Abstract
Purpose
Conventional staging methods are inadequate to identify patients with stage II colon cancer (CC) who are at high risk of recurrence after surgery with curative intent. ColDx is a gene expression, microarray-based assay shown to be independently prognostic for recurrence-free interval (RFI) and overall survival in CC. The objective of this study was to further validate ColDx using formalin-fixed, paraffin-embedded specimens collected as part of the Alliance phase III trial, C9581.
Patients and Methods
C9581 evaluated edrecolomab versus observation in patients with stage II CC and reported no survival benefit. Under an initial case-cohort sampling design, a randomly selected subcohort (RS) comprised 514 patients from 901 eligible patients with available tissue. Forty-nine additional patients with recurrence events were included in the analysis. Final analysis comprised 393 patients: 360 RS (58 events) and 33 non-RS events. Risk status was determined for each patient by ColDx. The Self-Prentice method was used to test the association between the resulting ColDx risk score and RFI adjusting for standard prognostic variables.
Results
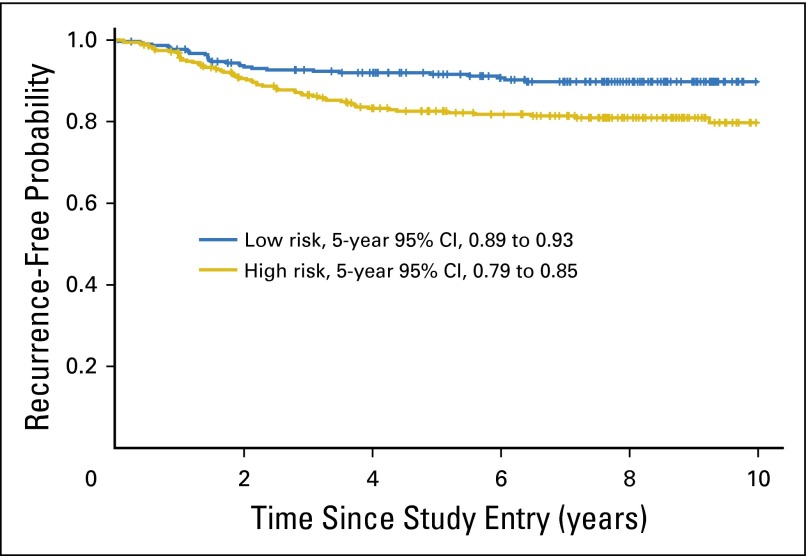
Fifty-five percent of patients (216 of 393) were classified as high risk. After adjustment for prognostic variables that included mismatch repair (MMR) deficiency, ColDx high-risk patients exhibited significantly worse RFI (multivariable hazard ratio, 2.13; 95% CI, 1.3 to 3.5; P < .01). Age and MMR status were marginally significant. RFI at 5 years for patients classified as high risk was 82% (95% CI, 79% to 85%), compared with 91% (95% CI, 89% to 93%) for patients classified as low risk.
Conclusion
ColDx is associated with RFI in the C9581 subsample in the presence of other prognostic factors, including MMR deficiency. ColDx could be incorporated with the traditional clinical markers of risk to refine patient prognosis.
INTRODUCTION
Stage I colon cancer (CC) is treated with surgery alone, and patients with stage III CC are routinely offered adjuvant chemotherapy; however, the optimal clinical management of patients with stage II CC is not clearly defined. Stage II CC accounts for approximately 25% of patients with CC, and the 5-year survival rate is approximately 75% to 80%, with 15% to 20% of patients experiencing disease recurrence.1 Current guidelines2,3 recommend adjuvant chemotherapy for patients with stage II CC with high-risk features such as T4 stage, high tumor grade, inadequate number of nodes sampled, positive or unknown margins, lymphovascular and perineural invasion, bowel obstruction, and perforation. However, a number of studies have suggested that these histopathological and clinical markers are limited in their prognostic power.4,5 These limitations have prompted efforts to identify molecular markers to guide the clinical management of patients with stage II CC. Although some molecular markers, such as loss of heterozygosity and p53 mutational status, require more evidence to establish their clinical utility as prognostic markers, assessment of microsatellite instability status has been adopted into clinical practice.3 Several gene signatures have also been shown to be independently associated with clinical outcome in patients with stage II CC.6-10
One such gene expression signature, the ColDx assay, can be applied to formalin-fixed paraffin-embedded (FFPE) tissue collected at the time of diagnosis and has previously been shown to be prognostic for recurrence-free interval (RFI) and overall survival (OS) both independent of, and superior to, standard pathologic features.6 The gene expression signature was developed in two stages. Stage one involved creation of a Colorectal Cancer Disease-Specific Array that included genes specifically expressed in human colorectal cancers.11 The Colorectal Cancer Disease-Specific Array content (61,528 total probe sets) was generated by a combination of high-throughput sequencing, public database mining, and experimental investigation and was optimized to enable profiling of RNA extracted from fresh and FFPE samples. A set of 215 human, stage II colorectal cancers with known outcome data were identified as a training set and used to develop a 634-probe set, partial least squares prognostic signature, specific for high risk of recurrence. The probe sets and statistical methods used were previously described, and statistically significant pathways were identified.6 The analytical properties of this assay have also been reported.12 The assay was developed and intended for use on FFPE, a commonly used, standard method of tumor tissue preservation.
The Cancer and Leukemia Group B (CALGB), now the Alliance for Clinical Trials in Oncology (Alliance), conducted a randomized phase III clinical trial (C9581) of 1,738 patients with stage II CC without high-risk features, which concluded that adjuvant edrecolomab did not significantly increase overall survival compared with observation only.13 The tissue and data from this clinical trial were archived for the purpose of biomarker analysis and thus provided a resource for validation of prognostic molecular markers in a large cohort of patients with stage II CC. The objective of this study was to use the C9581 trial samples to further assess the utility of the ColDx assay to classify patients with stage II CC as low or high risk of recurrence within 5 years postsurgery.
PATIENTS AND METHODS
Patients
Eligibility requirements for enrollment to the C9581 study have been reported previously.12 Institutional review board approval and patient informed consent were required at each participating institution. The study excluded patients with high-risk features, and the overall risk of recurrence observed in the study was 14.6% at 5 years.13 The eligibility requirements for the current study included: enrollment in C9581 with available FFPE tissue and patient consent for use of this tissue; a primary diagnosis of stage II adenocarcinoma of the colon; colon surgery with curative intent; negative surgical margins; no nonprotocol, postoperative therapy before recurrence; no preoperative therapy within 1 year of surgery; no local or regional recurrence only. Patients with rectal cancer were not eligible.
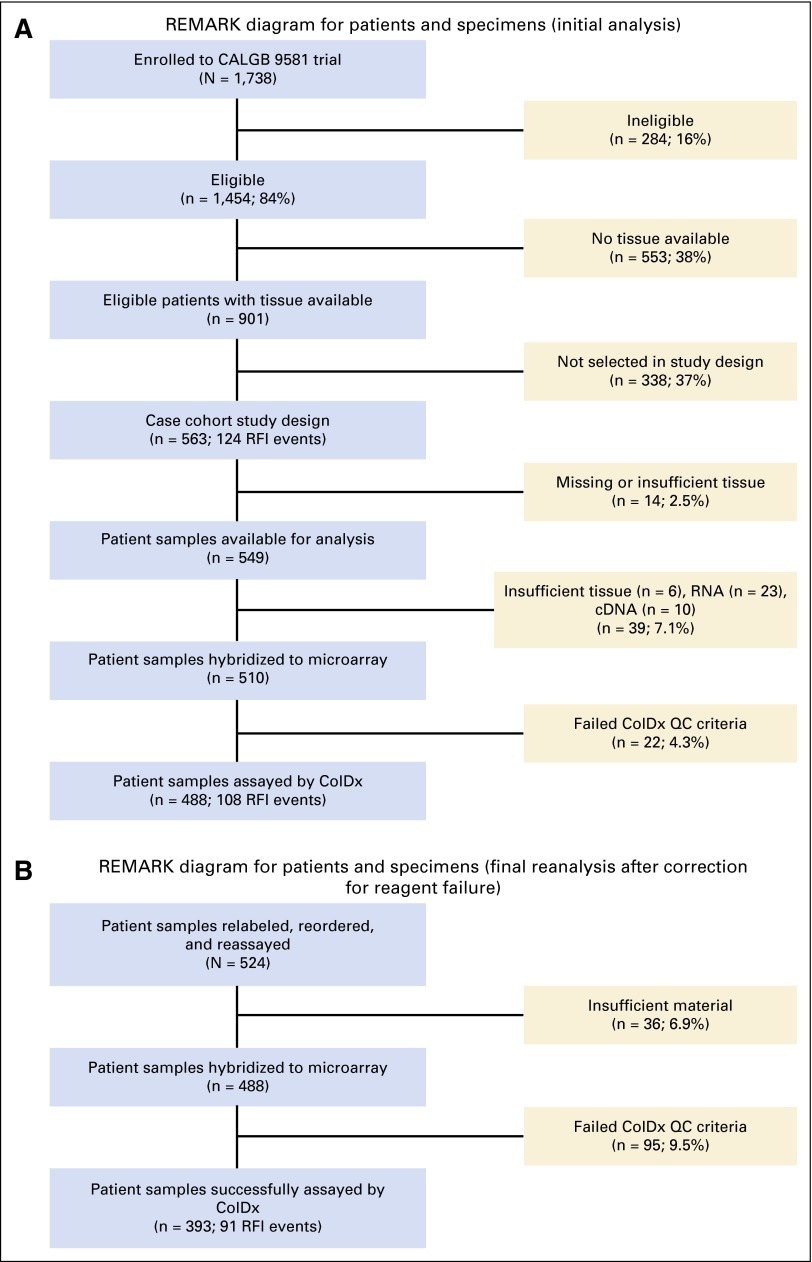
Of 1,454 patients who were eligible for the study, FFPE tissue was available for analysis from 901 patients. Among the 901 eligible patients with available tissue (Figs 1A and 1B), minimum follow-up time was 2.9 months and maximum follow-up time was 12.1 years. The incidence rate was 0.020 recurrences per person-year, with 124 (13.8%) RFI events and 777 (86.2%) censored observations. Median follow-up was 8.1 years (95% CI, 7.9 to 8.2 years) on the basis of the final clinical data set as of December 4, 2009.
Fig 1.
(A) REMARK diagram for patients and specimens (initial analysis). (B) REMARK diagram for patients and specimens (final analysis). CALGB, Cancer and Leukemia Group B; RFI, recurrence-free interval; QC, quality control; REMARK, REporting recommendations for tumor MARKer prognostic studies.
Data from eligible patients treated on C9581 with available samples were uniquely coded and analyzed in a blinded fashion for gene expression. All statistical analyses involving the clinical data were conducted by Alliance statisticians.
Pathology
During the conduct of C9581, FFPE primary tumor and normal colon were obtained for each case, with histology confirmed by central pathology review. Staging was conducted as previously described.13 Of the samples within the final study cohort for this investigation, 85% had a viable tumor volume of ≥ 50%. Specimens with a tumor volume of < 50% were macrodissected for inclusion within the study. DNA mismatch repair (MMR) status was assessed by immunohistochemistry for mut L homolog 1 (MLH1) and mut S homolog 2 (MSH2), also as previously described.7
Gene Expression Profiling
Total RNA was extracted from two 10-µm sections obtained from a single representative FFPE tissue block, amplified and hybridized to the Almac Colorectal Cancer disease-specific microarray in a Clinical Laboratory Improvement Amendments (CLIA), New York State–approved laboratory (Helomics Corporation, Pittsburgh, PA). The ColDx gene expression signature was used to classify patients as low or high risk for recurrence by computing a signature score as described previously.6
Statistical Methods
The primary end point for this study was RFI measured from study entry (randomization) to distant recurrence or death related to colon cancer. Patients without distant recurrence or who died of other causes including second malignancies were censored. Overall survival (any cause of death) measured from study entry was studied as a secondary end point.
A case-cohort design was used to compare RFI between patients with ColDx-predicted low- and high-risk scores.14,15 Under this design, a randomized subcohort (RS) was selected from 901 eligible patients after stratification on treatment.16 The recurrent cases that were not selected randomly were included in the analysis data set. The case-cohort design was used to maximize cost efficiency by limiting the number of nonrecurrent patient samples to be assayed. Power estimates were determined on the basis of the methods proposed by Cai and Zeng.17 The initial study cohort comprised a subcohort of 514 patients randomly sampled, including 75 patients with RFI events and 49 patients with RFI events outside the RS, resulting in a total of 563 patients (124 patients with RFI events).
For reasons described in the Appendix (online only), the study was amended. Power computations were revised on the basis of the following assumptions: 449 of 901 patients sampled in the RS using a sampling fraction of 0.4983; a proportion of samples marker positive, p1, of 0.20; a proportion of events in the full cohort, pD, of 0.1199 (108 of 901). Under these assumptions, a hazard ratio of 2.05 was detectable with approximately 80% power. Data from 393 patients were included in the final analysis.
Patients were categorized as low or high risk coded on the basis of a prognostic score of 0.4377, fixed before analysis involving patient outcomes. This threshold was selected after migration of the ColDx assay from the Affymetrix GeneChip System 3000 7G scanner to the Affymetrix microarray platform GeneChip System 3000Dx v.2.
The primary hypothesis that the ColDx classification was significantly associated with RFI independently of known prognostic factors was tested using a weighted Cox proportional hazards model adjusting for standard prognostic variables.15 This multivariable analysis included the coded prognostic score and the following potentially prognostic variables: age at study entry (years); sex (male, female); T stage (T3, T4); tumor grade (I-II, III-IV); tumor location (distal, proximal); number of nodes examined (continuous measure); MMR status (MMR-deficient, MMR-intact) determined by immunohistochemistry for MLH1 and MSH2, in 377 patients with complete data and 91 RFI events. Patients with missing data on any variable were omitted from the multivariable analysis. The likelihood ratio test was used to test the significance of the addition of the prognostic score to the reduced multivariable model. The association between RFI and only the coded prognostic score was also tested. Analyses of Schoenfeld residuals and regression methods were used to assess the proportional hazards assumption for each variable.
Data analysis was conducted in R using the Survival package by Therneau (Self-Prentice method)18,19 and in SAS, version 9.2, using the programming for case-cohort analysis proposed by Barlow et al.20 Two-sided P values less than .05 were considered significant.
The χ2 test was used to test the associations between ColDx and categorical prognostic factors; the t test was used to test associations with continuous factors. No adjustment was made for multiple comparisons in these analyses.
RESULTS
Sample Cohort
An initial cohort of 563 patients, with 124 RFI events, was selected for the study (comprising 514 patients with 75 RFI events in the RS and 49 patients with RFI events outside the RS). Tissue was available from 549 of these patients for analysis. A quality control (QC) failure arose in a subset of patient samples because of a reagent fault during the initial analysis, and the study was subsequently repeated with qualified reagents (Data Supplement).
Of the initial cohort of 549 tissue samples available for analysis, a total of 156 samples (28.4%) were excluded from the analysis because of insufficient tissue, total RNA, or poor quality. Median (range) time between resection and analysis was 13.2 years (11.2 to 16.0 years). The final evaluable data set in the repeated analysis contained 393 patients, of whom 91 experienced an RFI event (360 patients with 58 RFI events within the RS; 33 patients with RFI events outside the RS). Figures 1A and 1B depict the flow of patients through the study and reasons for exclusion.
The demographic characteristics of the C9581 cohort have been described.7,13 Patient and tumor characteristics for patients evaluated in the study were similar to those not included and to those in the subset of all eligible C9581 patients (Table 1).
Table 1.
Patient and Tumor Characteristics for 393 Patients in the Study Cohort, 1,061 Eligible Patients Not in the Study Cohort, and Total 1,454 Eligible Patients From the Parent Trial
| Characteristic | Value | Eligible Included in Study (n = 393)* | Eligible Patients Not Included in Study (n = 1,061)* | Parent C9581 Trial Eligible Patients (N = 1,454)* |
|---|---|---|---|---|
| No. (%)†‡ | No. (%)†‡ | No. (%)† | ||
| Age | Years | 64 (23) | 64 (13) | 64 (11) |
| Sex | Male | 209 (53) | 554 (52) | 763 (52) |
| Race/ethnicity | White | 357 (91) | 971 (91) | 1,328 (92) |
| (n = 392) | (n = 1,057) | (n = 1,449) | ||
| Treatment arm | Edrecolomab | 192 (49) | 528 (50) | 720 (50) |
| T stage | T4 | 18 (5) | 35 (3) | 53 (4) |
| Nodes examined | No. | 14.5 (17) | 14.7 (12) | 14.6 (9) |
| Perineural invasion | Present | 10 (2) | 27 (3) | 37 (3) |
| (n = 391) | (n = 1,056) | (n = 1,447) | ||
| Lymphovascular invasion | Present | 47 (11) | 115 (11) | 162 (11) |
| Tumor grade | High | 67 (17) | 148 (14) | 215 (15) |
| (n = 1,054) | (n = 1,447) | |||
| Obstruction or perforation | Present | 6 (2) | 20 (2) | 26 (2) |
| (n = 1,060) | (n = 1,453) | |||
| Tumor location | Proximal | 244 (62) | 634 (59) | 878 (60) |
| (n = 1,060) | (n = 1,453) | |||
| MMR status§ | Deficient | 94 (26) | 84 (18) | 178 (21) |
| (n = 377) | (n = 456) | (n = 833) |
Abbreviation: MMR, mismatch repair.
Sample size unless otherwise specified.
Denotes mean and standard deviation for age and nodes examined.
Percentages weighted to reflect those in the entire eligible cohort (N = 1,454).12
MMR as determined by immunohistochemistry on mut L homolog 1 (MLH1), mut S homolog 2 (MSH2).
Association Between ColDx and Prognostic Factors
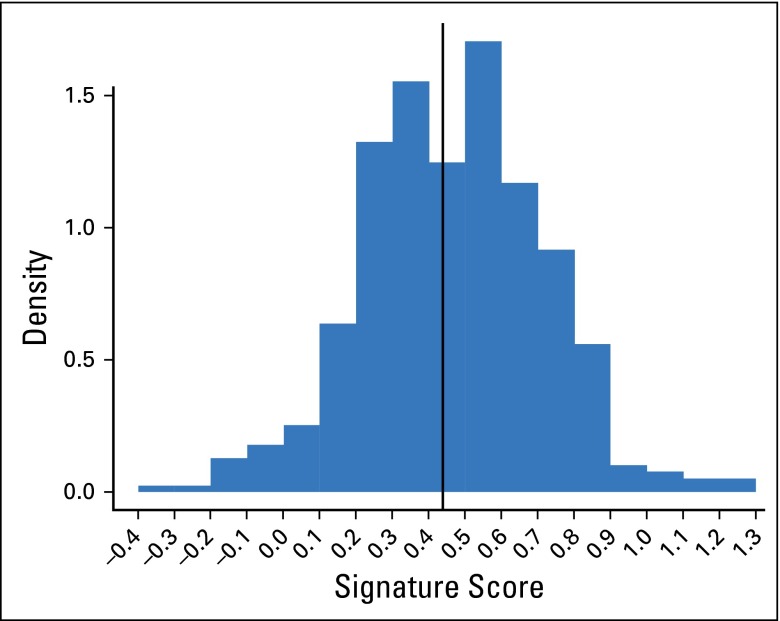
Results of preprocessing and derivation of the ColDx signature score were confirmed by the Alliance statisticians. Figure 2 provides a histogram of the distribution of signature scores. At the prespecified threshold, ColDx classified 55% of patients (216 of 393) as high risk and 45% (177 of 393) as low risk.
Fig 2.
Histogram of continuous ColDx signature score units (n = 393); high risk (≥ 0.4377) in 55% of patients. Data breaks occur between −0.4 and 1.3 at intervals of 0.1. For each interval, the density multiplied by 0.1 estimates the probability of the signature score being in that interval. The distribution mean and standard deviation are 0.46 and 0.24, respectively, with a median of 0.47 and range of −0.33 to 1.26.
Associations between the dichotomized signature score (high risk, low risk) and the potential prognostic variables in Table 1 were explored. ColDx was significantly associated with sex (P = .003), race (P < .001), and MMR status (P < .001) with higher proportions of men (57% v 49%), nonwhite race (73% v 52%), and MMR intact tumors (56% v 45%) classified as high risk, respectively. Associations with T stage, lymphovascular invasion, and tumor location were marginally significant (Appendix Table A1).
Association Between ColDx and Recurrence Risk
Results of univariable analyses of individual prognostic factors and RFI are given in Table 2. The primary test of hypothesis was the assessment of the ColDx score in the presence of conventional prognostic factors (Table 3). ColDx score was significantly associated with RFI after adjustment for other prognostic factors (hazard ratio [HR], 2.13; 95% CI, 1.3 to 3.5; P < .01) in multivariable analysis and was the prognostic factor most strongly associated with RFI (P < .01) in this data set. Age and MMR status were of borderline significance. The ColDx signature score contributed significantly to the reduced (prognostic variables only) model (likelihood ratio test, P < .001). No major deviations from proportional hazards were observed on the basis of the Schoenfeld residuals and regression models methods.
Table 2.
Univariable Analyses of Prognostic Factors
| Factor | HR (95% CI) | P |
|---|---|---|
| Age (continuous) | 1.02 (1.00 to 1.04) | .03 |
| Sex (male v female) | 0.97 (0.62 to 1.50) | .89 |
| T stage (T4 v T3) | 1.15 (0.37 to 3.57) | .80 |
| Nodes examined (continuous) | 0.97 (0.95 to 1.00) | .05 |
| Tumor grade (high v low) | 1.30 (0.75 to 2.25) | .34 |
| Tumor location (proximal v distal) | 1.11 (0.70 to 1.75) | .65 |
| MMR (deficient v intact) | 0.56 (0.31 to 1.01) | .05 |
NOTE. n = 393 with 91 RFI events for all variables except n = 377 with 91 RFI events for MMR.
Abbreviations: HR, hazard ratio; MMR, mismatch repair; RFI, recurrence-free interval.
Table 3.
Primary Analysis: Multivariable Model of ColDx Signature Score and Potentially Prognostic Covariates of RFI
| Factor | HR (95% CI) | P |
|---|---|---|
| ColDx signature score (high risk v low risk) | 2.13 (1.3 to 3.5) | < .01 |
| Age (continuous) | 1.02 (1.00 to 1.04) | .07 |
| Sex (male v female) | 0.94 (0.59 to 1.47) | .78 |
| T stage (T4 v T3) | 0.78 (0.20 to 3.0) | .72 |
| Nodes examined (continuous) | 0.98 (0.95 to 1.01) | .11 |
| Tumor grade (high v low) | 1.47 (0.82 to 2.6) | .19 |
| Tumor location (proximal v distal) | 1.24 (0.74 to 2.1) | .41 |
| MMR (deficient v intact) | 0.55 (0.29 to 1.02) | .05 |
NOTE. n = 377 with 91 RFI events.
Abbreviations: HR, hazard ratio; MMR, mismatch repair; RFI, recurrence-free interval.
ColDx high-risk patients also had a significantly shorter RFI relative to ColDx low-risk patients in an unadjusted analysis, with an HR of 2.03 (95% CI, 1.3 to 3.3; P < .01). The recurrence-free probability at 5 years for patients classified as high risk by the ColDx score was 82%, with a 95% CI of 79% to 85%, compared with 91% with a 95% CI of 89% to 93% for patients predicted as low risk (Fig 3).
Fig 3.
Weighted Kaplan-Meier plot of recurrence-free interval by ColDx signature score dichotomized at the prespecified cut point, 0.4377; 62 and 29 events observed for high and low risk, respectively.
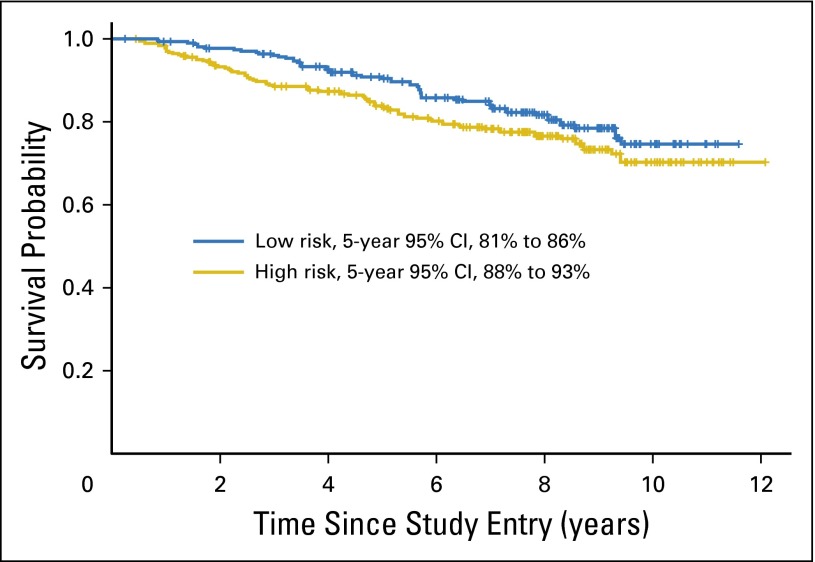
OS by ColDx score is illustrated in Figure 4. A marginally significant association was observed after adjustment for other prognostic factors in multivariable analysis (OS HR, 1.74; 95% CI, 0.97 to 3.1; P = .06).
Fig 4.
Weighted Kaplan-Meier plot of overall survival (all deaths regardless of cause) by ColDx signature score dichotomized at the prespecified cut point, 0.4377; 64 and 40 events observed for high and low risk, respectively.
DISCUSSION
Given the lower rates of recurrence among patients with stage II CC, there is reluctance to subject patients to potential toxicity and other treatment complications with little evidence of treatment benefit from adjuvant therapy. Current guidelines indicate that adjuvant therapy may be most appropriate for patients with high-risk disease characteristics.3 Substantial effort has been devoted to identifying a higher-risk patient subset under the assumption that patients at high risk of recurrence would be the most likely to benefit from adjuvant therapy. ColDx is one of several assays that have been reported to reliably stratify patients with colon cancer into groups at high and low risk of disease recurrence.
The ColDx assay has previously been shown to be an independent prognostic factor for RFI in stage II CC.6 In this second independent prospectively designed study, the ColDx assay was again demonstrated to be significantly associated with RFI in the stage II colon cancer C9581 cohort and to be the most significant prognostic factor in multivariable analysis, in the presence of other prognostic factors, inclusive of MMR status. The data and samples collected as part of the C9581 study protocol were obtained from patients who were uniformly treated and well characterized. As such, this cohort provided an invaluable resource for objective assessment of novel prognostic markers such as the ColDx assay.
However, a limitation of this study was the age of the tissue samples, which may have contributed to the high proportion of assay failures. (A maximum attrition rate of 10% was originally expected on the basis of experience in a number of prior studies.) On average, the length of time between patient resection and assay analysis was 13.2 years. In addition, results of the first assay series were affected by a reagent failure in a subset of patient samples, which was discovered only after the first statistical analysis was conducted. A plan to salvage the validation and maintain the integrity of the study was developed in conjunction with the National Cancer Institute Cancer Therapy Evaluation Program and incorporated into the protocol. Under this plan, specimens were returned to the Alliance Pathology Coordinating Office, relabeled, and reanalyzed by Helomics Corporation. The final statistical analysis was conducted on the resulting expression data. This process was transparent, and we are confident in the final results.
The study demonstrated that patients within the C9581 cohort predicted as high risk by the ColDx assay have a recurrence-free probability of 82% (18% probability of recurrence within 5 years) compared with 91% (9% probability of recurrence within 5 years) for patients predicted as low risk with an HR after multivariable analysis of 2.13. This compares favorably with the previously reported clinical performance of the assay, where it was demonstrated to predict high-risk patients with an HR of recurrence of 2.53 and an HR of cancer-related death of 2.21 in an independent validation data set.6 The ColDx assay was significantly associated with RFI in this study (P < .01), providing greater prognostic value than standard clinical markers, including age (P = .07), number of nodes examined (0.11), and, notably, MMR status (P = .05). Analysis of these clinical markers is currently the primary means by which clinicians determine the clinical management of patients with stage II colon cancer.
The prognostic performance of ColDx is likely derived from its relationship to biologic pathways previously reported to differentiate between good and poor prognosis in colon cancer via promotion of tumor growth, invasion, and metastasis that are not accounted for by traditional prognostic factors.6 In addition, although ColDx is a significant predictor of RFI in its own right, the addition of the ColDx score significantly contributes to a prognostic model including all clinical covariates (likelihood ratio test, P < .001), which demonstrates its additive contribution.
There have been several prognostic gene expression assays for stage II colon cancer.7-10 Only the 12-gene colon cancer recurrence score is comparable to ColDx, because it has also been developed to work with FFPE tissue and has undergone independent validation using the stage II C9581 sample cohort.7 The 12-gene colon cancer recurrence score was also found to be the most significant prognostic factor among patients studied from C9581 (HR, 1.68; 95% CI, 1.18 to 2.38; P = .004), with the HR calculated on a continuous scale for a 25-unit increase in the score. In the subset of 271 patients for whom data are available from both assays, there is low correlation between the continuous scores (R = 0.18; 95% CI, 0.06 to 0.29). Although there is some overlap, it does not seem that the signatures are measuring the same thing or identifying the same patients as high risk. Further analysis is being conducted in this patient subset.
Prognostic assays, such as ColDx, would have enhanced clinical utility if they were also predictive of response to therapy in colon cancer.21 Other than a marginal treatment effect in the Quick and Simple and Reliable (QUASAR) trial,22 there have never been convincing data showing benefit for adjuvant therapy in any study of stage II disease for CC, including C9581. A retrospective subset analysis of the stage II cohort in Intergroup 0035 suggested that fluorouracil therapy might be beneficial in patients with clinical high-risk factors, such as T4 lesions, obstruction, inadequate nodal sampling, and perforation.23 Although that hypothesis-generating observation has never been validated in a prospective manner, the practice has been embraced and incorporated into guidelines and clinical care. To support clinical utility, a direction for future research would be to show that prognostic gene expression signatures are also predictive for treatment efficacy.
In summary, the ColDx assay has now been prospectively validated for a second time as a prognostic marker for patients with stage II colon cancer and in this patient subset superior to current prognostic markers such as T stage, nodal sampling, and MMR status. This study is then an external validation of the prognostic value of the ColDX assay. Thus, ColDx assay results could be incorporated with the traditional clinical markers of risk to refine prognosis. Future studies could be designed to demonstrate the potential for treatment benefit among high-risk patients defined by the ColDx risk score.
Supplementary Material
Acknowledgment
We thank the following networks and groups and their patients who provided specimens to C9581: The Greenville Community Clinical Oncology Program of the Carolinas (Grant No. CA29165), Missouri Cancer Associates, Eastern Cooperative Oncology Group–American College of Radiology Imaging Network (Grant No. U10CA180820), National Cancer Institute of Canada Clinical Trials Group (Grant No. U10CA077202), and Southwest Oncology Group (Grant No. U10CA180888).
Appendix
Administrative Summary of Study Progress
Selection of initial study sample.
Five hundred fourteen patients were randomly selected from the available 901 patients meeting the entry criteria. Seventy-five patients meeting the recurrence end point were included in the random component of the study sample. Under the case-cohort design, the remaining 49 patients meeting the recurrence end point were also part of the study sample. A total of 563 patients were identified for potential inclusion in the study. Five hundred forty-nine patient samples were provided to Helomics Corporation. There were 14 blocks that were either missing or unable to be cut because they were insufficient.
Microarray analysis results.
Of the 549 samples provided by the Alliance, sufficient total RNA was extracted to allow for gene expression profiling of 520 samples. Among the 29 samples that were not profiled by gene expression, six were unable to be extracted because of lack of tissue availability and 23 because of insufficient RNA. Of these 520 samples, 510 samples were hybridized to a microarray generating raw data (CEL files). Ten samples were not hybridized to a microarray because of insufficient cDNA. Of these 510 samples with microarray data generated, 488 passed ColDx quality control (QC) metrics (a total of 22 QC failures).
An Excel file containing the following data fields was provided to the Alliance for the 510 samples that generated microarray data, including the 22 QC failures: Alliance Pathology Coordinating Office Number; Almac ID assigned to each sample by Almac (for correlation to raw CEL files); ColDx signature score; dichotomized clinical call (low risk, high risk, QC failure). The subgroup of 488 samples passing ColDx QC metrics comprised 449 samples, including 69 events in the randomized sample and 39 events outside of the randomized sample.
Statistical analysis of ColDx data.
Results of the statistical analysis were not expected. To verify that the results were correct, the laboratory and clinical data merge, recurrence-free interval end points, and statistical analyses were reviewed. Patient-level data were provided to Almac for exploration of potential issues related to QC, macrodissection, and other laboratory procedures. In this process, a strong batch effect related to a specific reagent lot number (1201176-B) was noted in a subset of samples. It was concluded that a lower-than-expected hazard ratio was observed overall because of the reagent failure in this subset. The reagent lot batch effect was observed at the global gene expression level (principal component analysis), and this effect persisted through to the signature score. Other technical factors were also evaluated and eliminated as causes.
A plan to salvage the validation and maintain the integrity of the study was developed in conjunction with National Cancer Institute-Cancer Therapy Evaluation Program and incorporated into the protocol. Under this plan, all specimens were to be returned to the Alliance PCO, relabeled, and reanalyzed by Helomics Corporation, in a blinded fashion. It was agreed that initial assessments of patients (514 in the randomly selected subcohort [RS], stratified by treatment) would not be used, and the final statistical analysis would be conducted on the expression data resulting from reanalysis of the specimens.
Residual material was identified for 524 samples, which were subsequently relabeled, reordered, and reassayed with reagents that passed QC; 36 had insufficient material and 95 failed ColDx QC. The final set of samples with new ColDx assay values comprised 393 patients (360 in the randomly selected subcohort, with 58 recurrence-free interval events and 33 events outside the RS).
Table A1.
Univariable Analyses of Prognostic Factors and ColDx Score (n = 393)
| Factor | Percent High Risk* | P† |
|---|---|---|
| Age (mean age; high v low risk) | 64 v 63 years | .43 |
| Sex (male v female) | 57% v 49% | .003 |
| Race (white v other) | 52% v 73% | < .001 |
| Treatment (edrecolomab v observation) | 53% v 54% | .66 |
| T stage (T4 v T3) | 65% v 53% | .04 |
| Nodes examined (mean No. of nodes; high v low risk) | 14 v 14 nodes | .89 |
| Perineural invasion (absent v present) | 53% v 60% | .46 |
| Lymphovascular invasion (absent v present) | 52% v 63% | .01 |
| Tumor grade (high v low) | 53% v 53% | .95 |
| Tumor location (proximal v distal) | 51% v 57% | .03 |
| Obstruction perforation (absent v present) | 53% v 67% | .17 |
| MMR (deficient v intact) | 45% v 56% | < .001 |
Abbreviation: MMR, mismatch repair.
Except for continuous variables age and number of nodes examined, where means are given.
χ2 test except for continuous variables, where the t test was used.
Footnotes
The research for CALGB 150705, B3 was supported, in part, by Grant No. U10 CA180821-01 from the National Cancer Institute to the Alliance for Clinical Trials in Oncology (Alliance; M.M.B., Chair) and Grant No. U10 CA180882-01 from the National Cancer Institute to the Alliance Statistics and Data Center (D.J. Sargent).
Results from CALGB 150705, B3 were presented at the American Society of Clinical Trials GI Symposium, January 16-18, 2014, San Francisco, as a poster in General Poster Session C: Cancers of the Colon and Rectum (Abstract 455).
The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.
Authors’ disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
AUTHOR CONTRIBUTIONS
Conception and design: Donna Niedzwiecki, Wendy L. Frankel, Alan P. Venook, Paula N. Friedman, Richard M. Goldberg, Robert J. Mayer, Jude Marie Mulligan, Timothy S. Davison, Peter Kerr, Patrick G. Johnston, Richard D. Kennedy, D. Paul Harkin, Monica M. Bertagnolli, Robert S. Warren
Administrative support: Thomas Anthony Colacchio
Provision of study materials or patients: Richard M. Goldberg, Thomas Anthony Colacchio
Collection and assembly of data: Donna Niedzwiecki, Richard M. Goldberg, Thomas Anthony Colacchio, Jude Marie Mulligan, Peter Kerr
Data analysis and interpretation: Donna Niedzwiecki, Alan P. Venook, Xing Ye, Richard M. Goldberg, Robert J. Mayer, Jude Marie Mulligan, Timothy S. Davison, Eamonn O’Brien, Richard D. Kennedy, Richard L. Schilsky, Robert S. Warren, Federico Innocenti
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Association Between Results of a Gene Expression Signature Assay and Recurrence-Free Interval in Patients With Stage II Colon Cancer in Cancer and Leukemia Group B 9581 (Alliance)
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Donna Niedzwiecki
No relationship to disclose
Wendy L. Frankel
No relationship to disclose
Alan P. Venook
Honoraria: Gilead Sciences, Genentech
Consulting or Advisory Role: Gilead Sciences, Genentech
Research Funding: Bayer Healthcare Pharmaceuticals, Onyx Pharmaceuticals, Genentech, Roche, Bristol-Myers Squibb, GlaxoSmithKline, Eli Lilly
Patents, Royalties, Other Intellectual Property: Royalties from Now-UptoDate for authoring and maintaining two chapters
Travel, Accommodations, Expenses: Halozyme Therapeutics, Genentech, Roche, Bristol-Myers Squibb, Merck Serono
Xing Ye
No relationship to disclose
Paula N. Friedman
Stock or Other Ownership: AbbVie, Abbott Laboratories, Bristol-Myers Squibb, CVS, Express Scripts, Exact Sciences, Foundation Medicine, Genomic Health, Illumina, Johnson & Johnson, Eli Lilly, Myriad Genetics, NanoString Technologies, Pfizer, Seattle Genetics, Teva Healthcare Pharmaceuticals, UnitedHealthcare
Richard M. Goldberg
Honoraria: Sanofi, Eli Lilly, Biothera, Baxter, Novo Nordisk, Kanghong Pharma, Momenta Pharmaceuticals, Immunocare Therapies, Amgen, Merck KGaA
Research Funding: Sanofi (Inst), Bayer AG (Inst), Immunomedics (Inst), Merck (Inst),
Travel, Accommodations, Expenses: Sanofi, Merck KGaA, Baxter, Amgen
Robert J. Mayer
Honoraria: Taiho Pharmaceutical
Consulting or Advisory Role: CASI Pharmaceuticals
Thomas Anthony Colacchio
No relationship to disclose
Jude Marie Mulligan
Employment: Almac Diagnostics
Patents, Royalties, Other Intellectual Property: Almac Diagnostics (Inst)
Timothy S. Davison
Employment: Almac Diagnostics
Patents, Royalties, Other Intellectual Property: Almac Diagnostics (Inst)
Eamonn O’Brien
Employment: Almac Diagnostics
Stock or Other Ownership: Abbott Diagnostics
Patents, Royalties, Other Intellectual Property: Almac Diagnostics (Inst)
Peter Kerr
Employment: Almac Diagnostics
Patents, Royalties, Other Intellectual Property: Almac Diagnostics (Inst)
Patrick G. Johnston
Stock or Other Ownership: Almac Diagnostics, Fusion Antibodies, CVG Therapeutics
Honoraria: Pfizer, Chugai Pharmaceutical
Consulting or Advisory Role: Chugai Pharmaceutical, Pfizer
Travel, Accommodations, Expenses: Chugai Pharmaceutical
Richard D. Kennedy
Employment: Almac Diagnostics
Patents, Royalties, Other Intellectual Property: Patent for the prognostic assay
D. Paul Harkin
Employment: Almac Diagnostics
Leadership: Almac Diagnostics
Patents, Royalties, Other Intellectual Property: Almac Diagnostics
Richard L. Schilsky
No relationship to disclose
Monica M. Bertagnolli
No relationship to disclose
Robert S. Warren
No relationship to disclose
Federico Innocenti
No relationship to disclose
REFERENCES
- 1.Edge S, Byrd D, Compton C, et al. AJCC Cancer Staging Manual. ed 7. New York, NY: Springer; 2010. [Google Scholar]
- 2.Benson AB, III, Schrag D, Somerfield MR, et al. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol. 2004;22:3408–3419. doi: 10.1200/JCO.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 3. National Comprehensive Cancer Network: NCCN Clinical Practice Guidelines in Oncology, Colon Cancer v2.2015. http://www.nccn.org. [DOI] [PubMed]
- 4. Van Cutsem E, Oliveria J, ESMO Guidelines Working Group: Primary colon cancer: ESMO clinical recommendations for diagnosis, adjuvant treatment and follow-up. Ann Oncol 20:49-50, 2009 (suppl 4) [DOI] [PubMed]
- 5.Vicuna B, Benson AB., III Adjuvant therapy for stage II colon cancer: Prognostic and predictive markers. J Natl Compr Canc Netw. 2007;5:927–936. doi: 10.6004/jnccn.2007.0080. [DOI] [PubMed] [Google Scholar]
- 6.Kennedy RD, Bylesjo M, Kerr P, et al. Development and independent validation of a prognostic assay for stage II colon cancer using formalin-fixed paraffin-embedded tissue. J Clin Oncol. 2011;29:4620–4626. doi: 10.1200/JCO.2011.35.4498. [DOI] [PubMed] [Google Scholar]
- 7. Venook AP, Niedzwiecki D, Lopatin M, et al: Biologic determinants of tumor recurrence in stage II colon cancer: Validation study of the 12-gene recurrence score in cancer and leukemia group B (CALGB) 9581. J Clin Oncol 31:1775-1781, 2013. [DOI] [PMC free article] [PubMed]
- 8. Salazar R, Roepman P, Capella G, et al: Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol 29:17-24, 2011. [DOI] [PubMed]
- 9.Agesen TH, Sveen A, Merok MA, et al. ColoGuideEx: A robust gene classifier specific for stage II colorectal cancer prognosis. Gut. 2012;61:1560–1567. doi: 10.1136/gutjnl-2011-301179. [DOI] [PubMed] [Google Scholar]
- 10.Zhang JX, Song W, Chen ZH, et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: A microRNA expression analysis. Lancet Oncol. 2013;14:1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 11. Kerr P, Black J, von Frese J, et al: Molecular profiling from FFPE tissue: Stratification of the stage II colorectal cancer population. Mol Cancer Ther 6:3751S, 2007 (abstr C138)
- 12.Plamadeala V, Huang S, McCreary SM, et al. Analytical performance of a formalin-fixed paraffin-embedded tissue-based 634-probe prognostic assay for predicting outcome of patients with stage II colon cancer. Appl Immunohistochem Mol Morphol. 2014;22:308–316. doi: 10.1097/PDM.0000000000000031. [DOI] [PubMed] [Google Scholar]
- 13.Niedzwiecki D, Bertagnolli MM, Warren RS, et al. Documenting the natural history of patients with resected stage II adenocarcinoma of the colon after randomization of adjuvant treatment with edrecolomab or observation: results from CALGB 9581. J Clin Oncol. 2011;29:3146–3152. doi: 10.1200/JCO.2010.32.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prentice RL. A case-cohort design for epidemiologic cohort studies and disease prevention trials. Biometrika. 1986;73:1–11. [Google Scholar]
- 15.Gray RJ. Weighted analyses for cohort sampling designs. Lifetime Data Anal. 2009;15:24–40. doi: 10.1007/s10985-008-9095-z. [DOI] [PubMed] [Google Scholar]
- 16.Borgan O, Langholz B, Samuelsen SO, et al. Exposure stratified case-cohort designs. Lifetime Data Anal. 2000;6:39–58. doi: 10.1023/a:1009661900674. [DOI] [PubMed] [Google Scholar]
- 17.Cai J, Zeng D. Sample size and power calculation for case-cohort studies. Biometrics. 2004;60:1015–1024. doi: 10.1111/j.0006-341X.2004.00257.x. [DOI] [PubMed] [Google Scholar]
- 18. Therneau TR: Package “survival.” Repository CRAN. http://r-forge.r-project.org.
- 19.Self S, Prentice RL. Asymptotic distribution theory and efficiency results for case-cohort studies. Ann Stat. 1988;16:64–81. [Google Scholar]
- 20.Barlow WE, Ichikawa L, Rosner D, et al. Analysis of case-cohort designs. J Clin Epidemiol. 1999;52:1165–1172. doi: 10.1016/s0895-4356(99)00102-x. [DOI] [PubMed] [Google Scholar]
- 21. Benson AB 3rd, Hamilton SR. Path toward prognostication and prediction: An evolving matrix. J Clin Oncol 29:4599-4601, 2011. [DOI] [PubMed]
- 22.Quasar Collaborative Group. Gray R, Barnwell J, et al. Adjuvant chemotherapy versus observation in patients with colorectal cancer: A randomised study. Lancet. 2007;370:2020–2029. doi: 10.1016/S0140-6736(07)61866-2. [DOI] [PubMed] [Google Scholar]
- 23.Moertel CG, Fleming TR, Macdonald JS, et al. Intergroup study of fluorouracil plus levamisole as adjuvant therapy for stage II/Dukes’ B2 colon cancer. J Clin Oncol. 1995;13:2936–2943. doi: 10.1200/JCO.1995.13.12.2936. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.