Abstract
Purpose
To identify diagnostic and prognostic markers of chronic graft-versus-host disease (cGVHD), the major cause of morbidity and mortality after allogeneic hematopoietic cell transplantation (HCT).
Patients and Methods
Using a quantitative proteomics approach, we compared pooled plasma samples obtained at matched time points after HCT (median, 103 days) from 35 patients with cGVHD and 18 without cGVHD (data are available via ProteomeXchange with identifier PXD002762). Of 105 proteins showing at least a 1.25-fold difference in expression, 22 were selected on the basis of involvement in relevant pathways and enzyme-linked immunosorbent assay availability. Chemokine (C-X-C motif) ligand 9 (CXCL9) and suppression of tumorigenicity 2 (ST2) also were measured on the basis of previously determined associations with GVHD. Concentrations of the four lead biomarkers were measured at or after diagnosis in plasma from two independent verification cohorts (n = 391) to determine their association with cGVHD. Their prognostic ability when measured at approximately day +100 after HCT was evaluated in plasma of a second verification cohort (n = 172).
Results
Of 24 proteins measured in the first verification cohort, nine proteins were associated with cGVHD, and only four (ST2, CXCL9, matrix metalloproteinase 3, and osteopontin) were necessary to compose a four-biomarker panel with an area under the receiver operating characteristic curve (AUC) of 0.89 and significant correlation with cGVHD diagnosis, cGVHD severity, and nonrelapse mortality. In a second verification cohort, this panel distinguished patients with cGVHD (AUC, 0.75), and finally, the panel measured at day +100 could predict cGVHD occurring within the next 3 months with an AUC of 0.67 and 0.79 without and with known clinical risk factors, respectively.
Conclusion
We conclude that the biomarker panel measured at diagnosis or day +100 after HCT may allow patient stratification according to risk of cGVHD.
INTRODUCTION
Chronic graft-versus-host disease (cGVHD) remains the most common long-term complication of allogeneic hematopoietic cell transplantation (HCT), reportedly occurring in 30% to 70% of patients surviving more than 100 days.1-3 It is also the leading cause of nonrelapse mortality (NRM) occurring more than 2 years after HCT for malignant disease.4,5 In 2014, the National Institutes of Health (NIH) Consensus Development Project on Criteria for Clinical Trials in cGVHD published an updated series of articles to help standardize the clinical approach to treating these patients, thereby promoting new interest in this important post-transplantation complication and motivating the development of well-defined biomarkers for future use as an aid in clinical trials.6-10
The development of acute graft-versus-host disease (aGVHD) biomarkers11-14 has increased interest in identifying biomarkers that provide meaningful information for cGVHD. Several studies have reported the discovery of cGVHD biomarkers,9,15 but verification studies of these biomarkers in independent cohorts are currently lacking. Here, we used in-depth proteomic profiling using gel-free, high-resolution tandem mass spectrometry (MS/MS). Following the recent recommendations from the NIH consensus,9 we then verified the concentrations of individual candidate proteins in samples from an independent cohort of 211 HCT patients using high-throughput immunoassays. The leading proteins were then tested in a second independent verification cohort of 180 HCT patients. Finally, plasma protein concentrations were measured in samples collected on day +100 after HCT from 172 of the 180 patients in verification cohort 2 to test their potential as prognostic markers on the basis of their ability to correctly identify patients who would subsequently develop cGVHD.
PATIENTS AND METHODS
Three cohorts of patients were included in this study, as previously described.16-18 This study was approved by the institutional review boards of the participating institutions, and informed consent was obtained from all patients or their legal guardians. Heparinized blood samples were collected prospectively at the onset of manifestations in patients with cGVHD or at matched time points in controls. Patients in the verification 2 cohort also had samples drawn at approximately day +100.
Concurrent controls were available in all three cohorts, and their characteristics are listed in Table 1. For the discovery cohort and verification cohort 1, controls were patients who did not have cGVHD. For the verification cohort 2, where excess controls were available, controls were selected to match patient cases on the basis of time since transplantation, transplantation center, prior aGVHD, conditioning intensity, and graft source. Chart review confirmed that controls did not develop cGVHD in the 3 months after the sample was taken. See the Data Supplement for additional information.
Table 1.
Patient and GVHD Characteristics
| Characteristic | Discovery Cohort (n = 53) | Independent Verification Cohort 1 (n = 211) | Independent Verification Cohort 2 (n = 180)*† | ||||||
|---|---|---|---|---|---|---|---|---|---|
| cGVHD (n = 35) | Controls (n = 18) | P | cGVHD (n = 178) | Controls (n = 33) | P | cGVHD (n = 87) | Controls (n = 93) | P | |
| Age, years | .03 | .55 | .44 | ||||||
| Median | 50 | 27 | 52 | 54 | 51 | 52 | |||
| Range | 6-67 | 1-66 | 19-79 | 22-72 | 21-78 | 20-71 | |||
| Sex, No. (%) | 1.00 | .23 | .50 | ||||||
| Female | 11 (31) | 6 (33) | 77 (43) | 18 (55) | 49 (56) | 57 (61) | |||
| Male | 24 (69) | 12 (67) | 101 (57) | 15 (45) | 38 (44) | 36 (39) | |||
| Donor type and match, No. (%) | .40 | .06 | .50 | ||||||
| Matched sibling | 29 (83) | 17 (94) | 66 (37) | 18 (55) | 38 (44) | 36 (39) | |||
| Other‡ | 6 (17) | 1 (6) | 112 (63) | 15 (45) | 49 (56) | 57 (61) | |||
| Stem-cell source, No. (%) | .11 | .05 | .62 | ||||||
| PBSC | 28 (80) | 10 (56) | 161 (90) | 26 (79) | 77 (89) | 80 (86) | |||
| Other | 7 (20) | 8 (44) | 17 (10) | 7 (21) | 10 (11) | 13 (14) | |||
| Conditioning regimen intensity, No. (%) | 1.00 | .88 | .83 | ||||||
| Myeloablative | 26 (74) | 13 (72) | 105 (59) | 19 (58) | 37 (43) | 41 (44) | |||
| Nonmyeloablative | 9 (26) | 5 (28) | 73 (41) | 14 (42) | 50 (57) | 52 (56) | |||
| Prior acute GVHD, No. (%) | < .001 | .70 | .24 | ||||||
| Yes | 18 (51) | 0 (0) | 135 (76) | 24 (73) | 38 (44) | 49 (53) | |||
| No | 17 (49) | 18 (100) | 43 (34) | 9 (37) | 49 (56) | 44 (47) | |||
| Time post-HCT to cGVHD diagnosis, days | NA | NA | NA | ||||||
| Median | 119 | NA | 210 | NA | 203 | NA | |||
| Range | 56-352 | NA | 38-1757 | NA | 86-461 | NA | |||
| Time post-HCT to sample acquisition, days | .03 | .84 | .63 | ||||||
| Median | 103 | 102 | 391 | 369 | 236 | 192 | |||
| Range | 50-372 | 94-189 | 192-1,852 | 161-3,641 | 86-522 | 133-482 | |||
| NIH global severity, No. (%) | NA | NA | NA | ||||||
| Mild | 2 (6) | NA | 13 (7) | NA | 15 (18) | NA | |||
| Moderate | 20 (57) | NA | 103 (58) | NA | 46 (55) | NA | |||
| Severe | 13 (37) | NA | 62 (35) | NA | 22 (27) | NA | |||
| Corticosteroid use at time of sampling, No. (%) | .29 | < .001 | < .001 | ||||||
| Yes | 4 (11) | 0 (0) | 108 (61) | 2 (6) | 51 (61) | 22 (24) | |||
| No | 31 (89) | 18 (100) | 70 (39) | 31 (94) | 36 (39) | 71 (76) | |||
Abbreviations: cGVHD, chronic graft-versus-host disease; GVHD, graft-versus-host disease; HCT, hematopoietic cell transplantations; NA, not applicable; NIH, National Institutes of Health; PBSC, peripheral-blood stem cell.
Matched for time since transplantation to sample collection, transplantation center, prior acute GVHD, conditioning intensity, and graft source from the U54 multicenter cohort (Fred Hutchinson Cancer Research Center, Dana-Farber Cancer Institute, University of Minnesota, H. Lee Moffitt Cancer Center, Vanderbilt University Medical Center, Roswell Park Cancer Institute, Washington University St Louis, and Cleveland Clinic).
Eight patients missing data for prior acute GVHD, and four patients missing data for NIH severity.
Other: There were only four cord blood cell transplantations among the patients and none among the controls.
RESULTS
Discovery Study Using Quantitative Proteomics
Pooled plasma from 35 patients with cGVHD was labeled with one isobaric tag for relative and absolute quantification (iTRAQ) and compared with pooled plasma from 18 patients without cGVHD, labeled with a different iTRAQ label (Appendix Fig A1, online only). The characteristics of patients in the discovery cohort are listed in Table 1. Patients in the cGVHD group were older and more likely to have had prior aGVHD than the controls without cGVHD. Otherwise, there were no statistically significant differences between the patients with cGVHD and without cGVHD according to sex, donor and HLA type, graft source, or conditioning intensity. Of note, samples were selected so that patients had minimal to no corticosteroid immunosuppression at the time of sampling in both patients with cGVHD and controls. Samples were collected at similar times for both patients with cGVHD and controls, with a less extended range for controls because the blood collection from controls occurred at regular visits, around day +100 and day +180 after HCT. Samples were thus obtained at a median of 103 days (range, 50 to 372 days) after HCT in the cGVHD group compared with 102 days (range, 94 to 189 days) after HCT in the control group (P = .03).
iTRAQ labels allow for comparison of relative concentrations of proteins between groups (see Patients and Methods). Overall, 1,998 proteins were identified and quantified, and 105 of these proteins were differentially expressed by at least a 1.25-fold difference that distinguished cGVHD patients from patients without cGVHD at the time of sampling (Data Supplement). Antibodies suitable for enzyme-linked immunosorbent assays were available for 22 of these proteins (Data Supplement), and CXCL9 and ST2 also were considered for validation based on previously noted associations with cGVHD or refractory GVHD.14,18
Biomarker Panel Development With Verification Cohort 1
We measured the concentrations of the 24 proteins mentioned earlier in samples from cohort 1, which consisted of 178 HCT patients with cGVHD and 33 HCT controls without cGVHD. The patient and GVHD characteristics for the two groups are listed in Table 1. Patient sex and median age at the time of transplantation were similar between the two groups. There was a trend toward over-representation of unrelated donors in the cGVHD group. As expected, the use of peripheral-blood stem cells (PBSCs) was a risk factor for cGVHD, whereas the use of full-intensity conditioning was not. The incidence of prior aGVHD was similar in both groups. According to the NIH global severity score, 7% of the patients had mild cGVHD, 58% had moderate cGVHD, and 35% had severe cGVHD. Plasma samples were collected at similar times from both cGVHD patients and controls, at a median of 391 days (range, 192 to 1,852 days) after HCT in the cGVHD group compared with 369 days (range, 161 to 3,641 days) after HCT in the control group (P = .84). Unlike the discovery cohort, 61% of the patients with cGVHD were receiving corticosteroid treatment at the time of sampling, compared with 6% of control patients (P < .001).
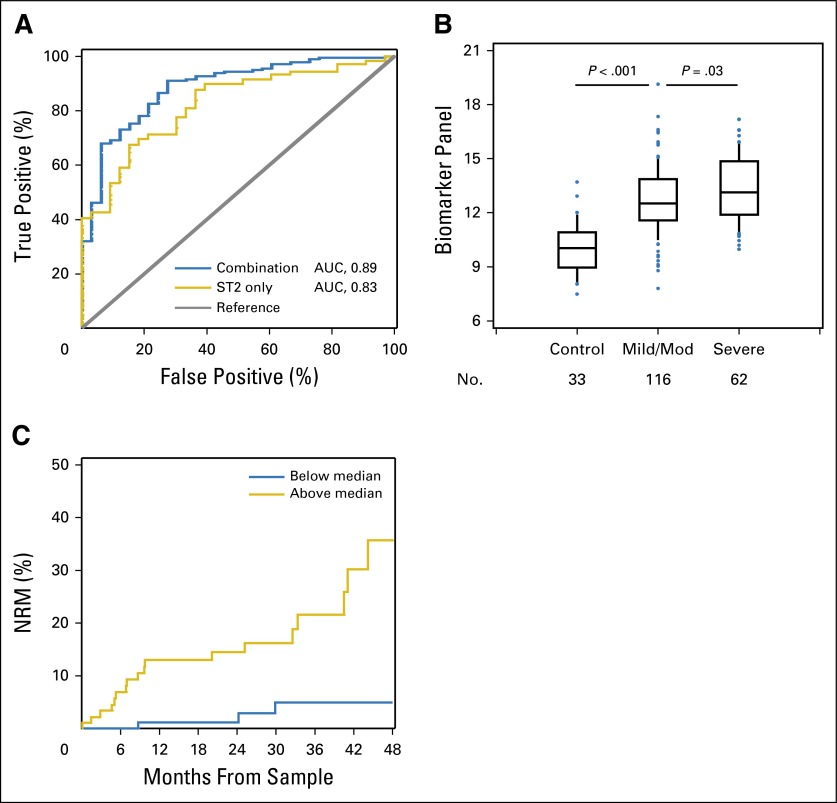
To create the biomarker panel, logistic regression was used to evaluate associations between cGVHD and biomarkers after log transformation. All analyses of individual biomarkers were adjusted for the following seven clinical variables: age, sex, donor (matched sibling v others), stem-cell source (PBSC v others), conditioning intensity (myeloablative v others), prior aGVHD, and time from HCT to sample collection. Notably, only donor type (P = .03) and PBSCs as the stem-cell source (P = .01) were significant in multivariable analysis. Of the 24 proteins tested, nine were associated with cGVHD with a P < .05 (Table 2). To determine the best combination model, we used unadjusted forward selection with a P = .05 significance threshold using these nine markers, confirmed by backward selection. The selection procedure identified four proteins, ST2, MMP3, CXCL9, and OPN, for inclusion in the combination panel. The other five proteins were not included in the panel because they did not show an additional effect in forward and backward selection models (Table 2). We then generated receiver operating characteristic curves for the best single biomarker and the combination model. The area under the curve (AUC) for the four-biomarker panel was 0.89, whereas the AUC for ST2 was 0.83 (Fig 1A). To better define the potential clinical utility of the four identified proteins, we also tested, in multivariable models with and without clinical covariates, the association of biomarker levels with cGVHD in this cohort (Table 2).
Table 2.
Association of Biomarker Levels With cGVHD in Verification Cohort 1 (178 patients and 33 controls)
| Model | OR* (95% CI) | P | AUC† |
|---|---|---|---|
| Single biomarkers | |||
| ST2 | 104 (17 to 623) | < .001 | 0.83 |
| MMP3 | 11.6 (4.1 to 33) | < .001 | 0.79 |
| TNFRSF10C | 68.0 (7.5 to 617) | < .001 | 0.72 |
| OPN | 11.7 (3.8 to 36) | < .001 | 0.71 |
| SELP | 22.8 (2.3 to 227) | .005 | 0.66 |
| CKIT | 0.06 (0.007 to 0.45) | .004 | 0.65 |
| COMP | 9.6 (1.7 to 53) | .006 | 0.64 |
| CXCL9 | 3.5 (1.2 to 10) | .02 | 0.63 |
| CD146 | 39.5 (1.4 to 999) | .03 | 0.62 |
| Best combination of biomarkers | |||
| ST2 | 25.4 (3.6 to 180) | < .001 | 0.89 |
| CXCL9 | 6.1 (1.5 to 24) | ||
| MMP3 | 5.2 (1.5 to 18) | ||
| OPN | 4.8 (1.2 to 19) | ||
| Single biomarkers with clinical covariates‡ | |||
| ST2 | 124 (18 to 857) | < .001 | 0.85 |
| MMP3 | 11.7 (3.8 to 36) | < .001 | 0.80 |
| TRAILR3 | 91.9 (8.2 to 999+) | < .001 | 0.77 |
| OPN | 14.5 (4.1 to 52) | < .001 | 0.79 |
| SELP | 31.5 (2.8 to 357) | .003 | 0.73 |
| CKIT | 0.06 (0.006 to 0.54) | .008 | 0.73 |
| COMP | 25.4 (3.2 to 200) | .008 | 0.74 |
| CXCL9 | 4.5 (1.4 to 14.3) | .009 | 0.71 |
| CD146 | 47.2 (1.3 to 999) | .03 | 0.70 |
| Best combination of biomarkers with clinical covariates‡ | |||
| ST2 | 70.2 (6.5 to 761) | < .001 | 0.92 |
| CXCL9 | 26.9 (3.9 to 186) | ||
| MMP3 | 7.9 (2.0 to 30.9) | ||
| OPN | 11.9 (2.1 to 68.5) |
Abbreviations: AUC, area under the curve; cGVHD, chronic graft-versus-host disease; OR, odds ratio.
OR per log increase in biomarker, estimated using logistic regression.
AUC calculated from logistic regression; includes clinical covariates in model where indicated.
AUC with clinical covariates only: 0.66.
Fig 1.
(A) Receiver operating characteristic (ROC) curves for the best single biomarker and the combination model in verification cohort 1. ROC curves for the best single biomarker (ST2; area under the curve [AUC], 0.83) and the combination model (AUC, 0.89) comparing patients with chronic graft-versus-host disease (cGVHD; n = 178) and time-matched controls without cGVHD (n = 33). AUC values are reported from multivariable models. (B) Increased biomarker panel levels are associated with increased cGVHD severity in verification cohort 1. Median value of the four-biomarker panel from controls (n = 33), patients with mild to moderate (Mod) cGVHD (n = 116), and patients with severe cGVHD (n = 62) in verification cohort 1. Data are illustrated as box and whisker plots, with the whiskers indicating the 90th and 10th percentiles. P values compare controls versus patients with mild to moderate cGVHD, and patients with mild to moderate cGVHD versus those with severe cGVHD, according to Wilcoxon two-sample tests. (C) Nonrelapse mortality (NRM) stratified by the biomarker panel in verification cohort 1. The cumulative incidence of NRM is plotted, divided according to the median value of the biomarker panel among patients with cGVHD. The NRM at 48 months was 5% (95% CI, 1% to 9%) for the group less than the median value and 36% (95% CI, 19% to 53%) for the group greater than the median value (P = .002; adjusted for age, sex, donor [matched sibling v others], stem-cell source [peripheral-blood stem cells v others], conditioning intensity [myeloablative v others], prior acute GVHD, and time from hematopoietic cell transplantation to sample collection).
Biomarker Panel and cGVHD Severity and NRM in Verification Cohort 1
We next evaluated whether the biomarker panel was associated with cGVHD severity. Few patients had mild cGVHD, and thus, these patients were combined with those who presented with moderate cGVHD. Using Wilcoxon two-sample tests, we compared the biomarker panel (defined using the following equation: 3.23 × log10 ST2 + 1.81 × log10 CXCL9 + 1.64 × log10 MMP3 + 1.58 × log10 OPN) between groups with different cGVHD severity (none, mild or moderate, and severe). The severity of cGVHD was correlated with the biomarker panel (P < .001 comparing none with any cGVHD, and P = .03 comparing mild or moderate with severe cGVHD [unadjusted], Fig 1B). In regression analysis with adjustment for clinical variables, P = .006 for mild or moderate versus severe cGVHD.
NRM was compared between groups with high versus low biomarker levels using a cutoff of the median value of the biomarker panel (median value obtained from the formula given earlier) among patients with cGVHD. Patients with cGVHD and biomarker panel levels greater than the median had higher NRM, with a hazard ratio of 7.0 (95% CI, 2.0 to 24.8) and P = .003 after adjustment for the previously mentioned clinical characteristics, compared with patients with cGVHD with lower biomarker panel levels (Fig 1C). Finally, because the cohort consisted entirely of patients with multiorgan involvement, the biomarker panel was not associated with any specific target organ (Data Supplement).
Verification Cohort 2 From a Prospective Multicenter Consortium
We then sought to verify the four-biomarker panel further in a second independent cohort of 180 patients from the Chronic GVHD Consortium. Patient and GVHD characteristics in the first and second cohorts are listed in Table 1 and are similar because patients had been matched accordingly. However, compared with the first cohort, fewer patients in this cohort received full-intensity conditioning and fewer patients had previously experienced aGVHD. Cohort 2 also included more patients with mild cGVHD (18% v 7% in cohort 1) and fewer patients with severe cGVHD (27% v 35% in cohort 1) according to the NIH global severity score. The median day of sample collection was earlier than in the previous cohort, at approximately 8 months, and did not differ between the cGVHD and control groups. Similar to verification cohort 1, 61% of the patients with cGVHD were being treated with corticosteroids at time of sampling, but unlike verification cohort 1, 24% of the controls also were receiving corticosteroid treatment at the time of sampling (P < .001), which was adjusted for in the analysis.
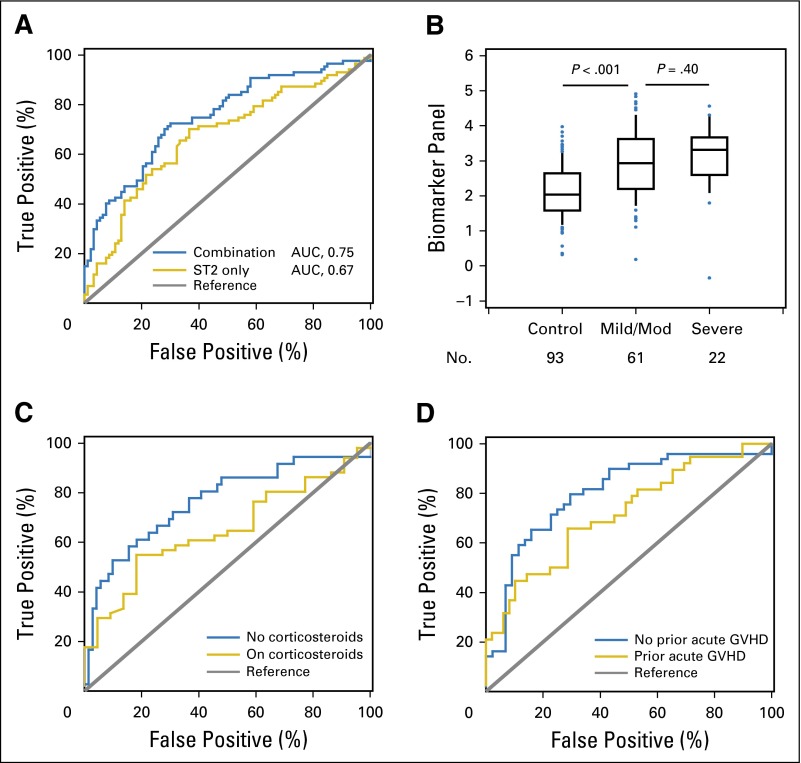
Three of the four biomarkers included in the panel (ST2, CXCL9, and MMP3) were confirmed to be associated with cGVHD in this cohort, whereas OPN was not. The biomarker panel including all four markers had an AUC of 0.75 in this cohort, whereas the AUC for ST2 was 0.67 (P < .001 and P < .001, respectively; Table 3 and Fig 2A). The combination of biomarkers with clinical covariates increased the AUC to 0.82 (P < .001; Table 3).
Table 3.
Association of Biomarker Levels With cGVHD in Verification Cohort 2 (87 patients and 93 controls)
| Model | OR* (95% CI) | P | AUC† |
|---|---|---|---|
| Single biomarkers | |||
| ST2 | 4.77 (2.0 to 11.6) | < .001 | 0.67 |
| CXCL9 | 2.69 (1.5 to 5.0) | < .001 | 0.64 |
| MMP3 | 3.36 (1.7 to 6.7) | .003 | 0.66 |
| OPN | 1.31 (0.4 to 4.6) | .66 | 0.53 |
| Combination of biomarkers | |||
| ST2 | 4.29 (1.5 to 12.2) | < .001 | 0.75 |
| CXCL9 | 3.48 (1.8 to 6.9) | ||
| MMP3 | 2.45 (1.1 to 5.5) | ||
| OPN | 0.35 (0.1 to 1.6) | ||
| Single biomarkers with clinical covariates‡ | |||
| ST2 | 3.76 (1.2 to 11.5) | .02 | 0.76 |
| CXCL9 | 3.81 (1.9 to 7.7) | < .001 | 0.79 |
| MMP3 | 1.97 (0.9 to 4.4) | .09 | 0.76 |
| OPN | 0.99 (0.2 to 4.2) | .98 | 0.74 |
| Combination of biomarkers with clinical covariates‡ | |||
| ST2 | 4.19 (1.2 to 14.4) | < .001 | 0.82 |
| CXCL9 | 4.47 (2.1 to 9.5) | ||
| MMP3 | 1.69 (0.7 to 4.0) | ||
| OPN | 0.40 (0.1 to 2.0) |
Abbreviations: AUC, area under the curve; cGVHD, chronic graft-versus-host disease; OR, odds ratio.
OR per log increase in biomarker, estimated using logistic regression.
AUC calculated from logistic regression; includes clinical covariates in model where indicated
AUC with clinical covariates only: 0.74.
Fig 2.
(A) Receiver operating characteristic (ROC) curves for the best single biomarker and the combination model in verification cohort 2. ROC curves for the best single biomarker (ST2; area under the curve [AUC], 0.67) and the combination model (AUC, 0.75) comparing patients with chronic graft-versus-host disease (cGVHD; n = 87) and time-matched controls without cGVHD (n = 93). AUC values are reported from multivariable models. (B) Association between biomarker panel and cGVHD severity in verification cohort 2. Median value of the four-biomarker panel from controls (n = 93), patients with mild to moderate (Mod) cGVHD (n = 61), and patients with severe cGVHD (n = 22) in verification cohort 2. Data are illustrated as box and whisker plots, with the whiskers indicating the 90th and 10th percentiles. P values compare controls versus patients with mild to moderate cGVHD and patients with mild to moderate cGVHD versus those with severe cGVHD, according to Wilcoxon two-sample tests. (C and D) The utility of the biomarker panel is influenced by immunosuppression therapy and prior acute graft-versus-host disease (GVHD). (C) ROC curves for the combination model in verification cohort 2 comparing patients on prednisone at time of sampling with cGVHD (n = 51) versus time-matched controls on prednisone without cGVHD (n = 22; AUC, 0.65) and comparing patients not on prednisone at time of sampling with cGVHD (n = 36) versus time-matched controls not on prednisone without cGVHD (n = 71; AUC, 0.77). (D) ROC curves for the combination model in the verification cohort 2 comparing patients with cGVHD and with prior acute GVHD (n = 38) versus time-matched controls without cGVHD but with prior acute GVHD (n = 49; AUC, 0.72) and comparing patients with cGVHD but without prior acute GVHD (n = 49) versus time-matched controls without cGVHD and without prior acute GVHD (n = 44; AUC, 0.79).
In this cohort, the biomarker panel was not significantly associated with a difference in severity of cGVHD (P = .40; Fig 2B). NRM could not be evaluated because only eight patients with cGVHD experienced NRM events during the period of observation. Previously reported biomarkers for both aGVHD and cGVHD have been shown to decrease after initiation of corticosteroid treatment,19,20 and therefore, we analyzed the effect of corticosteroid treatment and prior aGVHD on the ability of the biomarker panel to distinguish patients with cGVHD from those without cGVHD. The biomarker panel showed better performance when comparing patients who were not receiving treatment with corticosteroids (Fig 2C) and did not have prior aGVHD (Fig 2D). The difference in corticosteroid use in the controls is a result of the cohorts from which they were derived. Cohort 2 controls were a median of approximately 6.5 months after transplantation. Their corticosteroid use likely was from residual treatment of aGVHD. In contrast, cohort 1 controls were approximately 12 months after transplantation, explaining the lower corticosteroid exposure at the time of sampling. We reviewed the charts of all controls to confirm that none had active aGVHD or cGVHD at the time of sampling or within the subsequent 3 months. We also analyzed the influence of calcineurin inhibitors (CNIs) on biomarker expression in cohort 2 and found that the fold-difference in levels for patients on CNI versus those not on CNI was significantly decreased only for CXCL9 (0.67-fold, P = .03), whereas levels were not decreased for ST2, MMP3, and OPN. Because CNIs are also used to treat GVHD, we analyzed the effect of CNI treatment on the ability of the biomarker panel to distinguish patients with cGVHD from those without and found that the panel showed better performance when comparing patients who were not receiving CNIs with those who were receiving CNIs (AUC, 0.92; P < .001 v AUC, 0.71; P = .002, respectively). This same analysis is not meaningful in cohort 1 because only 27% of controls were on CNIs, whereas 67% of the patient cases were on CNIs at time of sample draw, consistent with the fact that the samples were drawn a median of 1 year after HCT. Similar to cohort 1, cohort 2 consisted entirely of patients with multiorgan involvement, and thus, the biomarker panel was not associated with involvement of any specific target organ (Data Supplement).
Day +100 Post-HCT Samples From Verification Cohort 2 and Association With Future cGVHD Occurrence
We next measured the plasma concentrations of the four biomarkers on approximately day +100 after HCT in 172 patients from cohort 2 for whom day +100 samples were available to evaluate their potential prognostic value before the diagnosis of cGVHD. The biomarker panel (particularly CXCL9 and ST2) at day +100 after transplantation was significantly associated with subsequent development of cGVHD within 3 months (median time from sample to diagnosis of 51 days; range, 14 to 91 days), with an AUC of 0.67 (P = .009; Table 4). The combination of biomarkers with clinical covariates increased the AUC to 0.79 (P = .001; Table 4). In this cohort, the AUC with the clinical covariates only was 0.72, and the AUC with aGVHD status only was 0.60 (data not shown). Interestingly, although corticosteroid use at day +100 was correlated with prior aGVHD, as expected, it was not prognostic for the future development of cGVHD (AUC, 0.52).
Table 4.
Association of Day 100 Biomarker Levels With Subsequent cGVHD (n = 172 patients)
| Model | Within 3 Months (37 events) | Within 6 Months (59 events) | Within 1 Year (81 events) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HR* (95% CI) | P | AUC† | HR* (95% CI) | P | AUC† | HR* (95% CI) | P | AUC† | |
| Single biomarkers | |||||||||
| ST2 | 1.45 (0.6 to 3.3) | .38 | 0.54 | 0.86 (0.4 to 1.7) | .66 | 0.53 | 1.04 (0.6 to 1.8) | .91 | 0.53 |
| CXCL9 | 2.88 (1.5 to 5.7) | < .001 | 0.66 | 1.44 (1.0 to 2.2) | .06 | 0.57 | 1.46 (1.0 to 2.1) | .03 | 0.59 |
| MMP3 | 0.89 (0.3 to 2.3) | .81 | 0.52 | 0.99 (0.5 to 2.1) | .98 | 0.51 | 1.15 (0.6 to 2.2) | .67 | 0.52 |
| OPN | 1.00 (0.2 to 5.4) | .99 | — | 2.03 (0.5 to 7.8) | .30 | 0.55 | 1.54 (0.5 to 4.8) | .46 | 0.52 |
| Combination of biomarkers | |||||||||
| ST2 | 1.85 (0.6 to 5.7) | .009 | 0.67 | 0.68 (0.3 to 1.6) | .27 | 0.61 | 0.87 (0.4 to 18) | .22 | 0.59 |
| CXCL9 | 2.81 (1.4 to 5.6) | 1.43 (0.9 to 2.2) | 1.45 (1.0 to 2.1) | ||||||
| MMP3 | 0.67 (0.2 to 2.2) | 1.21 (0.5 to 3.0) | 1.28(0.6 to 2.9) | ||||||
| OPN | 0.54 (0.1 to 3.5) | 2.43 (0.6 to 10.5) | 1.50 (0.4 to 5.1) | ||||||
| Single biomarkers with clinical covariates | |||||||||
| ST2 | 3.91 (1.3 to 11.6) | .01 | 0.76 | 1.66 (0.7 to 4.0) | .26 | 0.65 | 1.47 (0.7 to 3.1) | .30 | 0.61 |
| CXCL9 | 3.10 (1.5 to 6.3) | < .001 | 0.77 | 1.47 (1.0 to 2.2) | .05 | 0.66 | 1.49 (1.1 to 2.1) | .02 | 0.62 |
| MMP3 | 1.56 (0.5 to 5.4) | .49 | 0.73 | 2.30 (0.8 to 6.4) | .11 | 0.67 | 1.96 (0.8 to 4.9) | .15 | 0.62 |
| OPN | 2.66 (0.5 to 14.5) | .26 | 0.73 | 4.34 (1.1 to 17.6) | .04 | 0.68 | 2.47 (0.8 to 7.9) | .13 | 0.62 |
| Combination of biomarkers with clinical covariates | |||||||||
| ST2 | 3.22 (0.9 to 11.8) | .001 | 0.79‡ | 1.02 (0.4 to 2.7) | .06 | 0.70‡ | 1.07 (0.5 to 2.4) | .05 | 0.64‡ |
| CXCL9 | 2.92 (1.4 to 6.1) | 1.46 (1.0 to 2.2) | 1.49 (1.1 to 2.1) | ||||||
| MMP3 | 1.19 (0.3 to 4.3) | 1.89 (0.6 to 5.7) | 1.79 (0.7 to 4.8) | ||||||
| OPN | 0.68 (0.1 to 4.7) | 3.06 (0.7 to 13.8) | 1.88 (0.5 to 6.7) | ||||||
Abbreviations: AUC, area under the curve; cGVHD, chronic graft-versus-host disease; HR, hazard ratio.
HR per log increase in biomarker, estimated using Cox regression.
AUC calculated from logistic regression as described in Patients and Methods.
AUC with clinical covariates only: 0.72 within 3 months, 0.65 within 6 months, and 0.60 within 1 year.
DISCUSSION
The discovery of valid and reproducible biomarkers for cGVHD remains a significant challenge. Compared with aGVHD, cGVHD is clinically more heterogeneous and can involve many more target organs, often simultaneously. First, a major strength of this study was our ability to reproduce a correlation of the four-biomarker panel with cGVHD in a second verification cohort that included samples from eight different sites, which meets the current preferred requirement for the identification of biomarkers according to the 2014 NIH biomarker consensus.9 Unfortunately, the association of biomarkers with cGVHD severity and NRM in the first verification cohort was not seen in the second verification cohort, possibly because of the number of controls on corticosteroids at the time of sampling in cohort 2 as compared with that in cohort 1, which could mask the biomarker levels, and the low numbers of NRM events, respectively. Second, the fact that a biomarker panel at day +100 after HCT was associated with subsequent cGVHD and remained elevated at the time of onset suggests that serial measurements from day +100 after HCT and beyond may identify high-risk patients for earlier intervention. For example, patients at high risk for cGVHD could be monitored more closely or even receive pre-emptive treatment designed to avert onset or at least minimize cGVHD severity.
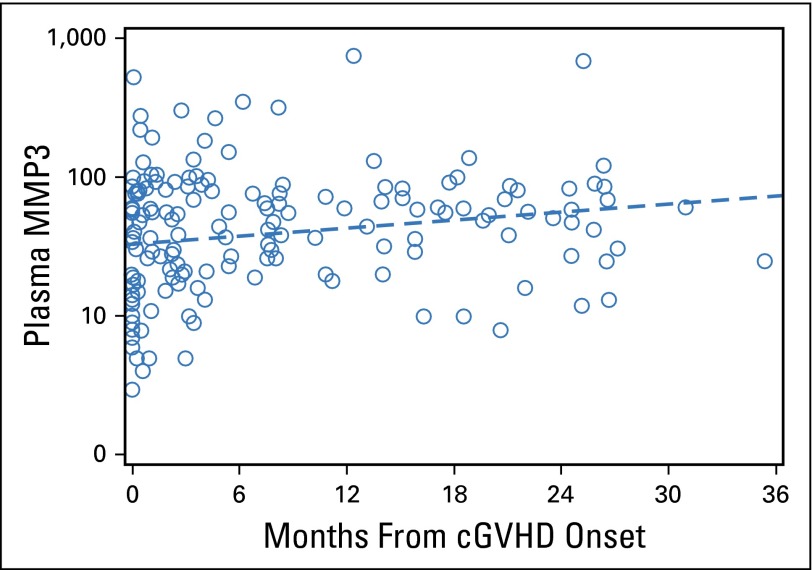
In addition, our discovery approach has highlighted some interesting biologic pathways that may represent novel therapeutic avenues relevant to the three proteins found to be increased in both verification cohorts. CXCL9 is a T-helper type 1 chemokine that has been shown to attract CXCR3+CD4+ T cells in target organs and can be detected in the blood.18,21,22 Some therapies used for aGVHD, such as bortezomib, inhibit T-cell chemotactic movements, decrease CXCR3 expression and CXCL9 secretion,23 and may prevent GVHD.24,25 Other CXCL9 and CXCR3 inhibitors are under development. Soluble ST2 is secreted by intestinal cells during aGVHD in experimental models, and ST2 blockade reduces the numbers of these cells.26,27 Further experiments are needed to determine the effects of ST2 blockade in cGVHD models. MMPs are prominent contributors to microenvironmental signals because these proteolytic enzymes degrade structural components of the extracellular matrix, permitting tissue remodeling.28 MMP3, in particular, has been shown to promote the epithelial-mesenchymal transition, potentially leading to tissue fibrosis.29 Opposite to the three other markers that were found at high levels early in the cGVHD course, MMP3 plasma concentrations seemed to increase over time from cGVHD onset (Appendix Fig A2, online only), potentially reflecting matrix remodeling in response to fibrosis. We could not explore the differences between lichenoid and sclerotic forms of cGVHD because the NIH skin score combines these two types of manifestation, and thus, a clear distinction is not possible. Notably, no other MMPs were found to be differentially regulated in this proteomics experiment. MMP3 inhibitors are expected to be available in the near future.30
Several limitations to the current study should be noted. First, although our approach used tandem MS/MS and can be considered unbiased, tandem MS/MS has a technical limitation in the detection of low-abundance proteins, particularly if trypsin digestion is applied at the beginning of the workflow (before fractionation), as in our iTRAQ experiments.31 This was not true for our previous approach using the Intact Protein Analysis System with trypsin digestion occurring after extensive fractionation.12 Second, the timing of sample acquisition for biomarker assessment is critical. Once immunosuppression has been initiated, the biomarker pattern may change, as has been observed with B-cell activating factor plasma concentrations in patients treated with corticosteroids.19 Newly diagnosed and established patients with cGVHD were studied together, although the pathologic processes culminating in a new diagnosis may differ from those present in established disease. Similarly, all phenotypes of cGVHD were grouped together into cGVHD, although the organ involvement and prognosis of cGVHD are heterogeneous. If different cGVHD phenotypes reflect different underlying biology, this aggregation may impede identification of important biomarkers. The biomarker panel performed better when applied to similar groups of patients and controls who were not receiving corticosteroid treatment at the time of sample collection. Third, the relevance of the AUCs with clinical covariates in the verification cohort 2 should be interpreted with caution because the samples were selected to be matched for some covariates. The same consideration is applicable to the day +100 analysis that aimed to determine whether the biomarker panel could be used to anticipate the development of cGVHD before the clinical onset of the disease. Specifically, as a result of our case-control design, we cannot estimate the positive predictive value of the biomarker factors as prognostic factors for cGVHD because they would be used in an actual clinical setting of unselected patients. Thus, this biomarker panel should be evaluated in a prospective study with serial collection of samples ideally starting at day +100 after HCT or even earlier (day +80) before it is accepted for risk stratification in clinical trials.
We conclude that the combination of plasma concentrations of four proteins might represent a biomarker panel for noninvasive identification of cGVHD as verified in two independent cohorts. The biomarker panel was associated with cGVHD severity and prediction of NRM in one cohort. Importantly, when measured as early as day +100 after HCT, the biomarker panel in association with key clinical covariates was a good predictor of the risk of cGVHD occurrence in the next 3 months. Future directions should include prospective and serial evaluations of the panel to define its clinical use as a prognostic biomarker panel in an unselected population.
Supplementary Material
Acknowledgment
We thank the clinicians at all of the institutions who participated in accrual of samples; the Fred Hutchinson Cancer Research Center data managers, particularly Kate Chilson, for excellent management of the database and biobank; and the members of the Paczesny laboratory.
Appendix
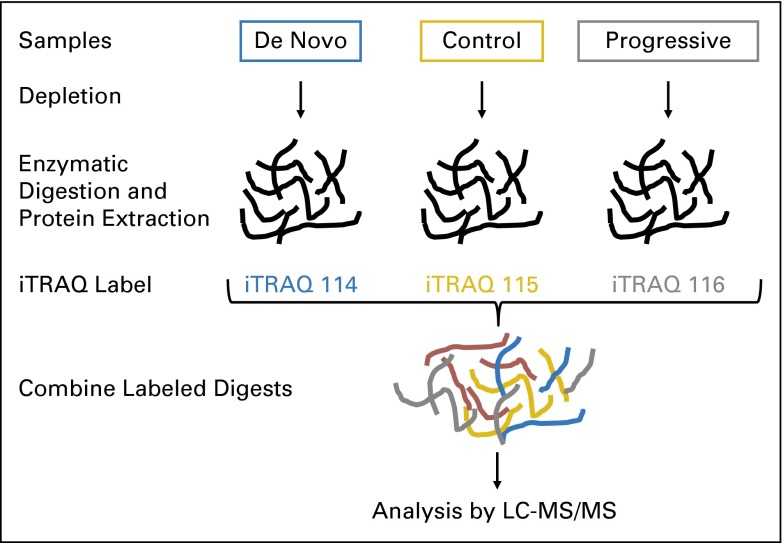
Fig A1.
Isobaric tags for relative and absolute quantification (iTRAQ) proteomics workflow in the chronic graft-versus-host disease (cGVHD) study. Three pools of plasma were compared in the same proteomic experiment. Pool 1 contained plasma from 17 patients with de novo cGVHD available at the onset of clinical symptoms, pool 2 contained plasma from 18 patients without cGVHD (collected at similar time points as cGVHD samples), and pool 3 contained plasma from 18 patients with progressive cGVHD. The three pooled plasmas were then individually immunodepleted of the 20 common hyperabundant proteins followed by enzymatic digestion with trypsin and protein extraction. Each pool was then labeled for quantification by iTRAQ (32) as follows: de novo cGVHD with iTRAQ 114, control with iTRAQ 115, and progressive cGVHD with iTRAQ 116. All the pools were then combined for analysis by liquid chromatography–tandem mass spectrometry (LC-MS/MS).
Fig A2.
Plasma MMP3 concentrations increase from time of chronic graft-versus-host disease (cGVHD) onset in patients with cGVHD in verification cohort 1 (n = 178). Dashed line is estimated fit from linear regression model (P = .009).
Footnotes
Supported by the National Marrow Donor Program through the Amy Strelzer Manasevit Scholar Grant No. 200513 (S.P.) and National Institutes of Health (NIH) Grants No. R01CA174667 (S.P.), R01CA118953 (S.J.L.), and U54CA163438 (S.J.L.). The Chronic Graft-Versus-Host Disease Consortium (Grant No. U54 CA163438) is part of the National Center for Advancing Translational Science (NCATS) Rare Diseases Clinical Research Network (RDCRN). RDCRN is an initiative of the Office of Rare Disease Research, NCATS, funded through a collaboration between NCATS and the National Cancer Institute. The Fred Hutchinson Cancer Research Center Proteomics Facility is partially funded by Cancer Center Support Grant No. P30 CA015704 from the NIH.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Authors’ disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
AUTHOR CONTRIBUTIONS
Conception and design: Stephanie J. Lee, Sophie Paczesny
Financial support: Stephanie J. Lee, Sophie Paczesny
Administrative support: Stephanie J. Lee, Sophie Paczesny
Provision of study materials or patients: Paul J. Martin, Mary E. Flowers, John A. Hansen, Mukta Arora, Corey Cutler, Madan Jagasia, Joseph Pidala, Betty K. Hamilton, George L. Chen, Iskra Pusic, Stephanie J. Lee, Sophie Paczesny
Collection and assembly of data: Jeffrey Yu, Kushi Kushekhar, Mohammad Abu Zaid, Qing Zhang, Philip R. Gafken, Yuko Ogata, Stephanie J. Lee, Sophie Paczesny
Data analysis and interpretation: Jeffrey Yu, Barry E. Storer, Qing Zhang, Philip R. Gafken, Yuko Ogata, Paul J. Martin, Mary E. Flowers, John A. Hansen, Mukta Arora, Corey Cutler, Madan Jagasia, Joseph Pidala, Betty K. Hamilton, George L. Chen, Iskra Pusic, Stephanie J. Lee, Sophie Paczesny
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Biomarker Panel for Chronic Graft-Versus-Host Disease
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Jeffrey Yu
No relationship to disclose
Barry E. Storer
No relationship to disclose
Kushi Kushekhar
No relationship to disclose
Mohammad Abu Zaid
No relationship to disclose
Qing Zhang
No relationship to disclose
Philip R. Gafken
No relationship to disclose
Yuko Ogata
No relationship to disclose
Paul J. Martin
Consulting or Advisory Role: Pfizer, Enlivex Therapeutics, Janssen
Research Funding: Astellas Pharma (Inst), Neovii Biotech (Inst)
Mary E. Flowers
No relationship to disclose
John A. Hansen
No relationship to disclose
Mukta Arora
Consulting or Advisory Role: Takeda, Neovii Biotech
Corey Cutler
No relationship to disclose
Madan Jagasia
Consulting or Advisory Role: Gilead Sciences, Therakos
Research Funding: Janssen Pharmaceuticals
Joseph Pidala
No relationship to disclose
Betty K. Hamilton
No relationship to disclose
George L. Chen
No relationship to disclose
Iskra Pusic
No relationship to disclose
Stephanie J. Lee
Honoraria: Therakos
Consulting or Advisory Role: Bristol-Myers Squibb, Kadmon, EMD Serono (I)
Research Funding: Millennium (Inst), Amgen (Inst), Bristol-Myers Squibb (I)
Travel, Accommodations, Expenses: Therakos, EMD Serono (I)
Sophie Paczesny
Patents, Royalties, Other Intellectual Property: Inventor on a patent on “Methods of detection of graft-versus-host disease” (US- 13/573,766) (Inst)
REFERENCES
- 1.Flowers ME, Parker PM, Johnston LJ, et al. Comparison of chronic graft-versus-host disease after transplantation of peripheral blood stem cells versus bone marrow in allogeneic recipients: Long-term follow-up of a randomized trial. Blood. 2002;100:415–419. doi: 10.1182/blood-2002-01-0011. [DOI] [PubMed] [Google Scholar]
- 2.Zecca M, Prete A, Rondelli R, et al. Chronic graft-versus-host disease in children: Incidence, risk factors, and impact on outcome. Blood. 2002;100:1192–1200. doi: 10.1182/blood-2001-11-0059. [DOI] [PubMed] [Google Scholar]
- 3.Anasetti C, Logan BR, Lee SJ, et al. Peripheral-blood stem cells versus bone marrow from unrelated donors. N Engl J Med. 2012;367:1487–1496. doi: 10.1056/NEJMoa1203517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Socié G, Stone JV, Wingard JR, et al. Long-term survival and late deaths after allogeneic bone marrow transplantation. N Engl J Med. 1999;341:14–21. doi: 10.1056/NEJM199907013410103. [DOI] [PubMed] [Google Scholar]
- 5.Lee SJ, Klein JP, Barrett AJ, et al. Severity of chronic graft-versus-host disease: Association with treatment-related mortality and relapse. Blood. 2002;100:406–414. doi: 10.1182/blood.v100.2.406. [DOI] [PubMed] [Google Scholar]
- 6.Jagasia MH, Savani BN, Stricklin G, et al. Classic and overlap chronic graft-versus-host disease (cGVHD) is associated with superior outcome after extracorporeal photopheresis (ECP) Biol Blood Marrow Transplant. 2009;15:1288–1295. doi: 10.1016/j.bbmt.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SJ, Wolff D, Kitko C, et al. Measuring therapeutic response in chronic graft-versus-host disease: National Institutes of Health consensus development project on criteria for clinical trials in chronic graft-versus-host disease: IV. The 2014 Response Criteria Working Group report. Biol Blood Marrow Transplant. 2015;21:984–999. doi: 10.1016/j.bbmt.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavletic SZ, Vogelsang GB, Lee SJ. 2014 National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease: Preface to the series. Biol Blood Marrow Transplant. 2015;21:387–388. doi: 10.1016/j.bbmt.2014.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Paczesny S, Hakim FT, Pidala J, et al. National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease: III. The 2014 Biomarker Working Group report. Biol Blood Marrow Transplant. 2015;21:780–792. doi: 10.1016/j.bbmt.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin PJ, Lee SJ, Przepiorka D, et al. National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease: VI. The 2014 Clinical Trial Design Working Group report. Biol Blood Marrow Transplant. 2015;21:1343–1359. doi: 10.1016/j.bbmt.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paczesny S, Krijanovski OI, Braun TM, et al. A biomarker panel for acute graft-versus-host disease. Blood. 2009;113:273–278. doi: 10.1182/blood-2008-07-167098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paczesny S, Braun TM, Levine JE, et al. Elafin is a biomarker of graft-versus-host disease of the skin. Sci Transl Med. 2010;2:13ra2. doi: 10.1126/scitranslmed.3000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrara JL, Harris AC, Greenson JK, et al. Regenerating islet-derived 3-alpha is a biomarker of gastrointestinal graft-versus-host disease. Blood. 2011;118:6702–6708. doi: 10.1182/blood-2011-08-375006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vander Lugt MT, Braun TM, Hanash S, et al. ST2 as a marker for risk of therapy-resistant graft-versus-host disease and death. N Engl J Med. 2013;369:529–539. doi: 10.1056/NEJMoa1213299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pidala J, Sarwal M, Roedder S, et al. Biologic markers of chronic GVHD. Bone Marrow Transplant. 2014;49:324–331. doi: 10.1038/bmt.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chronic GVHD Consortium Rationale and design of the chronic GVHD cohort study: Improving outcomes assessment in chronic GVHD. Biol Blood Marrow Transplant. 2011;17:1114–1120. doi: 10.1016/j.bbmt.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arora M, Cutler CS, Jagasia MH, et al: Incidence, risk factors, and prognosis of late immune-mediated disorders after allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant 21:S54, 2015. [Google Scholar]
- 18.Kitko CL, Levine JE, Storer BE, et al. Plasma CXCL9 elevations correlate with chronic GVHD diagnosis. Blood. 2014;123:786–793. doi: 10.1182/blood-2013-08-520072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarantopoulos S, Stevenson KE, Kim HT, et al. High levels of B-cell activating factor in patients with active chronic graft-versus-host disease. Clin Cancer Res. 2007;13:6107–6114. doi: 10.1158/1078-0432.CCR-07-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levine JE, Paczesny S, Mineishi S, et al. Etanercept plus methylprednisolone as initial therapy for acute graft-versus-host disease. Blood. 2008;111:2470–2475. doi: 10.1182/blood-2007-09-112987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Croudace JE, Inman CF, Abbotts BE, et al. Chemokine-mediated tissue recruitment of CXCR3+ CD4+ T cells plays a major role in the pathogenesis of chronic GVHD. Blood. 2012;120:4246–4255. doi: 10.1182/blood-2012-02-413260. [DOI] [PubMed] [Google Scholar]
- 22.Hakim F, Memon S. Elevated expression of interferon-induced genes and damage associated molecular pattern receptor genes in chronic graft versus host disease. Biol Blood Marrow Transplant. 2014;20:S226. [Google Scholar]
- 23.Liu W, Ren HY, Dong YJ, et al. Bortezomib regulates the chemotactic characteristics of T cells through downregulation of CXCR3/CXCL9 expression and induction of apoptosis. Int J Hematol. 2012;96:764–772. doi: 10.1007/s12185-012-1195-6. [DOI] [PubMed] [Google Scholar]
- 24.Koreth J, Stevenson KE, Kim HT, et al. Bortezomib, tacrolimus, and methotrexate for prophylaxis of graft-versus-host disease after reduced-intensity conditioning allogeneic stem cell transplantation from HLA-mismatched unrelated donors. Blood. 2009;114:3956–3959. doi: 10.1182/blood-2009-07-231092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koreth J, Stevenson KE, Kim HT, et al. Bortezomib-based graft-versus-host disease prophylaxis in HLA-mismatched unrelated donor transplantation. J Clin Oncol. 2012;30:3202–3208. doi: 10.1200/JCO.2012.42.0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Ramadan AM, Griesenauer B, et al. ST2 blockade reduces sST2-producing T cells while maintaining protective mST2-expressing T cells during graft-versus-host disease. Sci Transl Med. 2015;7:308ra160. doi: 10.1126/scitranslmed.aab0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reichenbach DK, Schwarze V, Matta BM, et al. The IL-33/ST2 axis augments effector T-cell responses during acute GVHD. Blood. 2015;125:3183–3192. doi: 10.1182/blood-2014-10-606830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radisky DC, Levy DD, Littlepage LE, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436:123–127. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vandenbroucke RE, Libert C. Is there new hope for therapeutic matrix metalloproteinase inhibition? Nat Rev Drug Discov. 2014;13:904–927. doi: 10.1038/nrd4390. [DOI] [PubMed] [Google Scholar]
- 31.Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.