer cell survival by protecting cells from HIF-1alpha-mediated apoptosis. Br J Cancer. 2011;104:1151–9. doi: 10.1038/bjc.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kroeze SG, Vermaat JS, van Brussel A, et al. Expression of nuclear FIH independently predicts overall survival of clear cell renal cell carcinoma patients. Eur J Cancer. 2010;46:3375–82. doi: 10.1016/j.ejca.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 97.Makino Y, Cao R, Svensson K, et al. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature. 2001;414:550–4. doi: 10.1038/35107085. [DOI] [PubMed] [Google Scholar]
- 98.Maynard MA, Evans AJ, Hosomi T, Hara S, Jewett MA, Ohh M. Human HIF-3alpha4 is a dominant-negative regulator of HIF-1 and is down-regulated in renal cell carcinoma. FASEB J. 2005;19:1396–406. doi: 10.1096/fj.05-3788com. [DOI] [PubMed] [Google Scholar]
- 99.Tanaka T, Wiesener M, Bernhardt W, Eckardt KU, Warnecke C. The human HIF (hypoxia-inducible factor)-3alpha gene is a HIF-1 target gene and may modulate hypoxic gene induction. Biochem J. 2009;424:143–51. doi: 10.1042/BJ20090120. [DOI] [PubMed] [Google Scholar]
- 100.Maynard MA, Evans AJ, Shi W, Kim WY, Liu FF, Ohh M. Dominant-negative HIF-3 alpha 4 suppresses VHL-null renal cell carcinoma progression. Cell Cycle. 2007;6:2810–6. doi: 10.4161/cc.6.22.4947. [DOI] [PubMed] [Google Scholar]
- 101.Zhang P, Yao Q, Lu L, Li Y, Chen PJ, Duan C. Hypoxia-inducible factor 3 is an oxygen-dependent transcription activator and regulates a distinct transcriptional response to hypoxia. Cell Rep. 2014;6:1110–21. doi: 10.1016/j.celrep.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 102.Klatte T, Kroeger N, Rampersaud EN, et al. Gain of chromosome 8q is associated with metastases and poor survival of patients with clear cell renal cell carcinoma. Cancer. 2012;118:5777–82. doi: 10.1002/cncr.27607. [DOI] [PubMed] [Google Scholar]
- 103.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11:335–47. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–13. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bertout JA, Majmundar AJ, Gordan JD, et al. HIF2alpha inhibition promotes p53 pathway activity, tumor cell death, and radiation responses. Proc Natl Acad Sci U S A. 2009;106:14391–6. doi: 10.1073/pnas.0907357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Burrows AE, Smogorzewska A, Elledge SJ. Polybromo-associated BRG1-associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proc Natl Acad Sci U S A. 2010;107:14280–5. doi: 10.1073/pnas.1009559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Biswas S, Charlesworth PJ, Turner GD, et al. CD31 angiogenesis and combined expression of HIF-1alpha and HIF-2alpha are prognostic in primary clear-cell renal cell carcinoma (CC-RCC), but HIFalpha transcriptional products are not: implications for antiangiogenic trials and HIFalpha biomarker studies in primary CC-RCC. Carcinogenesis. 2012;33:1717–25. doi: 10.1093/carcin/bgs222. [DOI] [PubMed] [Google Scholar]
- 108.Klatte T, Seligson DB, Riggs SB, et al. Hypoxia-inducible factor 1 alpha in clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:7388–93. doi: 10.1158/1078-0432.CCR-07-0411. [DOI] [PubMed] [Google Scholar]
- 109.Lidgren A, Hedberg Y, Grankvist K, Rasmuson T, Vasko J, Ljungberg B. The expression of hypoxia-inducible factor 1alpha is a favorable independent prognostic factor in renal cell carcinoma. Clin Cancer Res. 2005;11:1129–35. [PubMed] [Google Scholar]
- 110.Hudson CC, Liu M, Chiang GG, et al. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol Cell Biol. 2002;22:7004–14. doi: 10.1128/MCB.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sternberg CN, Davis ID, Mardiak J, et al. Pazopanib in locally advanced or metastatic renal cell carcinoma: results of a randomized phase III trial. J Clin Oncol. 2010;28:1061–8. doi: 10.1200/JCO.2009.23.9764. [DOI] [PubMed] [Google Scholar]
- 112.Choueiri TK, Fay AP, Gagnon R, et al. The role of aberrant VHL/HIF pathway elements in predicting clinical outcome to pazopanib therapy in patients with metastatic clear-cell renal cell carcinoma. Clin Cancer Res. 2013;19:5218–26. doi: 10.1158/1078-0432.CCR-13-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Choueiri TK, Vaziri SA, Jaeger E, et al. von Hippel-Lindau gene status and response to vascular endothelial growth factor targeted therapy for metastatic clear cell renal cell carcinoma. J Urol. 2008;180:860–5. doi: 10.1016/j.juro.2008.05.015. discussion 865–6. [DOI] [PubMed] [Google Scholar]
- 114.Kondo K, Yao M, Yoshida M, et al. Comprehensive mutational analysis of the VHL gene in sporadic renal cell carcinoma: relationship to clinicopathological parameters. Genes Chromosomes Cancer. 2002;34:58–68. doi: 10.1002/gcc.10054. [DOI] [PubMed] [Google Scholar]
- 115.Atkins DJ, Gingert C, Justenhoven C, et al. Concomitant deregulation of HIF1alpha and cell cycle proteins in VHL-mutated renal cell carcinomas. Virchows Arch. 2005;447:634–42. doi: 10.1007/s00428-005-1262-y. [DOI] [PubMed] [Google Scholar]
- 116.Yao M, Yoshida M, Kishida T, et al. VHL tumor suppressor gene alterations associated with good prognosis in sporadic clear-cell renal carcinoma. J Natl Cancer Inst. 2002;94:1569–75. doi: 10.1093/jnci/94.20.1569. [DOI] [PubMed] [Google Scholar]
- 117.Schraml P, Struckmann K, Hatz F, et al. VHL mutations and their correlation with tumour cell proliferation, microvessel density, and patient prognosis in clear cell renal cell carcinoma. J Pathol. 2002;196:186–93. doi: 10.1002/path.1034. [DOI] [PubMed] [Google Scholar]
- 118.Staller P, Sulitkova J, Lisztwan J, Moch H, Oakeley EJ, Krek W. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature. 2003;425:307–11. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- 119.Vanharanta S, Shu W, Brenet F, et al. Epigenetic expansion of VHL-HIF signal output drives multiorgan metastasis in renal cancer. Nat Med. 2013;19:50–6. doi: 10.1038/nm.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pollard PJ, Loenarz C, Mole DR, et al. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1alpha. Biochem J. 2008;416:387–94. doi: 10.1042/BJ20081238. [DOI] [PubMed] [Google Scholar]
- 121.Beyer S, Kristensen MM, Jensen KS, Johansen JV, Staller P. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J Biol Chem. 2008;283:36542–52. doi: 10.1074/jbc.M804578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chan DA, Sutphin PD, Nguyen P, et al. Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci Transl Med. 2011;3:94ra70. doi: 10.1126/scitranslmed.3002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rankin EB, Fuh KC, Castellini L, et al. Direct regulation of GAS6/AXL signaling by HIF promotes renal metastasis through SRC and MET. Proc Natl Acad Sci U S A. 2014;111:13373–8. doi: 10.1073/pnas.1404848111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tong WH, Sourbier C, Kovtunovych G, et al. The glycolytic shift in fumarate-hydratase-deficient kidney cancer lowers AMPK levels, increases anabolic propensities and lowers cellular iron levels. Cancer Cell. 2011;20:315–27. doi: 10.1016/j.ccr.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Preston RS, Philp A, Claessens T, et al. Absence of the Birt-Hogg-Dube gene product is associated with increased hypoxia-inducible factor transcriptional activity and a loss of metabolic flexibility. Oncogene. 2011;30:1159–73. doi: 10.1038/onc.2010.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–74. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McCormick RI, Blick C, Ragoussis J, et al. miR-210 is a target of hypoxia-inducible factors 1 and 2 in renal cancer, regulates ISCU and correlates with good prognosis. Br J Cancer. 2013;108:1133–42. doi: 10.1038/bjc.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Razorenova OV, Finger EC, Colavitti R, et al. VHL loss in renal cell carcinoma leads to up-regulation of CUB domain-containing protein 1 to stimulate PKC{delta}-driven migration. Proc Natl Acad Sci U S A. 2011;108:1931–6. doi: 10.1073/pnas.1011777108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li B, Qiu B, Lee DS, et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513:251–5. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Knaup KX, Monti J, Hackenbeck T, et al. Hypoxia regulates the sperm associated antigen 4 (SPAG4) via HIF, which is expressed in renal clear cell carcinoma and promotes migration and invasion in vitro. Mol Carcinog. 2014;53:970–8. doi: 10.1002/mc.22065. [DOI] [PubMed] [Google Scholar]
- 131.Shoji K, Murayama T, Mimura I, et al. Sperm-associated antigen 4, a novel hypoxia-inducible factor 1 target, regulates cytokinesis, and its expression correlates with the prognosis of renal cell carcinoma. Am J Pathol. 2013;182:2191–203. doi: 10.1016/j.ajpath.2013.02.024. [DOI] [PubMed] [Google Scholar]
- 132.Grabmaier K, MC AdW, Verhaegh GW, Schalken JA, Oosterwijk E. Strict regulation of CAIX(G250/MN) by HIF-1alpha in clear cell renal cell carcinoma. Oncogene. 2004;23:5624–31. doi: 10.1038/sj.onc.1207764. [DOI] [PubMed] [Google Scholar]
- 133.Gimm T, Wiese M, Teschemacher B, et al. Hypoxia-inducible protein 2 is a novel lipid droplet protein and a specific target gene of hypoxia-inducible factor-1. FASEB J. 2010;24:4443–58. doi: 10.1096/fj.10-159806. [DOI] [PubMed] [Google Scholar]
- 134.Petrella BL, Lohi J, Brinckerhoff CE. Identification of membrane type-1 matrix metalloproteinase as a target of hypoxia-inducible factor-2 alpha in von Hippel-Lindau renal cell carcinoma. Oncogene. 2005;24:1043–52. doi: 10.1038/sj.onc.1208305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Salama MF, Carroll B, Adada M, Pulkoski-Gross M, Hannun YA, Obeid LM. A novel role of sphingosine kinase-1 in the invasion and angiogenesis of VHL mutant clear cell renal cell carcinoma. FASEB J. 2015;29:2803–13. doi: 10.1096/fj.15-270413. [DOI] [PMC free article] [PubMed] [Google Scholar]