Abstract
Purpose
The goal of this study was to analyze the association of copy number gain of 1q in favorable-histology Wilms tumors (FHWTs) with event-free survival (EFS) and overall survival (OS) within each tumor stage and with 1p and 16q copy number loss and/or loss of heterozygosity.
Methods
Unilateral FHWTs from 1,114 patients enrolled in National Wilms Tumor Study-5 that were informative for 1p and 16q microsatellite markers (previously determined) and informative for 1q gain, 1p loss, and 16q loss using multiplex ligation-dependent probe amplification were analyzed.
Results
Eight-year EFS was 86% (95% CI, 84% to 88%) for the entire cohort. Of 1,114 patients, 317 tumors (28%) displayed 1q gain. Eight-year EFS was 77% for those with 1q gain and 90% for those lacking 1q gain (P < .001). Eight-year OS was 88% for those with 1q gain and 96% for those lacking 1q gain (P < .001). Within each disease stage, 1q gain was associated with inferior EFS (stage I, 85% v 95%; P = .0052; stage II, 81% v 87%; P = .0775; stage III, 79% v 89%; P = .01; stage IV, 64% v 91%; P = .001). OS was significantly inferior in patients with stage I (P < .0015) and stage IV disease (P = .011). With multivariable analysis, 1q gain was associated with an increased relative risk of relapse of 2.4 (P < .001), whereas 1p loss was not, despite significance on univariable analysis.
Conclusion
Gain of 1q is associated with inferior survival in unilateral FHWTs and may be used to guide risk stratification in future studies.
INTRODUCTION
The current overall survival rates of all patients with Wilms tumor (WT) is approximately 90%. Nevertheless, certain subsets of patients have survival estimates well below this figure, including those with favorable histology who relapse, those with anaplastic histology, and those with bilateral disease. Moreover, WT therapy comes at a cost; 24% of survivors are affected by severe chronic health conditions, including cardiac and pulmonary toxicities, infertility, and secondary malignancies.1 Further improvement in clinical outcomes can be achieved by identifying novel biologic prognostic factors that would improve our ability to better tailor therapy.
Clinical prognostic factors currently used to stratify therapy for WT include tumor histology, disease stage, patient age, tumor weight, and completeness of lung nodule response after 6 weeks of chemotherapy.2 The only molecular prognostic factor that has been used in clinical studies for the risk stratification for WT is the combined loss of heterozygosity (LOH) of chromosomes 1p and 16q. A study evaluating patients enrolled in National Wilms Tumor Study (NWTS)-3 and NWTS-4 found that LOH at either 1p or 16q correlated with an adverse prognosis.3 Further studies from national groups in the United Kingdom, Germany, and Italy confirmed this association.4-8 NWTS-5 prospectively demonstrated that LOH at either 1p or 16q showed a trend toward an increased risk of relapse or death; post hoc analysis revealed that the greatest effect was seen with combined LOH at both loci.9 Although LOH at 1p and 16q is specific for predicting relapse, it is not sensitive, because only 4.6% (76/1,656) of patients with favorable-histology WTs (FHWTs) had tumors with combined LOH for 1p and 16q, and combined LOH for 1p and 16q was present in only 9.4% (20/213) relapses in the NWTS-5 cohort.9
Gain of chromosome 1q is one of the most common cytogenetic abnormalities in WT, observed in approximately 30% of tumors.10 Retrospective studies involving convenience sample sets demonstrated an association between 1q copy number (CN) or expression gain and tumor recurrence.11-16 The Children’s Cancer and Leukaemia Group confirmed that 1q gain was associated with inferior event-free survival (EFS) and overall survival (OS) independent of high tumor stage or anaplasia in 331 patients.17 An analysis of 212 FHWTs from patients treated in NWTS-4 demonstrated an 8-year EFS of 76% and 93% for patients with and without 1q gain, respectively (P = .0024).18 Moreover, 8-year OS was 89% (95% CI, 78% to 95%) with 1q gain versus 98% (95% CI, 94% to 99%) without 1q gain (P = .0075). The studies mentioned all lacked sufficient power to detect survival differences within each disease stage. The goal of the current study was to allow for the assessment of the prognostic significance of 1q gain in FHWTs in relation to other established prognostic factors, including tumor stage and LOH at 1p and 16q.
METHODS
Clinical Samples
Patients were registered prospectively in NWTS-5 after providing informed consent (accrual August 1995 to June 2002). Requirements for inclusion were stage I to IV unilateral WT, an assignment to receive chemotherapy, favorable histology confirmed by central pathology review, full eligibility and clinical follow-up in NWTS-5, availability of DNA from pretherapy tumor samples, and evaluable 1p and 16q LOH data obtained prospectively by microsatellite analysis during NWTS-5. All patients received either vincristine and dactinomycin or vincristine, dactinomycin, and doxorubicin, with or without radiation.19 Patients with stage 1 very low risk WT (nephrectomy < 550 g, age < 24 months) who did not receive initial adjuvant chemotherapy were not included.
Multiplex Ligation-Dependent Probe Amplification
Multiplex ligation-dependent probe amplification (MLPA) was performed as previously described using a synthetic probe mixture directed at four loci on each 1p, 16q, and 1q, and with nine control probes.18,20 For quality control purposes, 120 tumors were analyzed in duplicate at a separate time using the same synthetic probe panel. In addition, 64 tumors were analyzed using the P380 Wilms Tumor probemix (MRC Holland, Amsterdam, the Netherlands), which includes a minimum of five probesets for each 1q, 1p, and 16q. The calls of gain or loss were made for each locus without knowledge of the calls made from the original probesets and using the same criteria. For all patients examined, the calls were the same for each locus. Lastly, 18 tumors were assessed by the Genome Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA) and confirmed to be concordant with the MLPA calls for the presence of 1q gain.21
Analysis
After separation by capillary electrophoresis, peaks corresponding to each probe were identified by GeneMapper analysis (Applied Biosystems, Foster City, CA). Samples in which the smallest peak was < 100 relative fluorescent intensity units were not analyzed. The raw peak area for each probe in each sample was divided by the average raw peak area for all probes in that sample. This normalized peak area was then divided by the normalized peak area of the reference samples. Control probes with a coefficient of variation > 20% were removed. Only samples with at least three control probes remaining were considered evaluable and included in the study. Test probes > 1.25 were considered gained, and those < 0.75 were considered lost. Gain or loss for a chromosomal region was scored if at least two markers were gained or lost, respectively. Scoring was performed without knowledge of outcome or of 1p and 16q LOH status.
Statistical Analysis
The two end points were 8-year EFS and OS. EFS and OS curves were estimated using the Kaplan-Meier method22 and compared using the log-rank test.23 Relative risks (RRs) were calculated using the Cox proportional hazards model.24 Tests of correlation of 1q gain status and patient or disease characteristics were performed using the standard χ2 test for contingency tables.
RESULTS
Overall Cohort
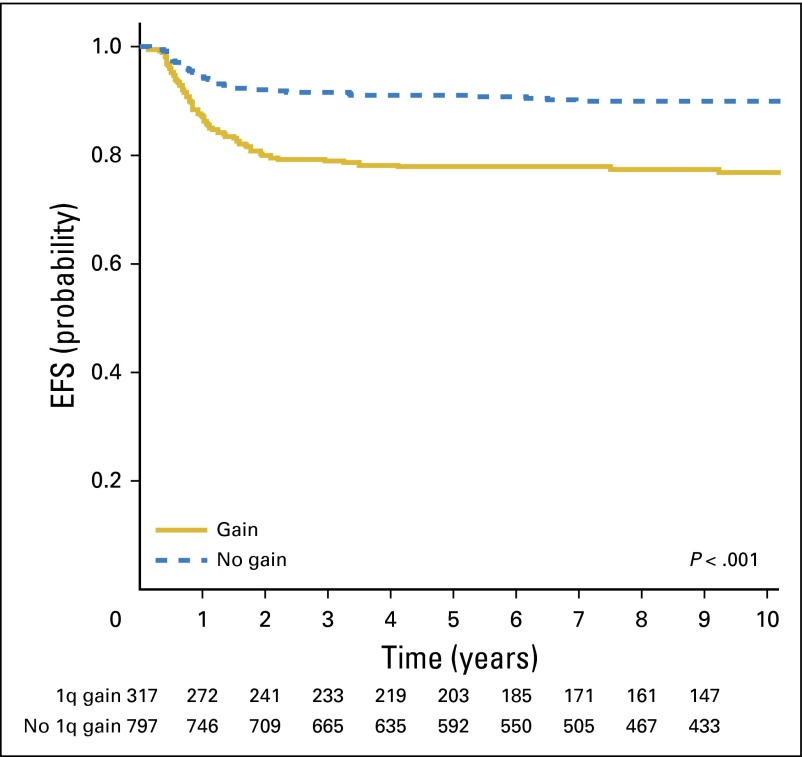
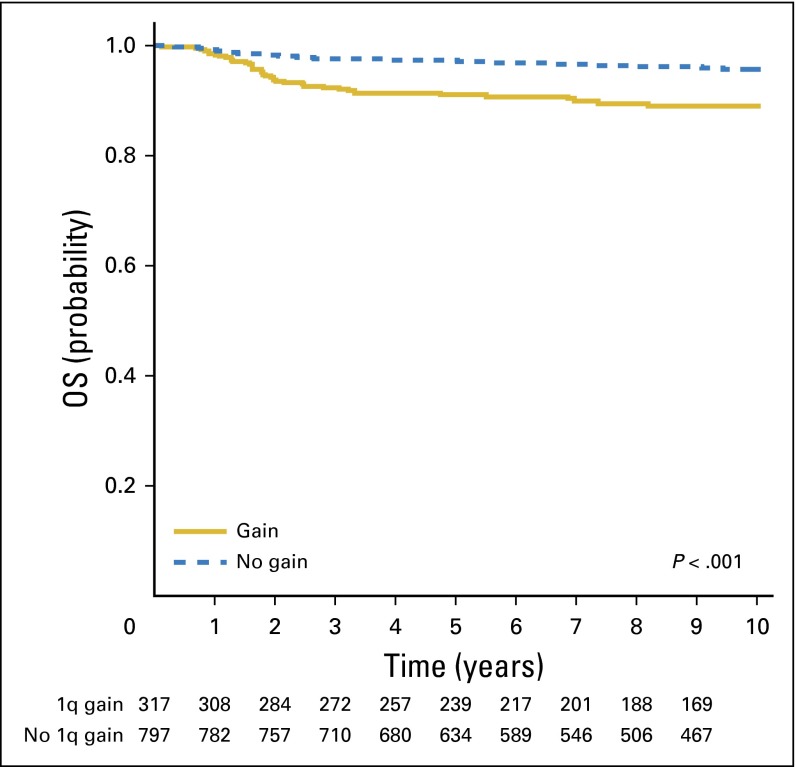
A total of 1,114 patients with FHWTs met the inclusion criteria and passed the quality control steps resulting in evaluable MLPA data. The stage distribution for patients with stages II, III, and IV disease in these 1,114 patients (Table 1) was similar to that of the 1,484 NWTS-5 patients previously reported9; in the current study, some patients with stage I < 24 months of age with tumors < 550 g who received adjuvant chemotherapy after closure of the very low risk therapeutic arm were included in the analysis. The 8-year EFS and OS estimates for all 1,114 patients were 86% (95% CI, 84% to 88%) and 94% (95% CI, 92% to 95%), respectively. A gain of 1q was present in 317 of 1,114 tumors (28%). Patients with gain of 1q were older (median age, 51.5 months) than were those without gain of 1q (median age, 36.5 months; P < .001 [Wilcoxon test]). The 8-year EFS estimate was 77% (95% CI, 72% to 81%) for those with 1q gain and 90% (95% CI, 88% to 92%) for those without 1q gain (P < .001; Fig 1). The 8-year OS estimate was 88% (95% CI, 83% to 91%) for those with 1q gain and 96% (95% CI, 94% to 97%) for those without 1q gain (P < .001; Fig 2). There was no difference in the pattern of relapse between those tumors with and without 1q gain (lung only, 25/61 [41%] in patients with 1q gain and 34/65 [52%] in those lacking 1q gain; operative bed, 16% and 18% with and without 1q gain, respectively; abdomen/pelvis, 15% and 20% with and without 1q gain, respectively; other sites, 28% and 9% with and without 1q gain, respectively).
Table 1.
Eight-Year EFS and OS Stratified by Disease Stage and 1q Status
| Disease Stage | No. (% of stage group) | 8-Year EFS (95% CI) | P (EFS) | 8-Year OS (95% CI) | P (OS) |
|---|---|---|---|---|---|
| Stage I (n = 241, 21.6%) | |||||
| 1q gain | 46 (20) | 85 (72 to 98) | 90 (80 to 100) | ||
| No 1q gain | 195 (80) | 95 (91 to 99) | .0052 | 98 (96 to 100) | .0015 |
| Stage II (n = 382, 34.3%) | |||||
| 1q gain | 98 (26) | 81 (71 to 91) | 94 (87 to 100) | ||
| No 1q gain | 284 (74) | 87 (83 to 92) | .0775 | 97 (94 to 99) | .1917 |
| Stage III (n = 358, 32.1%) | |||||
| 1q gain | 115 (32) | 79 (70 to 87) | 91 (85 to 97) | ||
| No 1q gain | 243 (68) | 89 (84 to 94) | .0100 | 95 (91 to 98) | .3335 |
| Stage IV (n =133, 11.9%) | |||||
| 1q gain | 58 (44) | 64 (48 to 79) | 74 (60 to 88) | ||
| No 1q gain | 75 (56) | 91 (83 to 99) | .0004 | 92 (84 to 99) | .0110 |
Abbreviations: EFS, event-free survival; OS, overall survival.
Fig 1.
Event-free survival (EFS) stratified for 1q gain.
Fig 2.
Overall survival (OS) stratified for 1q gain.
For the correlation between 1q gain and histologic pattern, prechemotherapy tumors were classified during central pathologic review as epithelial, stromal, or blastemal if more than two thirds of the tumor demonstrated this histology. The remainder were classified as mixed. There was no significant difference in the distribution of histologic patterns between tumors with 1q gain (42% blastemal, 6% epithelial, 50% mixed, and 2% stromal) and tumors lacking 1q gain (35% blastemal, 8% epithelial, 55% mixed, and 2% stromal).
Outcomes by Disease Stage
Stage distribution was as follows: stage I, 241 (22%); stage II, 382 (35%); stage III, 358 (32%); and stage IV, 133 (12%). Patients with gain of 1q were less likely to have stage I disease (14.5% v 24.5%) and more likely to have stage IV disease (18.3% v 9.4%) than were those without gain of 1q (P < .001). Whereas the frequency of 1q gain for the entire cohort was 28%, the frequency of 1q gain according to stage was as follows: stage I, 20%; stage II, 26%; stage III, 32%; and stage IV, 44%. Gain of 1q was associated with significant inferior EFS in patients with stages I, III, and IV disease and with clearly inferior OS in patients with stages I and IV disease (Table 1). For patients with stage III disease, there was no significant difference in the distribution of 1q gain on the basis of the reason for stage III disease. For patients with 1q gain, 35% had a tumor in regional lymph nodes, 22% had a tumor at the surgical margin, 13% had both a positive margin and lymph node, 9% received a biopsy only before therapy, 16% had tumor rupture/spill, and 3% had either piecemeal or incomplete excision. For patients lacking 1q gain, the distribution was 30%, 27%, 13%, 12%, 13%, and 3%, respectively.
CN Loss for 1p and/or 16q
CN loss of 1p was identified in 103 tumors (9%) and was associated with decreased 8-year EFS (P = .01). CN loss of 16q was seen in 146 patients (13%) and was associated with decreased 8-year EFS (P = .008). There were no significant differences in the distribution of stage by 1p CN loss (P = .20) or by 16q CN loss (P = .37). With univariable analysis, the presence of either 1p loss or 16q loss was prognostic (P = .007), with the lowest EFS seen in tumors with both 1p and 16q CN loss. With multivariable analysis (Cox proportional hazards model), 1q gain was prognostic for EFS (RR, 2.4; P < .001), but after adjusting for the effect of 1q gain, neither 1p loss nor 16q loss were significantly associated with EFS. When multivariable analysis was repeated to include disease stage, 1q gain remained significantly associated with EFS (RR, 2.2; P < .001). Multivariable analysis assumes no interaction between the variables analyzed. However, among those patients with gain of 1q, loss of 1p was much more common (73/317, 23%) than in patients without gain of 1q (30/797, 4%; P < .001). Likewise, among those tumors with gain of 1q, loss of 16q was much more common (82/317, 26%) than in tumors without gain of 1q (64/797, 8%; P < .001). Therefore, the outcome of patients with and without 1q gain in their tumors was separately analyzed for impact of 1p and 16q loss. This demonstrated no significantly increased risk of relapse associated with 1p and 16q in the group of patients with 1q gain. However, in the group of patients lacking 1q gain, the 8-year EFS was 91% for those without 1p and/or 16q loss and 84% for those with 1p and/or 16q loss (P = .03; Table 2).
Table 2.
Eight-Year EFS Stratified by 1q Status and LOH 1p/16q Status
| 1q Status | 1p or 16q Call | No. of Patients | 8-Year EFS (95% CI) |
|---|---|---|---|
| No gain | No loss | 715 | 91 (88 to 93) |
| No gain | Loss | 82 | 84 (74 to 93) |
| Gain | No loss | 174 | 77 (69 to 84) |
| Gain | Loss | 143 | 78 (70 to 87) |
Abbreviation: EFS, event-free survival; LOH, loss of heterozygosity.
Correlation Between 1p and 16q CN and LOH
Because MLPA was performed on the same extracted DNA sample on which the original LOH analysis was originally performed, we were uniquely able to compare CN loss with LOH. For 1p, 1,070 of 1,114 (96%) CN calls correlated with LOH calls; for 16q, 1,067 of 1,114 (96%) CN calls correlated with LOH calls (Table 3). Copy-neutral LOH was identified for 1p and 16q in 25 (2%) and 32 (3%) tumors, respectively. Failure to detect LOH in tumors with CN loss was identified for 1p and 16q in 19 (1.7%) and 15 (1.3%) tumors, respectively. This may be attributable to the continuous improvement in LOH analysis methodology throughout the course of NWTS-5.
Table 3.
Correlation Between Copy Number Loss and LOH for 1p and 16q
| Copy Loss | No Copy Loss | Total | |
|---|---|---|---|
| 1p evaluation | |||
| 1p LOH, yes | 84 | 25 | 109 |
| 1p LOH, no | 19 | 986 | 1,005 |
| Total | 103 | 1,011 | 1,114 |
| 16q evaluation | |||
| 16q LOH, yes | 131 | 32 | 163 |
| 16q LOH, no | 15 | 936 | 951 |
| Total | 146 | 968 | 1,114 |
Abbreviation: LOH, loss of heterozygosity.
DISCUSSION
This study confirms the association of 1q gain with inferior EFS within each disease stage of FHWT. The presence of 1q gain in 28% of FHWTs creates the potential to influence risk stratification of a substantial number of patients. By contrast, LOH at both chromosomes 1p and 16q, used for risk stratification in the most recently completed Children’s Oncology Group (COG) renal tumor studies, was observed in only 5% of patients. Gain of 1q was more prevalent within tumors of higher stage disease, suggesting that this cytogenetic change is associated with tumor aggressiveness and metastatic potential. It is noteworthy that 1q gain has been linked with adverse outcome in other tumor types, including neuroblastoma, pediatric ependymoma, medulloblastoma, Ewing sarcoma, and several adult tumors.25-30
The relevant genes on 1q that confer adverse prognosis remain to be determined, despite numerous studies and a variety of applied technologies. The vast majority of tumors with 1q gain show gain of the entire long arm. Detailed discussions of the cytogenetic mechanisms responsible for 1q gain in many WTs (translocations and 1q isochromosome formation) have been previously reviewed.10,18
Following is a review of our findings by stage:
Stage I: Patients with stage I FHWTs and 1q gain had significantly lower EFS (85% v 95%) and OS (90% v 98%) compared with patients without 1q gain. The lower survival rates observed with 1q gain may provide an opportunity to augment therapy to avoid relapses and cancer-related deaths in the future. However, 85% of patients with 1q gain did not relapse with the current two-drug therapy; therefore, increasing treatment of this group would subject 16% of patients with stage I disease (19% with 1q gain × 85% without relapse) to unnecessary therapy. If therapy augmentation were to be studied, it would be desirable to use anti-WT agents without known long-term toxicities, such as the camptothecins.29 Another potential approach would be to augment salvage therapy for the 15% with 1q gain who experience recurrence. This latter approach should likely be undertaken regardless of initial approach because the salvage therapy used in NWTS-5 was effective for only 33% of this group. We did not include observation-only very low risk FHWTs in this study, but note that only two of 39 patients had 1q gain and neither relapsed; similar findings are reported for these patients in the current AREN study.30
Stage II: Patients with stage II FHWTs and 1q gain had lower EFS (81% v 87%) and OS (94% v 97%) compared with patients without 1q gain, although neither was statistically significant. Strategies for therapy or salvage augmentation similar to that described for patients with stage I can thus be considered. However, again, 21% of patients with stage II (26% with 1q gain × 81% without relapse) would not benefit from this. To further develop the optimal strategies for future studies, analysis of both stages I and II FHWTs enrolled in the recent COG studies are under way.
Stage III: Patients with stage III FHWTs and 1q gain had significantly lower EFS than did those without 1q gain (79% v 89%), but the difference in OS (91% v. 95%) was not statistically significant. The distribution of 1q gain did not differ on the basis of the reason for stage III disease. Although patients with stage III disease had relatively good outcomes, their current treatment included doxorubicin and flank or abdominal radiation therapy (XRT), which are associated with an increased risk of late effects.1 The outstanding OS for patients with stage III FHWTs who lack 1q gain may provide an opportunity to reduce therapy (doxorubicin and/or XRT) in the context of a carefully monitored clinical trial.
Stage IV: EFS (64% v 91%) and OS (74% v 92%) were strikingly worse in patients with stage IV FHWTs with 1q gain. Although this suggests an opportunity for therapeutic augmentation for those with 1q gain, a new treatment approach adopted for stage IV FHWTs in the recently completed COG AREN0533 study used risk-adapted therapy on the basis of completeness of lung nodule response after 6 weeks of therapy.31 Patients with complete lung nodule response after chemotherapy were treated without lung XRT, and patients with incomplete lung nodule response received additional cyclophosphamide and etoposide. The prognostic value of 1q gain with this new approach, as well as whether 1q gain correlates with completeness of lung nodule response, needs to be determined before planning future protocols.
This study clearly demonstrates that the presence of 1q gain is not independent of 1p and 16q loss, as is expected, given that t(1;16) and i(1q) are common cytogenetic features identified in WT.10,18 Both can result in loss of 1p and/or 16q and the simultaneous gain of 1q. Which individual or combination of CN changes results in the adverse outcome is not clear. Our data suggest that the single most powerful predictor of outcome is 1q gain and that in the presence of 1q gain, neither 1p nor 16q loss is significant. However, in the absence of 1q gain, 1p and/or 16q loss do retain some prognostic significance and may therefore be helpful in further stratifying 1q-gain-negative patients.
The methods historically used to determine LOH (microsatellite analysis) do not measure gain. MLPA is able to directly and cost effectively measure both copy loss and gain and is now widely used in clinical testing. It can be easily multiplexed to include 1p and 16q loss as well as 1 q gain and does not rely on the concomitant availability and analysis of a source of constitutional DNA. We demonstrated a strong concordance between LOH and CN loss for 1p and 16q, with true lack of concordance in only approximately 2% of tumors. Another 2% to 3% showed copy-neutral LOH, a recognized mechanism of chromosomal aberration. This suggests that direct CN loss (as measured using MLPA) may replace microsatellite analysis.
In conclusion, these findings validate the strong association between outcome and 1q gain that affects all stages to some degree, providing support for the use of 1q gain to stratify therapy in future FHWT therapeutic trials in a stage-specific manner. Although the overall prognosis for FHWT is reasonably good compared with some childhood cancers, a significant percentage of patients will relapse with current standard therapeutic approaches.2 Conversely, a large number of patients may be receiving toxic therapy that is not warranted. The addition of 1q gain to the existing prognostic framework of risk factors has the potential to refine the selection of patients as candidates for either therapy intensification or reduction.
Footnotes
Listen to the podcast by Dr Geoerger at www.jco.org/podcasts
Supported by grants from the National Institutes of Health to E.J.P. (Grant No. R21CA155556), to the National Wilms Tumor Study Group (Grant No. CA-42326), to the National Wilms Tumor Study Group Late Effects Study (Grant No. CA-54498), and to the Children’s Oncology Group (Grant Nos. U10CA180886, U10CA180899, U10CA098543, U10CA098413, and U24CA114766).
Authors’ disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
AUTHOR CONTRIBUTIONS
Conception and design: Eric J. Gratias, Jeffrey S. Dome, Elizabeth A. Mullen, Elizabeth J. Perlman
Financial support: Elizabeth J. Perlman
Provision of study materials or patients: Jeffrey S. Dome
Collection and assembly of data: Jeffrey S. Dome, Lawrence J. Jennings, James Anderson, Elizabeth J. Perlman
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Association of Chromosome 1q Gain With Inferior Survival in Favorable-Histology Wilms Tumor: A Report From the Children’s Oncology Group
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Eric J. Gratias
Employment: Evicore Healthcare
Jeffrey S. Dome
Patents, Royalties, Other Intellectual Property: Rockland Immunochemicals/St Jude Children's Research Hospital
Lawrence J. Jennings
No relationship to disclose
Yueh-Yun Chi
No relationship to disclose
Jing Tian
No relationship to disclose
James Anderson
Employment: Merck
Stock or Other Ownership: Merck
Consulting or Advisory Role: Merck, Amgen, SFD Pharma
Paul Grundy
No relationship to disclose
Elizabeth A. Mullen
Consulting or Advisory Role: Advanced Medical
James I. Geller
No relationship to disclose
Conrad V. Fernandez
No relationship to disclose
Elizabeth J. Perlman
No relationship to disclose
REFERENCES
- 1.Termuhlen AM, Tersak JM, Liu Q, et al. Twenty-five year follow-up of childhood Wilms tumor: A report from the Childhood Cancer Survivor Study. Pediatr Blood Cancer. 2011;57:1210–1216. doi: 10.1002/pbc.23090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dome JS, Perlman EJ, Graf N. Risk stratification for Wilms tumor: Current approach and future directions. Am Soc Clin Oncol Ed Book. 2014:215–223. doi: 10.14694/EdBook_AM.2014.34.215. [DOI] [PubMed] [Google Scholar]
- 3.Grundy PE, Telzerow PE, Breslow N, et al. Loss of heterozygosity for chromosomes 16q and 1p in Wilms’ tumors predicts an adverse outcome. Cancer Res. 1994;54:2331–2333. [PubMed] [Google Scholar]
- 4.Grundy RG, Pritchard J, Scambler P, et al. Loss of heterozygosity on chromosome 16 in sporadic Wilms’ tumour. Br J Cancer. 1998;78:1181–1187. doi: 10.1038/bjc.1998.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klamt B, Schulze M, Thäte C, et al. Allele loss in Wilms tumors of chromosome arms 11q, 16q, and 22q correlate with clinicopathological parameters. Genes Chromosomes Cancer. 1998;22:287–294. doi: 10.1002/(sici)1098-2264(199808)22:4<287::aid-gcc4>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 6.Wittmann S, Zirn B, Alkassar M, et al. Loss of 11q and 16q in Wilms tumors is associated with anaplasia, tumor recurrence, and poor prognosis. Genes Chromosomes Cancer. 2007;46:163–170. doi: 10.1002/gcc.20397. [DOI] [PubMed] [Google Scholar]
- 7.Messahel B, Williams R, Ridolfi A, et al. Allele loss at 16q defines poorer prognosis Wilms tumour irrespective of treatment approach in the UKW1-3 clinical trials: A Children’s Cancer and Leukaemia Group (CCLG) Study. Eur J Cancer. 2009;45:819–826. doi: 10.1016/j.ejca.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Spreafico F, Gamba B, Mariani L, et al. Loss of heterozygosity analysis at different chromosome regions in Wilms tumor confirms 1p allelic loss as a marker of worse prognosis: A study from the Italian Association of Pediatric Hematology and Oncology. J Urol. 2013;189:260–266. doi: 10.1016/j.juro.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Grundy PE, Breslow NE, Li S, et al. Loss of heterozygosity for chromosomes 1p and 16q is an adverse prognostic factor in favorable-histology Wilms tumor: A report from the National Wilms Tumor Study Group. J Clin Oncol. 2005;23:7312–7321. doi: 10.1200/JCO.2005.01.2799. [DOI] [PubMed] [Google Scholar]
- 10.Bown N, Cotterill SJ, Roberts P, et al. Cytogenetic abnormalities and clinical outcome in Wilms tumor: A study by the U.K. cancer cytogenetics group and the U.K. Children’s Cancer Study Group. Med Pediatr Oncol. 2002;38:11–21. doi: 10.1002/mpo.1258. [DOI] [PubMed] [Google Scholar]
- 11.Lu YJ, Hing S, Williams R, et al. Chromosome 1q expression profiling and relapse in Wilms’ tumour. Lancet. 2002;360:385–386. doi: 10.1016/S0140-6736(02)09596-X. [DOI] [PubMed] [Google Scholar]
- 12.Hing S, Lu Y-J, Summersgill B, et al. Gain of 1q is associated with adverse outcome in favorable histology Wilms’ tumors. Am J Pathol. 2001;158:393–398. doi: 10.1016/S0002-9440(10)63982-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Natrajan R, Williams RD, Hing SN, et al. Array CGH profiling of favourable histology Wilms tumours reveals novel gains and losses associated with relapse. J Pathol. 2006;210:49–58. doi: 10.1002/path.2021. [DOI] [PubMed] [Google Scholar]
- 14.Natrajan R, Little SE, Sodha N, et al. Analysis by array CGH of genomic changes associated with the progression or relapse of Wilms’ tumour. J Pathol. 2007;211:52–59. doi: 10.1002/path.2087. [DOI] [PubMed] [Google Scholar]
- 15.Williams RD, Hing SN, Greer BT, et al. Prognostic classification of relapsing favorable histology Wilms tumor using cDNA microarray expression profiling and support vector machines. Genes Chromosomes Cancer. 2004;41:65–79. doi: 10.1002/gcc.20060. [DOI] [PubMed] [Google Scholar]
- 16.Huang C-C, Gadd S, Breslow N, et al. Predicting relapse in favorable histology Wilms tumor using gene expression analysis: A report from the Renal Tumor Committee of the Children’s Oncology Group. Clin Cancer Res. 2009;15:1770–1778. doi: 10.1158/1078-0432.CCR-08-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segers H, van den Heuvel-Eibrink MM, Williams RD, et al. Gain of 1q is a marker of poor prognosis in Wilms’ tumors. Genes Chromosomes Cancer. 2013;52:1065–1074. doi: 10.1002/gcc.22101. [DOI] [PubMed] [Google Scholar]
- 18.Gratias EJ, Jennings LJ, Anderson JR, et al. Gain of 1q is associated with inferior event-free and overall survival in patients with favorable histology Wilms tumor: A report from the Children’s Oncology Group. Cancer. 2013;119:3887–3894. doi: 10.1002/cncr.28239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gratias EJ, Dome JS. Current and emerging chemotherapy treatment strategies for Wilms tumor in North America. Paediatr Drugs. 2008;10:115–124. doi: 10.2165/00148581-200810020-00006. [DOI] [PubMed] [Google Scholar]
- 20.Schouten JP, McElgunn CJ, Waaijer R, et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Walz AL, Ooms A, Gadd, S, et al: Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable histology Wilms tumors. Cancer Cell 27:286-297, 2015. [DOI] [PMC free article] [PubMed]
- 22.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 23.Peto R, Peto J. Asymptotically efficient rank invariant test procedures. J R Stat Soc [Ser A] 1972;135:185–207. [Google Scholar]
- 24.Cox DR. Regression models and life-tables. J R Stat Soc Series B Stat Methodol. 1972;34:187–220. [Google Scholar]
- 25.Kilday JP, Mitra B, Domerg C, et al. Copy number gain of 1q25 predicts poor progression-free survival for pediatric intracranial ependymomas and enables patient risk stratification: A prospective European clinical trial cohort analysis on behalf of the Children’s Cancer Leukaemia Group (CCLG), Societe Francaise d’Oncologie Pediatrique (SFOP), and International Society for Pediatric Oncology (SIOP) Clin Cancer Res. 2012;18:2001–2011. doi: 10.1158/1078-0432.CCR-11-2489. [DOI] [PubMed] [Google Scholar]
- 26.Lo KC, Ma C, Bundy BN, et al. Gain of 1q is a potential univariate negative prognostic marker for survival in medulloblastoma. Clin Cancer Res. 2007;13:7022–7028. doi: 10.1158/1078-0432.CCR-07-1420. [DOI] [PubMed] [Google Scholar]
- 27.Pezzolo A, Rossi E, Gimelli S, et al. Presence of 1q gain and absence of 7p gain are new predictors of local or metastatic relapse in localized resectable neuroblastoma. Neuro Oncol. 2009;11:192–200. doi: 10.1215/15228517-2008-086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackintosh C, Ordóñez JL, García-Domínguez DJ, et al. 1q gain and CDT2 overexpression underlie an aggressive and highly proliferative form of Ewing sarcoma. Oncogene. 2012;31:1287–1298. doi: 10.1038/onc.2011.317. [DOI] [PubMed] [Google Scholar]
- 29.Miwa T, Hirose Y, Sasaki H, et al. Single-copy gain of chromosome 1q is a negative prognostic marker in pediatric nonependymal, nonpilocytic gliomas. Neurosurgery. 2011;68:206–212. doi: 10.1227/NEU.0b013e3181fd2c2e. [DOI] [PubMed] [Google Scholar]
- 30. doi: 10.1097/SLA.0000000000001716. Fernandez CV, Perlman EJ, Mullen EA, et al: Clinical outcome and biological predictors of relapse following nephrectomy only for very low risk Wilms tumor (VLR WT): A report from Children’s Oncology Group AREN0532. Ann Surg (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dix DB, Gratias EJ, Seibel N, et al: Treatment of stage IV favorable histology Wilms tumor with incomplete lung metastasis response after chemotherapy: A report from Children’s Oncology Group study AREN0533. J Clin Oncol 32, 2014 (suppl 5s; abstr 10001) [DOI] [PMC free article] [PubMed]