Abstract
Pituitary FSH synthesis is regulated by TGFβ superfamily ligands; most notably the activins and inhibins. Bone morphogenetic proteins (BMPs) also regulate FSHβ subunit (Fshb) expression in immortalized murine gonadotrope-like LβT2 cells and in primary murine or ovine primary pituitary cultures. BMP2 signals preferentially via the BMP type I receptor, BMPR1A, to stimulate murine Fshb transcription in vitro. Here, we used a Cre-lox approach to assess BMPR1A’s role in FSH synthesis in mice in vivo. Gonadotrope-specific Bmpr1a knockout animals developed normally and had reproductive organ weights comparable to those of controls. Knockouts were fertile, with normal serum gonadotropins and pituitary gonadotropin subunit mRNA expression. Cre-mediated recombination of the floxed Bmpr1a allele was efficient and specific, as indicated by PCR analysis of diverse tissues and isolated gonadotrope cells. Furthermore, BMP2 stimulation of inhibitor of DNA binding 3 expression was impaired in gonadotropes isolated from Bmpr1a knockout mice, confirming the loss of functional receptor protein in these cells. Treatment of purified gonadotropes with small molecule inhibitors of BMPR1A (and the related receptors BMPR1B and ACVR1) suppressed Fshb mRNA expression, suggesting that an autocrine BMP-like molecule might regulate FSH synthesis. However, deletion of Bmpr1a and Acvr1 in cultured pituitary cells did not alter Fshb expression, indicating that the inhibitors had off-target effects. In sum, BMPs or related ligands acting via BMPR1A or ACVR1 are unlikely to play direct physiological roles in FSH synthesis by murine gonadotrope cells.
Keywords: Pituitary, FSH, bone morphogenetic protein, activin receptor-like kinase, Cre-lox
Introduction
The pituitary glycoprotein follicle-stimulating hormone (FSH) is an essential regulator of ovarian function and testicular development in mammals. FSH synthesis by pituitary gonadotrope cells is primarily stimulated by gonadotropin-releasing hormone (GnRH) from the hypothalamus and intra-pituitary autocrine/paracrine factors, the best studied of which are the activins (Bernard et al., 2010). Activins are members of the transforming growth factor β (TGFβ) superfamily and signal via complexes of serine/threonine kinase receptors, homolog of Drosophila mothers against decapentaplegic (SMAD) proteins, and forkhead box L2 to regulate transcription of the FSH β subunit gene (Fshb) (Weiss et al., 1995, Suszko et al., 2003, Bailey et al., 2004, Bernard, 2004, Gregory et al., 2005, Safwat et al., 2005, Suszko et al., 2005, Bernard et al., 2006, Lamba et al., 2006, Lamba et al., 2009, Lamba et al., 2010, Tran et al., 2011, Wang & Bernard, 2012, Bernard & Tran, 2013). In response to FSH signaling, the gonads synthesize and secrete inhibins, which act in endocrine fashion to suppress FSH synthesis by competitively binding to activin receptors (Lewis et al., 2000, Chapman et al., 2002).
Additional members of the TGFβ superfamily, the bone morphogenetic proteins (BMPs), have also been implicated in regulating Fshb expression both alone and in combination with other factors. For example, BMP6 and BMP7 (at 1 µg/ml) stimulate ovine Fshb promoter-reporter activity in pituitaries of transgenic mice or in transiently transfected murine gonadotrope-like LβT2 cells (Huang et al., 2001). In contrast, antisera against BMP7 suppress FSH release from murine, rat, and ovine primary pituitary cultures, suggesting a role for endogenous intra-pituitary BMP7 in FSH regulation. BMP15 similarly stimulates ovine Fshb promoter activity in LβT2 cells and FSH secretion from rat primary pituitary cell cultures, but at concentrations as low as 10–100 ng/ml (Otsuka & Shimasaki, 2002). BMP2 and BMP4 are ten-fold more potent in stimulating murine Fshb promoter-reporter activity than BMP6 or BMP7 in LβT2 cells (Lee et al., 2007). Moreover, BMP2 and activins synergistically stimulate murine, porcine, and ovine Fshb promoter-reporters as well as endogenous murine Fshb mRNA expression in this cell line. Similarly, BMP4 potentiates activin A and GnRH induction of Fshb mRNA expression and FSH release in LβT2 cells (Nicol et al., 2008), whereas BMP6 synergistically stimulates murine Fshb promoter-reporter activity with GnRH (Takeda et al., 2012). In contrast to these results, exogenous BMP4 and BMP6 suppress FSH secretion and Fshb mRNA expression in primary pituitary cultures from ewes and also counteract the stimulatory effects of activin A in this system (Faure et al., 2005, Young et al., 2008). Collectively, these data suggest that BMPs can regulate FSH at the level of the pituitary gland, though their effects may be ligand-, context-, and species-specific.
In LβT2 cells, BMP2 signals via the type I receptor, BMPR1A (also known as activin receptor-like kinase 3 or ALK3) to regulate murine Fshb transcription (Ho & Bernard, 2009). Though a second type I receptor, BMPR1B (ALK6), is also present in these cells (Lee et al., 2007, Nicol et al., 2008), it is expressed at low levels and BMP2 signaling is intact when this receptor is knocked down using short interfering RNAs (siRNAs) (Ho & Bernard, 2009). Similarly, in sheep, BMPR1A, but not BMPR1B, is expressed in gonadotrope cells (Faure et al., 2005), suggesting that BMP2 and BMP4 likely signal via BMPR1A in these animals as well. Here, we tested the hypothesis that signaling via BMPR1A is required for FSH synthesis in vivo by selectively ablating the receptor in murine gonadotropes using a Cre-lox approach.
Materials and Methods
Reagents
Human recombinant (rh−) BMP2 (355-BM) and activin A (338-AC) were from R&D Systems (Minneapolis, MN, USA). RQ1 RNase-Free DNase (M6101), random primers (C1181), MMLV-reverse transcriptase (M1701) and RNasin (N2511) were from Promega (Madison, WI, USA). SB431542 (S4317), pancreatin (P3292), and collagenase (Type I-C0130) were from Sigma (St. Louis, MO, USA). Media 199 (M199; 31100-035), Hanks’ Balanced Salt Solution (HBSS) without calcium/magnesium (14170-112), TRIzol Reagent, and SYBRgreen Supermix for qPCR were from Invitrogen (Burlington, ON, Canada). EvaGreen 2X qPCR MasterMix-S was from Applied Biological Materials Inc. (ABM, Richmond, BC, Canada). Oligonucleotides were purchased from IDT (Coralville, IA, USA). Gentamycin (450-135-XL), 100X antibiotic-antimycotic (450-115-EL) and deoxynucleotide triphosphates (dNTPs) were from Wisent (St-Bruno, Quebec, Canada). LDN 193189 hydrochloride (1509) was purchased from Axon MedChem (Reston, VA, USA) and compound C (171260) was from Calbiochem (EMD Chemicals Inc, Darmstadt, Germany).
Animals
The mouse strains used here have been described previously: Bmpr1a+/− (Mishina et al., 1995), Bmpr1afl/fl (Mishina et al., 2002), Acvr1fl/fl (Dudas et al., 2004) and GnRH-receptor-IRES-Cre (GRIC) (Wen et al., 2008). In the latter model, Cre recombinase is expressed as part of a bicistronic mRNA with the endogenous Gnrhr mRNA. Bmpr1a+/− males were crossed with GnrhrGRIC/GRIC females. Resulting Bmpr1a+/−;GnrhrGRIC/+ females were then crossed with Bmpr1afl/fl males to generate control (Bmpr1afl/+;GnrhrGRIC/+) and experimental animals (Bmpr1afl/−;GnrhrGRIC/+). The GRIC allele is active in the male germline (Wen et al., 2010); therefore, to avoid global recombination of the floxed allele, GRIC was always introduced from the female parent. The Bmpr1a+/−;GnrhrGRIC/+ × Bmpr1afl/fl cross generated animals of four genotypes at the expected frequencies (1:1:1:1). For genetic labeling of gonadotropes, GnrhrGRIC/GRIC mice were crossed with Rosa26-loxSTOPlox-EYFP (hereafter R26-YFP) reporter mice (Srinivas et al., 2001) acquired from Jackson labs. To label gonadotropes in control and experimental mice, we crossed Bmpr1a+/−;GnrhrGRIC/GRIC females with Bmpr1afl/fl;R26-YFP/R26-YFP males. Acvr1fl/fl and Bmpr1afl/fl mice were crossed to generate Acvr1fl/+;Bmpr1afl/+ and eventually Acvr1fl/fl;Bmpr1afl/flmice for in vitro recombination experiments. All animals were housed on a 12L:12D light cycle and were given ad libitum access to food and water. All mouse work was conducted in accordance with federal and institutional guidelines and with the approval of the McGill Animal Care and Use Committee (animal use protocol #5204).
DNA extraction and genotyping
Genomic DNA was extracted from tail biopsies (~0.5 cm) using 0.5 ml of lysis buffer [100 mmol l−1 Tris HCl (pH 8.5), 5 mmol l−1 EDTA (pH 8.0), 200 mmol l−1 NaCl, 0.2% (v/v) SDS, and 100 µg/ml proteinase K]. Tails were incubated at 55°C overnight in a water bath. Samples were then vortexed and centrifuged at 12,000 rpm for 10 min. The supernatant was collected and mixed by inversion with 0.5 ml isopropanol. Precipitated DNA was collected with a micropipette tip and dissolved in 40 µl of 10 mmol l−1Tris (pH 8.0). For comparison of recombination across different tissues, DNA was extracted from approximately 5 mg of the indicated tissues using the Gentra Puregene Blood Kit following the manufacturer's instructions (Qiagen). Wild-type, null, floxed, and recombined Bmpr1a alleles were detected by PCR using the protocols described in (Mishina et al., 1995, Mishina et al., 2002). The GRIC allele was detected using the primer set indicated in Table 1.
Table 1.
Genotyping and RT-qPCR primers
| Genotyping |
| Cre |
| Fwd: GGACATGTTCAGGGATCGCCAGGC |
| Rev: GCATAACCAGTGAAACAGCATTGCTG |
| Acvr1 |
| Fwd: CCCCCATTGAAGGTTTAGAGAGAC |
| Rev: CTAAGAGCCATGACAGAGGTTG |
| Bmpr1a |
| Fwd1: CTCTGAATTTCTAGTCCACATCTGC |
| Fwd2: AGACTGCCTTGGGAAAAGCGC |
| Rev: GGACTATGGACACACAATGGC |
| RT-qPCR |
| Acvr1 |
| Fwd: GCTGCATAGCAGATTTGGGC |
| Rev: CACTTCCGGAGCCATGTAGC |
| Bmpr1a |
| Fwd: CATCAGATTACTGGGAGCCTG |
| Rev: GGTATCCTCTGGTGCTAAAGTC |
| Bmpr1b |
| Fwd: GTCTCAGAGCTCGGGAAGTG |
| Rev: ATAGCGGCCTTTTCCAATCT |
| Fshb |
| Fwd: GTGCGGGCTACTGCTACACT |
| Rev: CAGGCAATCTTACGGTCTCG |
| Id1 |
| Fwd: GGTACTTGGTCTGTCGGAGC |
| Rev: GCAGGTCCCTGATGTAGTCG |
| Id3 |
| Fwd: TTAGCCAGGTGGAAATCCTG |
| Rev: TCAGTGGCAAAAGCTCCTCT |
| Lhb |
| Fwd: ACTGTGCCGGCCTGTCAACG |
| Rev: AGCAGCCGGCAGTACTCGGA |
| Rpl19 |
| Fwd: CGGGAATCCAAGAAGATTGA |
| Rev: TTCAGCTTGTGGATGTGCTC |
Hormone assays
Serum LH and FSH were measured by multiplex ELISA at the Ligand Assay and Analysis Core (LAAC) of the Center for Research in Reproduction at the University of Virginia. Both hormones were measured in singlet from 10 µl serum. The reportable ranges for LH and FSH were 0.24–30.0 ng/ml and 2.40–300.0 ng/ml, respectively.
Fluorescence activated cell sorting of genetically-labeled gonadotropes
Pituitaries from mice with YFP-labeled gonadotropes were enzymatically dispersed as described in (Ho et al., 2011) and single cell suspensions prepared in PBS. Cells were then passed through a 70 µm nozzle at 70 psi into a Becton Dickinson FACSAria Sorter in the McGill University Flow Cytometry Core Facility. Sorting was performed using FACSDiva software (v. 6.0). Gating was established on a forward and side scatter plot (for relative cell size and granularity, respectively) to exclude debris and cell clusters. A control without YFP labeled cells was run to establish a negative baseline profile using a single parameter histogram. The pituitary cells were then run and gated to sort YFP+ and YFP− cells. YFP was excited with a 488 nm argon ion laser and detected with a 530/30 band pass filter and a LP 502 dichroic mirror. Depending on the preparation, we obtained roughly 0.8–15 × 103 YFP+ and 4.6–18 × 104 YFP− cells per pituitary.
Gonadotrope cell culture
Purified gonadotropes were plated at a density of 4–5 ×104 cells per well in 96-well plates in Medium 199 with 10% fetal bovine serum (FBS), 1X antibiotic-antimycotic, and 120 µg/ml gentamicin. Cells were incubated at 37°C/5% CO2 for at least 16 h before treatment. Cells were treated in medium M199 plus 2% (v/v) FBS overnight with the indicated ligands and/or inhibitors. RNA and DNA were then collected with QIAGEN AllPrep DNA/RNA mini kits (cat. 80204) following the manufacturer’s instructions. DNA was analyzed as described above. RNA was analyzed by quantitative RT-PCR (below).
Primary pituitary cultures
Primary cultures were performed as previously described (Fortin et al., 2013). Briefly, pituitaries were collected from 10 week old Bmpr1afl/fl;Acvr1fl/fl male and female mice in M199 medium supplemented with 10% (v/v) fetal bovine serum (FBS). Pituitaries were washed three times in Hank’s Balanced Salt Solution (HBSS) with 150 µmol l−1 of CaCl2, cut several times with a scalpel, and digested in collagenase (1.5 mg/ml) (Sigma #C-0130; diluted in HBSS with 30 mg/ml BSA, pH 7.4, 40 µl/pituitary) at 37°C for 2 hours. The suspension was then washed with 5 ml calcium-free HBSS, centrifuged for 5 min at 1000 × g, and resuspended in pancreatin solution (Sigma P3292; 4.5 mg/ml in calcium-free HBSS; 40 µl/pituitary). Pancreatin digestion was performed in a 37°C water bath with manual agitation for 8 to 10 min. Finally, the cell suspension was washed three times in 5 ml M199 media containing 10% FBS and cells were seeded at density of 5 × 105/well in 48-well plates.
Adenoviral transduction and treatment of primary pituitary cultures
Primary cultures were transduced with adenoviruses as previously described (Fortin et al., 2014). Pituitary cultures were prepared as described above. After 24 hours, viral transductions were performed using adenoviruses that express enhanced green fluorescent protein (eGFP) or Cre-IRES-eGFP (Baylor College of Medicine Vector Development Laboratory, Houston, TX. USA) at a multiplicity of infection of 60 in M199 medium containing 10% (v/v) FBS. The following day, virus-containing medium was removed and replaced with medium containing 2% (v/v) FBS with 25 ng/ml activin A, 25 ng/ml BMP2, 1 µmol l−1 SB431542 or 1µmol l−1 LDN-193189 or DMSO as vehicle. All treatments were performed for 24 hours in duplicate and the experiment was repeated 5–7 times. Cells were harvested and RNA and DNA were extracted using the Qiagen Allprep DNA/RNA kit. RNA was eluted in 23 µl RNase-free water and reverse transcribed. The resulting cDNA was analyzed by qPCR.
Quantitative RT-PCR
Fshb and Lhb mRNA levels (normalized to Rpl19) in whole pituitary RNA were measured using the relative standard curve method with LβT2 cell RNA as standard, as previously described (Lamba et al., 2009). Fshb, Lhb, Id3, Bmpr1a, Bmpr1b, Acvr1a, and Id mRNA levels (normalized to Rpl19) in isolated gonadotropes or mixed pituitary cultures were measured using the 2−ΔΔCt method as described in (Ho & Bernard, 2010). Primer sequences are indicated in Table 1.
Statistics
Data from control and experimental mice and/or cell cultures were compared with t-tests or analyses of variance as indicated using Systat. Post-hoc pair-wise comparisons were made with Bonferroni corrections. Data were log transformed when variance were unequal. Significance was assessed relative to p< 0.05.
Results
Gonadotrope-specific Bmpr1a knockout mice
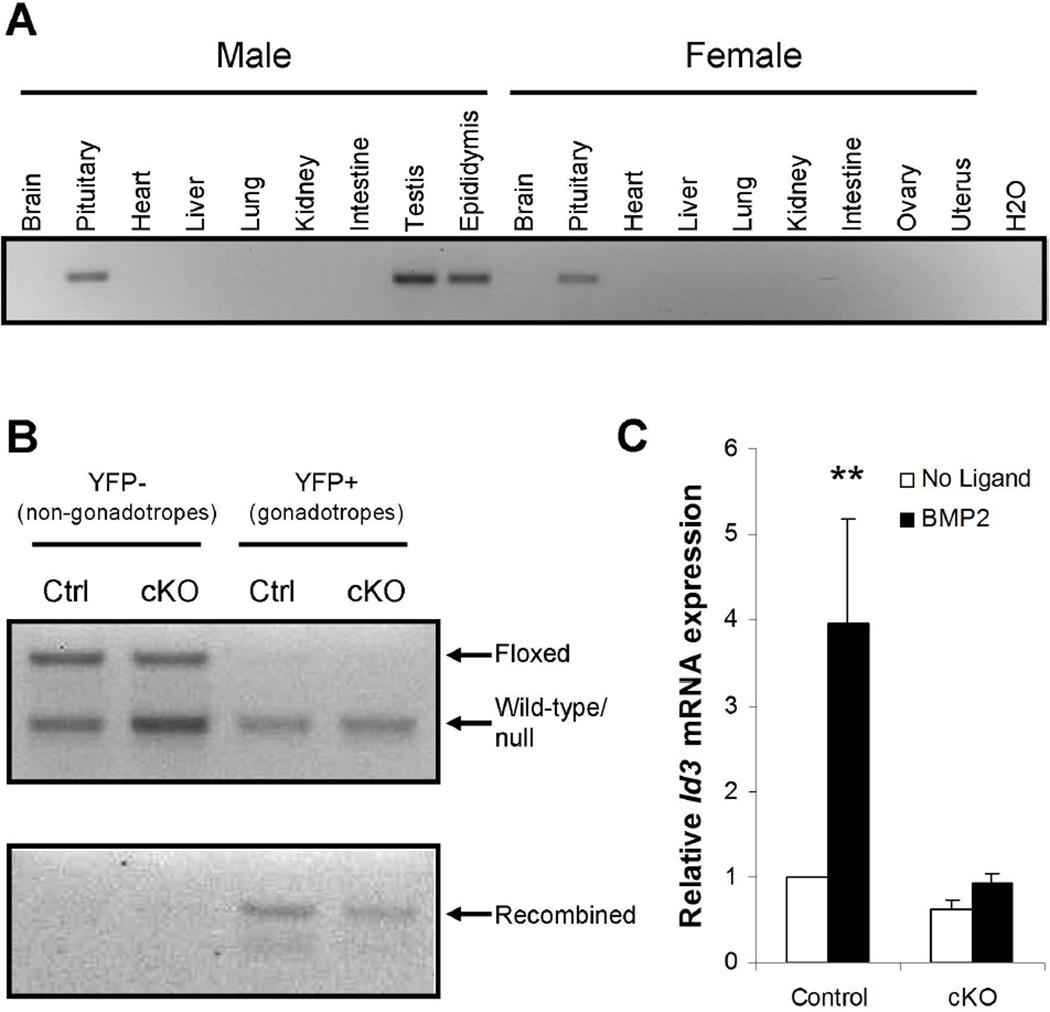
We generated gonadotrope-specific Bmpr1a knockout mice by crossing ‘floxed’ Bmpr1a animals with mice expressing Cre recombinase from the endogenous Gnrhr locus (so-called GRIC mice, see Materials and Methods). Because Bmpr1a+/− mice are viable and fertile, we elected to cross in a single null allele such that only one floxed Bmpr1a allele required recombination in our model. As a result, the experimental mice (hereafter conditional knockouts or cKO) had the following genotype: Bmpr1afl/−;GnrhrGRIC/+. Control littermates from the Bmpr1a+/−;GnrhrGRIC/+ × Bmpr1afl/fl cross used to generate cKO mice had the following genotype: Bmpr1afl/+;GnrhrGRIC/+. These animals allowed us to control for potential effects of the GRIC allele. Recombination of a single floxed allele in gonadotropes of these animals was not a concern because, as mentioned above, Bmpr1a+/− mice are fertile. Mice of the two other genotypes derived from this cross (Bmpr1afl/+;Gnrhr+/+ or Bmpr1afl/−;Gnrhr+/+) were not analyzed. The GRIC allele is active in pituitary gonadotropes and in male germ cells. Therefore, as anticipated, we observed recombination of the floxed Bmpr1a allele in pituitaries, testes, and epididymides, but not in other tissues, including ovaries and uteri of adult cKOs (Fig. 1A).
Fig. 1.
To further demonstrate the efficacy of recombination, we genetically labeled and purified gonadotropes from control and cKO mice. Here, an enhanced yellow fluorescent protein (eYFP) reporter allele, which is activated by Cre recombinase-mediated removal of a transcriptional stop cassette, was introduced into the genetic background of the control and cKO strains. Their pituitaries were extracted and enzymatically dissociated. eYFP-labeled (YFP+) cells were then separated from eYFP-negative (YFP−) cells by FACS. DNA was extracted from the purified cell populations and analyzed by PCR. The data confirm efficient recombination of the floxed Bmpr1a allele in YFP+, but not YFP- cells in males of both genotypes (Fig. 1B; recall that both control and cKO mice harbor a single floxed Bmpr1a allele; therefore, recombination is predicted in both genotypes). Similar results were obtained in females (data not shown).
We previously showed that BMP2 signals via BMPR1A to regulate Id3 expression in LβT2 cells and in primary pituitary cell cultures (Ho et al., 2011). Therefore, to confirm functional deletion of BMPR1A, we treated purified gonadotropes from control and cKO mice in primary culture with 25 ng/ml BMP2 for 24 h. BMP2-stimulated Id3 mRNA expression was significantly reduced in cKO relative to control gonadotropes (Fig. 1C).
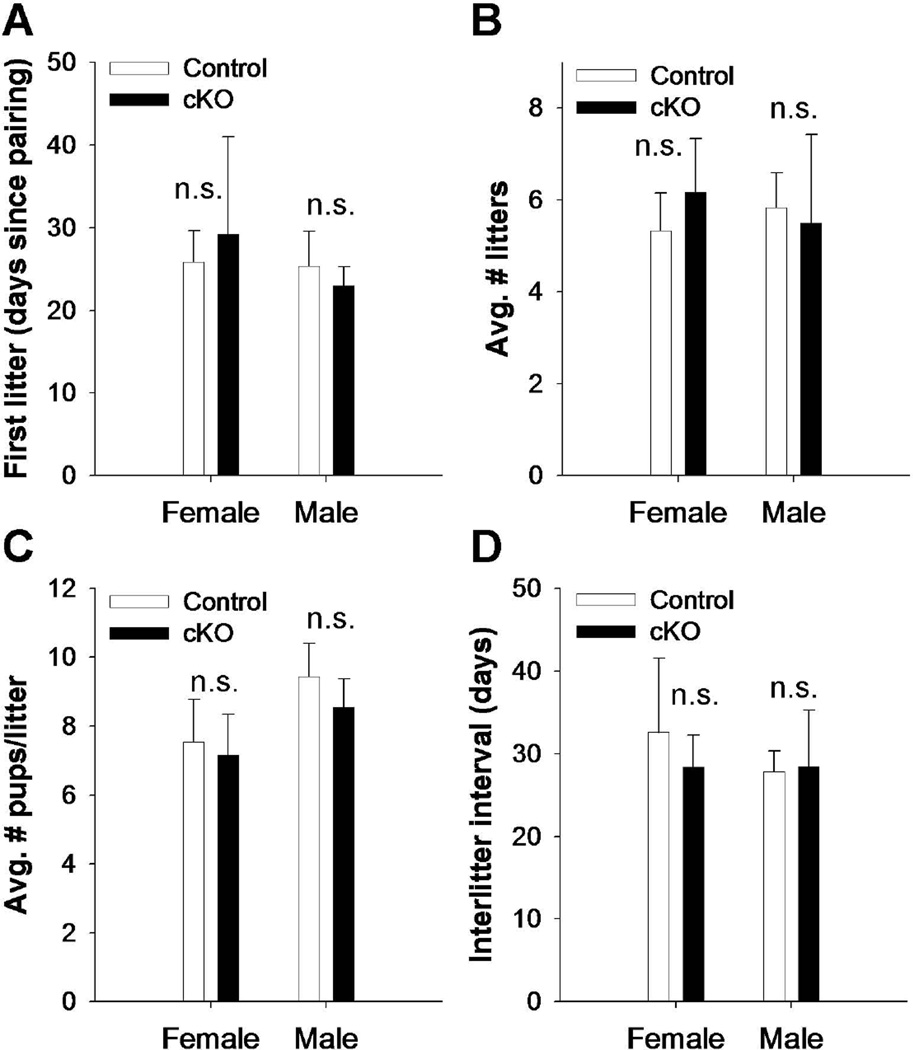
Fertility is normal in Bmpr1a cKO mice
Eight to 10 week old control and cKO males and females were paired with wild-type C57BL/6 (Charles River) opposite sex partners for a period of six months to assess their fertility. As shown in Fig. 2, there were no significant differences between genotypes in the number of days from pairing to delivery of the first litter, the average numbers of litters per animal, the average number of pups per litter, or the inter-litter interval.
Fig. 2.
Reproductive tissues and hormone levels are normal in Bmpr1a cKO mice
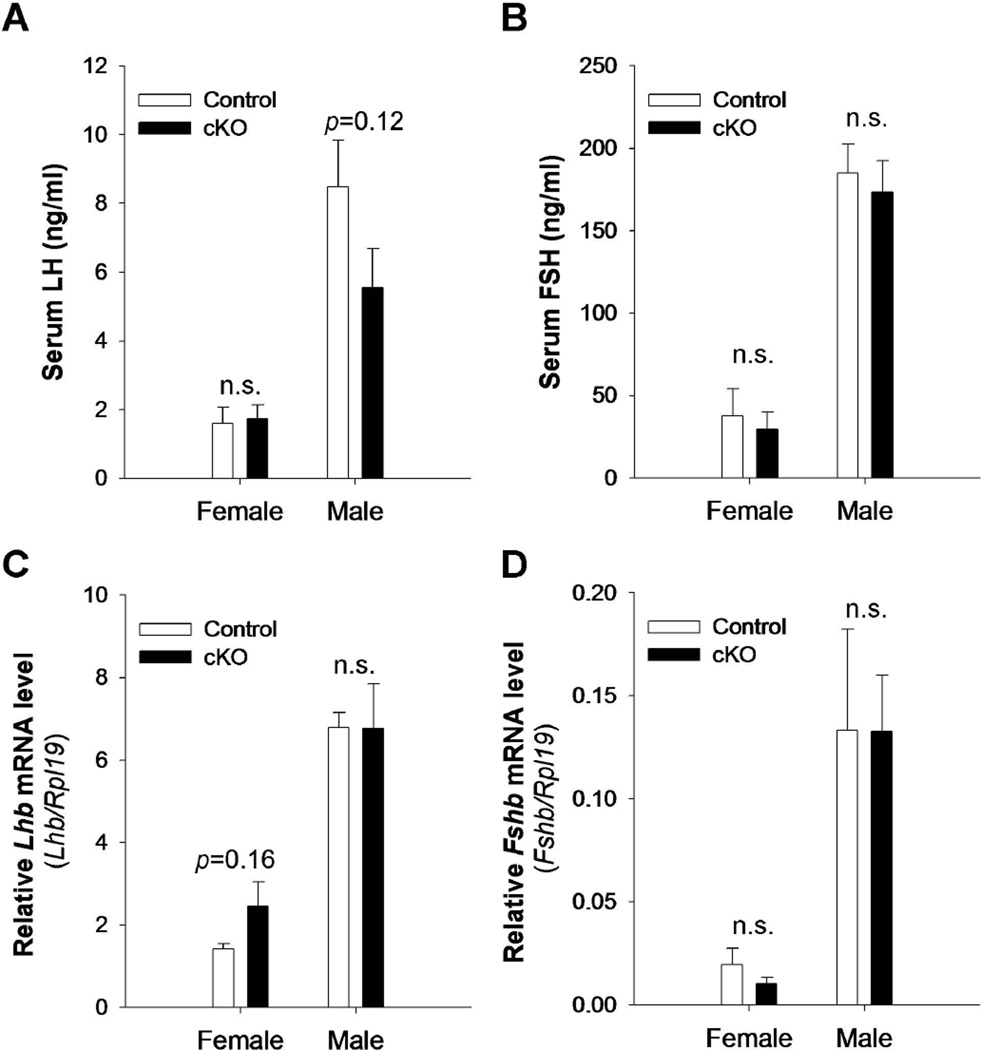
We next measured reproductive organ weights (Table 2), serum gonadotropin levels (Figs. 3A and B), and pituitary Lhb and Fshb mRNA levels (Figs. 3C and D) in post-pubertal (6-week old) control and cKO mice of both sexes. Again, we did not observe any differences between genotypes in any of these parameters, with the exception of testicular weight, which was slightly elevated in cKO mice.
Table 2.
Body and reproductive organ weights
| Sex | Genotype | N | Body wt (g) | Ovarian wt (g)* | Uterine wt (g) | |||
| F | Control | 10 | 17.65 ± 0.26 | p>0.1 | 0.0084 ± 0.001 | p>0.1 | 0.110 ± 0.013 | p>0.1 |
| F | cKO | 10 | 18.27 ± 0.45 | 0.011 ± 0.001a | 0.092 ± 0.013 | |||
| Sex | Genotype | N | Body wt (g) | Testicular wt (g)* | ||||
| M | Control | 11 | 21.36 ± 0.7 | p>0.1 | 0.168 ± 0.006 | p<0.01 | ||
| M | cKO | 14 | 22.68 ± 0.8 | 0.206 ± 0.008 |
Notes: Data are means ± SEM.
, Paired organ weights. Data were analyzed with unpaired t-tests. p values are presented for each comparison.
Fig. 3.
BMP2 does not stimulate Fshb mRNA expression in purified gonadotropes
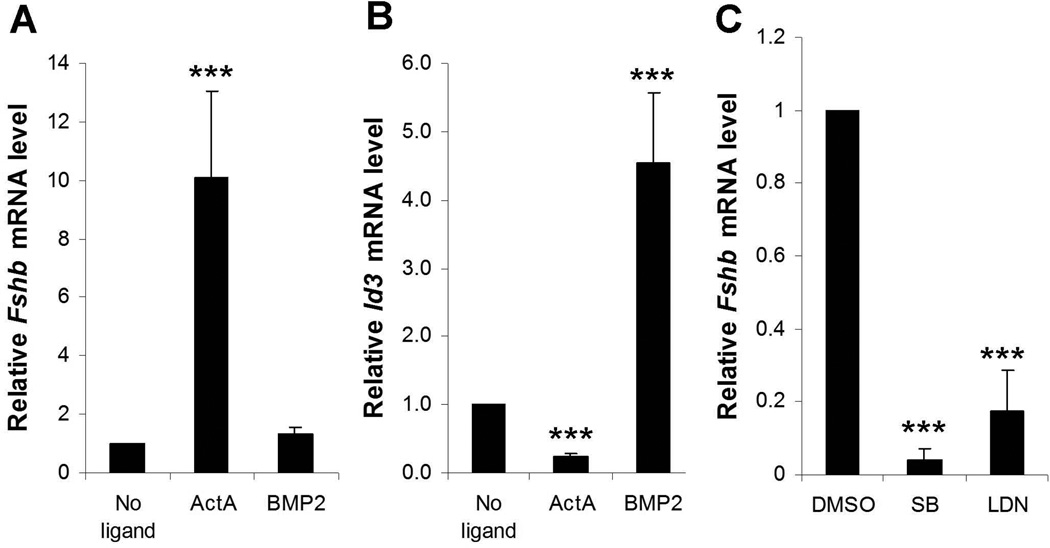
Previous analyses of BMP2-regulated Fshb expression were conducted almost entirely in the LβT2 cell model (Lee et al., 2007, Ho & Bernard, 2009, 2010, Ho et al., 2011). We therefore asked whether BMP2 similarly regulates Fshb expression in isolated murine gonadotropes of wild-type mice. Whereas activin A robustly stimulated Fshb mRNA in these cells, BMP2 had no effect (Fig. 4A). In contrast, BMP2 stimulated and activin A inhibited Id3 mRNA expression in these cultures (Fig. 4B). Thus, BMP2 was active in primary gonadotropes, but did not regulate the Fshb gene therein.
Fig. 4.
Inhibitors of BMP type I receptors suppress Fshb expression in primary gonadotropes
Activin B is generally regarded as the primary autocrine/paracrine stimulator of Fshb transcription (Corrigan et al., 1991, Sallon et al., 2010). Consistent with this idea, treatment of isolated gonadotropes from wild-type mice with a small molecule inhibitor (SB431542) (Inman et al., 2002) of the known activin B type I receptors (ACVR1B and ACVR1C) (Tsuchida et al., 2004, Bernard et al., 2006) dramatically reduced Fshb expression (Fig. 4C). To determine whether autocrine/paracrine BMP-like molecules (other than BMP2) might also play a role in Fshb expression, we also treated gonadotropes with LDN-193189 (Cuny et al., 2008). This molecule blocks the activities of ACVR1, BMPR1A, and BMPR1B, the three best characterized BMP type I receptors (for convenience, these receptors will be referred to as ALK2/3/6). LDN-193189 also suppressed Fshb mRNA levels (Fig. 4C). We obtained similar results with a second ALK2/3/6 inhibitor, compound C [also known as dorsomorphin; (Yu et al., 2008)] (data not shown).
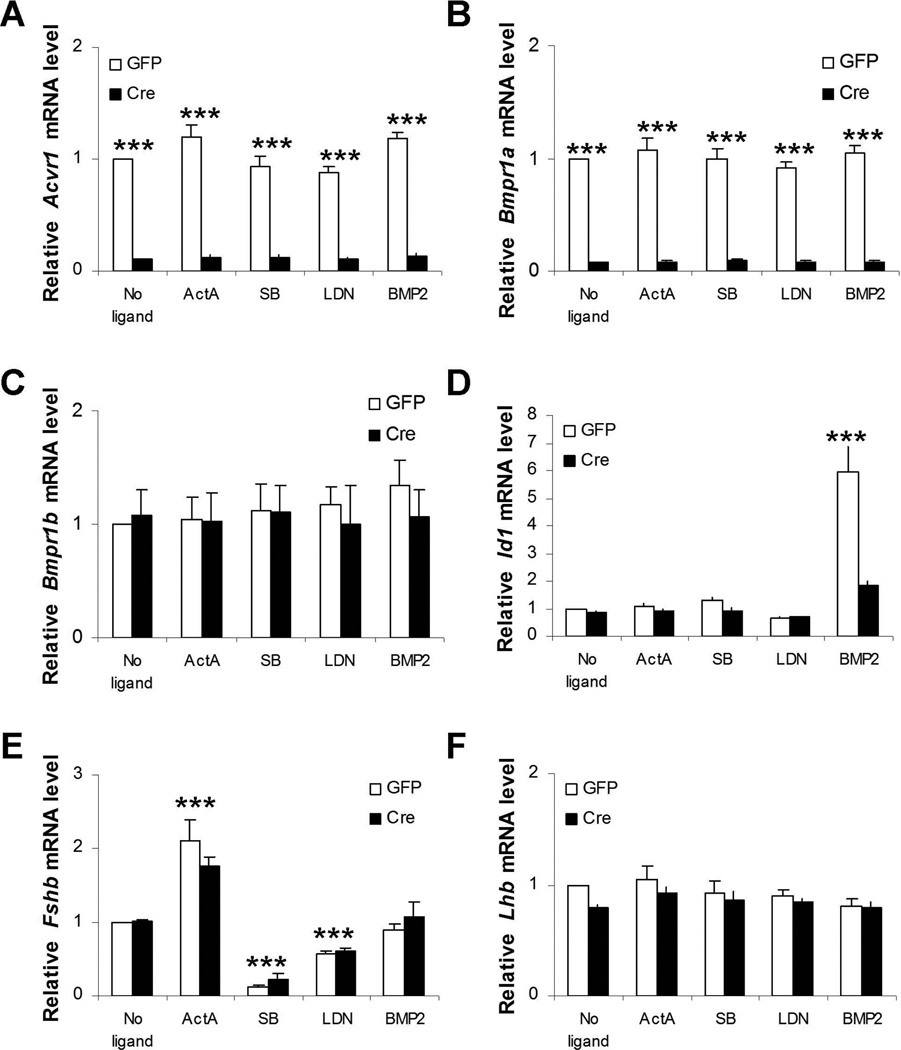
Fshb production by primary pituitary cultures is ACVR1/BMPR1A-independent
Though these data suggested a role for endogenous BMP-like molecules (other than BMP2) in FSH synthesis, LDN-193189 and compound C are known to antagonize other kinases in cells (Vogt et al., 2011). To assess potential ‘off-target’ effects, we examined inhibitor activity in pituitary cells expressing or lacking BMPR1A and ACVR1. It should be noted that BMPR1B/ALK6 is expressed at very low levels [if at all (Yi et al., 2001)] in the pituitary and in these cultures. Though we did not perform absolute quantification, the Ct values for Bmpr1b were 3–5 cycles higher than for Acvr1 or Bmpr1a; a difference of 8 to 32 fold. Similarly, a recent RNA-seq analysis indicates that Bmpr1a and Acvr1 mRNA levels are ~32 and ~16 fold greater than Bmpr1b in purified murine gonadotropes (Qiao et al., 2016). We prepared pituitary cultures from mice harboring floxed alleles for both BMPR1A (ALK3) and ACVR1 (ALK2) (Bmpr1afl/fl;Acvr1fl/fl). In these experiments, the cultures were prepared from whole pituitaries rather than from purified gonadotropes (it was not feasible to perform these experiments in purified gonadotropes). Half of the cells were infected with a control adenovirus expressing GFP. The other half was infected with a Cre-expressing adenovirus to recombine the floxed alleles. As shown in Fig. 5A and B, Acvr1 and Bmpr1a mRNA levels were significantly depleted in cells transduced with the Cre adenovirus. There was no compensatory increase in Bmpr1b mRNA expression (Fig. 5C). Stimulation of Id1 expression by BMP2 (Hollnagel et al., 1999) was significantly impaired in Cre adenovirus treated cells, demonstrating the loss of functional type I receptor proteins (Fig. 5D). The loss of the receptors, however, did not alter (reduce) basal Fshb mRNA expression (Fig. 5E). Thus, endogenous TGFβ superfamily ligands regulate Fshb expression independently of these receptors. Importantly, both LDN-193189 and compound C impaired Fshb mRNA expression in mixed pituitary cultures (though to a lesser extent than in purified gonadotropes; compare Fig. 5E with Fig. 4C) whether the Bmpr1a and Acvr1 alleles were recombined or not. Therefore, these inhibitors appear to suppress Fshb expression in BMPR1A/ACVR1-independent fashion. Again, activin A, but not BMP2, stimulated Fshb mRNA levels in these cultures (Fig. 5E). None of the treatments affected Lhb expression (Fig. 5F).
Fig. 5.
Discussion
The data presented here indicate that BMPR1A is dispensable for normal gonadotropin synthesis and fertility in mice in vivo. Though these observations do not definitively rule out a role for BMP signaling in FSH regulation, they do suggest that any biologically relevant BMP-like ligands signal via an alternative type I receptor in the absence of BMPR1A (i.e., there is some form of compensation) and/or that these ligands actually prefer a receptor other than BMPR1A.
We focused on BMPR1A for in vivo analyses based on earlier results with BMP2 and BMP4 in LβT2 cells (Lee et al., 2007, Ho & Bernard, 2009, 2010, Ho et al., 2011) and with BMP4 in ovine pituitary cultures (Faure et al., 2005, Young et al., 2008). Both of these ligands preferentially signal via BMPR1A and BMPR1B compared to the other type I receptors in the family (ten Dijke et al., 1994, Liu et al., 1995). BMPR1B is expressed at low levels, if at all, in gonadotropes (Lee et al., 2007, Nicol et al., 2008, Qiao et al., 2016) and BMP2 preferentially signals via BMPR1A in LβT2 cells to regulate both Fshb and Id3 transcription (Ho & Bernard, 2009, Ho et al., 2011). The latter data were collected in the context of transient transfection experiments, which may have precluded compensatory mechanisms from developing. In the in vivo knockout model presented here, Cre expression occurs as early as embryonic day 12.75 (Wen et al., 2010) and we only examined mice in adulthood. As a result, it is possible that another receptor(s) could have compensated for the absence of BMPR1A during this protracted time frame. That said, Bmpr1b mRNA levels are low in isolated murine gonadotropes under normal conditions (Qiao et al., 2016) and do not increase in Bmpr1a cKO mice (data not shown).
BMP2 and BMP4 can also signal via the type I receptor ACVR1 (ALK2) (Liu et al., 1995, Macias-Silva et al., 1998), which is expressed in murine gonadotropes (Qiao et al., 2016). Nonetheless, BMP2 signaling in isolated gonadotropes from Bmpr1a cKO mice was greatly impaired (e.g., Fig. 1C), arguably ruling out a compensatory role for ACVR1 or another type I receptor, at least for this particular BMP ligand.
To determine whether or not ACVR1 can compensate for the loss of BMPR1A or is perhaps the preferred type I receptor for the biologically relevant BMP ligands in vivo, one could conditionally ablate the receptor (Kaartinen & Nagy, 2001, Dudas et al., 2004) in gonadotropes either alone or in combination with BMPR1A (Yoon et al., 2005, Orvis et al., 2008, Edson et al., 2010). We have not systematically analyzed such mice to the extent that we have with Bmpr1a cKOs. However, we did generate a small number of GnrhrGRIC/+;Bmpr1afl/fl;Acvr1fl/fl, which lack both receptors in gonadotropes. Thus far, double knockout females produce litters of normal size. This would suggest that FSH (and LH) synthesis is unimpaired, though we have not assessed this directly. Indeed, in light of these preliminary observations and the results of the in vitro recombination experiments presented in Fig. 5, a thorough analysis of the double knockouts appears to us to be unjustified.
We pursued BMP2 and BMPR1A (by extension) because BMP2 and BMP4 were 10-fold more potent than BMP6 or BMP7 in stimulating Fshb transcription in LβT2 cells (Lee et al., 2007). As BMP2 and BMP4 are expressed at relatively low levels in gonadotropes (Lee et al., 2007, Qiao et al., 2016), we argued that they would most likely regulate Fshb in paracrine fashion. However, in light of our observations here with conditional Bmpr1a knockout mice and previous data implicating endogenous BMP7 in FSH regulation (Huang et al., 2001), it is fair to question whether a singular focus on BMP2 and its canonical receptor BMPR1A might have been limiting. That is, perhaps autocrine BMP7 is the primary BMP ligand regulating FSH in vivo. Indeed, Bmp7 is among the most highly expressed BMP ligands in isolated gonadotropes (Qiao et al., 2016). However, ACVR1 is the preferred BMP7 type I receptor (Macias-Silva et al., 1998) and the recombination experiments in Fig. 5 rule out a role for an endogenous ligand that signals via this receptor in Fshb synthesis, at least in primary culture.
In sum, though in vitro analyses in the gonadotrope-like LβT2 cell line suggested a role for BMP2 as a regulator of Fshb transcription both alone and in synergy with activins, the data presented here demonstrate that the ligand fails to stimulate Fshb expression in isolated primary gonadotropes and that its primary type I receptor, BMPR1A, is not required for normal FSH production or fertility in mice. Small molecule BMPR1A, BMPR1B, and ACVR1 inhibitors suppressed Fshb expression in primary cells, suggesting a role for endogenous BMPs or related ligands in FSH synthesis. However, it now appears that the inhibitors’ effects were likely independent of BMP signaling as the genetic deletion of BMPR1A and ACVR1 together did not alter Fshb production in the same cells. In light of these data, we suggest that studies on the direct regulation of FSH synthesis by TGFβ superfamily ligands should focus on activins and related proteins that signal via ACVR1B (ALK4), TGFBR1 (ALK5), and/or ACVR1C (ALK7).
Acknowledgments
The authors thank Ken McDonald from the McGill University Flow Cytometry Core Facility for technical assistance with cell sorting. The LAAC of the Center for Research in Reproduction at the University of Virginia is supported by the Eunice Kennedy Shriver NICHD/NIH (SCCPIR) Grant U54-HD28934.
Grant support
Canadian Institutes of Health Research operating grant MOP-133394 and Natural Sciences and Engineering Research Council of Canada Discovery grant 2015-05178 to DJB. National Institutes of Health R01DE020843 to YM.
Footnotes
Declaration of interest
The authors have no conflicts of interest to disclose.
Author contributions
XZ performed all of the in vivo experiments; prepared, treated, and analyzed the pituitary cultures; and edited the manuscript. YW performed several of the RT-qPCR analyses and edited the manuscript. LO wrote sections of the manuscript and edited the final version. UB, VK, and YM produced the floxed and Cre strains, provided advice regarding experimental design, and edited the manuscript. DJB designed the experiments, analyzed the data, generated the final figures and tables, wrote the original draft of manuscript, and edited and formatted the final version of the manuscript.
References
- Bailey JS, Rave-Harel N, McGillivray SM, Coss D, Mellon PL. Activin regulation of the follicle-stimulating hormone beta-subunit gene involves Smads and the TALE homeodomain proteins Pbx1 and Prep1. Mol Endocrinol. 2004;18:1158–1170. doi: 10.1210/me.2003-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard DJ. Both SMAD2 and SMAD3 mediate activin-stimulated expression of the follicle-stimulating hormone beta subunit in mouse gonadotrope cells. Mol Endocrinol. 2004;18:606–623. doi: 10.1210/me.2003-0264. [DOI] [PubMed] [Google Scholar]
- Bernard DJ, Tran S. Mechanisms of activin-stimulated FSH synthesis: the story of a pig and a FOX. Biol Reprod. 2013;88:78. doi: 10.1095/biolreprod.113.107797. [DOI] [PubMed] [Google Scholar]
- Bernard DJ, Lee KB, Santos MM. Activin B can signal through both ALK4 and ALK7 in gonadotrope cells. Reprod Biol Endocrinol. 2006;4:52. doi: 10.1186/1477-7827-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard DJ, Fortin J, Wang Y, Lamba P. Mechanisms of FSH synthesis: what we know, what we don't, and why you should care. Fertil Steril. 2010;93:2465–2485. doi: 10.1016/j.fertnstert.2010.03.034. [DOI] [PubMed] [Google Scholar]
- Chapman SC, Bernard DJ, Jelen J, Woodruff TK. Properties of inhibin binding to betaglycan, InhBP/p120 and the activin type II receptors. Mol Cell Endocrinol. 2002;196:79–93. doi: 10.1016/s0303-7207(02)00227-7. [DOI] [PubMed] [Google Scholar]
- Corrigan AZ, Bilezikjian LM, Carroll RS, Bald LN, Schmelzer CH, Fendly BM, Mason AJ, Chin WW, Schwall RH, Vale W. Evidence for an autocrine role of activin B within rat anterior pituitary cultures. Endocrinology. 1991;128:1682–1684. doi: 10.1210/endo-128-3-1682. [DOI] [PubMed] [Google Scholar]
- Cuny GD, Yu PB, Laha JK, Xing X, Liu JF, Lai CS, Deng DY, Sachidanandan C, Bloch KD, Peterson RT. Structure-activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg Med Chem Lett. 2008;18:4388–4392. doi: 10.1016/j.bmcl.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas M, Sridurongrit S, Nagy A, Okazaki K, Kaartinen V. Craniofacial defects in mice lacking BMP type I receptor Alk2 in neural crest cells. Mechanisms of Development. 2004;121:173–182. doi: 10.1016/j.mod.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Edson MA, Nalam RL, Clementi C, Franco HL, Demayo FJ, Lyons KM, Pangas SA, Matzuk MM. Granulosa cell-expressed BMPR1A and BMPR1B have unique functions in regulating fertility but act redundantly to suppress ovarian tumor development. Mol Endocrinol. 2010;24:1251–1266. doi: 10.1210/me.2009-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure MO, Nicol L, Fabre S, Fontaine J, Mohoric N, McNeilly A, Taragnat C. BMP-4 inhibits follicle-stimulating hormone secretion in ewe pituitary. J Endocrinol. 2005;186:109–121. doi: 10.1677/joe.1.05988. [DOI] [PubMed] [Google Scholar]
- Fortin J, Boehm U, Deng CX, Treier M, Bernard DJ. Follicle-stimulating hormone synthesis and fertility depend on SMAD4 and FOXL2. Faseb J. 2014;28:3396–3410. doi: 10.1096/fj.14-249532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J, Kumar V, Zhou X, Wang Y, Auwerx J, Schoonjans K, Boehm U, Boerboom D, Bernard DJ. NR5A2 regulates Lhb and Fshb transcription in gonadotrope-like cells in vitro, but is dispensable for gonadotropin synthesis and fertility in vivo. PLoS One. 2013;8:e59058. doi: 10.1371/journal.pone.0059058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory SJ, Lacza CT, Detz AA, Xu S, Petrillo LA, Kaiser UB. Synergy between activin A and gonadotropin-releasing hormone in transcriptional activation of the rat follicle-stimulating hormone-beta gene. Mol Endocrinol. 2005;19:237–254. doi: 10.1210/me.2003-0473. [DOI] [PubMed] [Google Scholar]
- Ho CC, Bernard DJ. Bone morphogenetic protein 2 signals via BMPR1A to regulate murine follicle-stimulating hormone beta subunit transcription. Biol Reprod. 2009;81:133–141. doi: 10.1095/biolreprod.108.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CC, Bernard DJ. Bone morphogenetic protein 2 acts via inhibitor of DNA binding proteins to synergistically regulate follicle-stimulating hormone beta transcription with activin A. Endocrinology. 2010;151:3445–3453. doi: 10.1210/en.2010-0071. [DOI] [PubMed] [Google Scholar]
- Ho CC, Zhou X, Mishina Y, Bernard DJ. Mechanisms of bone morphogenetic protein 2 (BMP2) stimulated inhibitor of DNA binding 3 (Id3) transcription. Mol Cell Endocrinol. 2011;332:242–252. doi: 10.1016/j.mce.2010.10.019. [DOI] [PubMed] [Google Scholar]
- Hollnagel A, Oehlmann V, Heymer J, Ruther U, Nordheim A. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem. 1999;274:19838–19845. doi: 10.1074/jbc.274.28.19838. [DOI] [PubMed] [Google Scholar]
- Huang HJ, Wu JC, Su P, Zhirnov O, Miller WL. A novel role for bone morphogenetic proteins in the synthesis of follicle-stimulating hormone. Endocrinology. 2001;142:2275–2283. doi: 10.1210/endo.142.6.8159. [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- Kaartinen V, Nagy A. Removal of the floxed neo gene from a conditional knockout allele by the adenoviral Cre recombinase in vivo. Genesis. 2001;31:126–129. doi: 10.1002/gene.10015. [DOI] [PubMed] [Google Scholar]
- Lamba P, Santos MM, Philips DP, Bernard DJ. Acute regulation of murine follicle-stimulating hormone beta subunit transcription by activin A. J Mol Endocrinol. 2006;36:201–220. doi: 10.1677/jme.1.01961. [DOI] [PubMed] [Google Scholar]
- Lamba P, Fortin J, Tran S, Wang Y, Bernard DJ. A novel role for the forkhead transcription factor FOXL2 in activin A-regulated follicle-stimulating hormone beta subunit transcription. Mol Endocrinol. 2009;23:1001–1013. doi: 10.1210/me.2008-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba P, Wang Y, Tran S, Ouspenskaia T, Libasci V, Hebert TE, Miller GJ, Bernard DJ. Activin A regulates porcine follicle-stimulating hormone beta-subunit transcription via cooperative actions of SMADs and FOXL2. Endocrinology. 2010;151:5456–5467. doi: 10.1210/en.2010-0605. [DOI] [PubMed] [Google Scholar]
- Lee KB, Khivansara V, Santos MM, Lamba P, Yuen T, Sealfon SC, Bernard DJ. Bone morphogenetic protein 2 and activin A synergistically stimulate follicle-stimulating hormone {beta} subunit transcription. J Mol Endocrinol. 2007;38:315–330. doi: 10.1677/jme.1.02196. [DOI] [PubMed] [Google Scholar]
- Lewis KA, Gray PC, Blount AL, MacConell LA, Wiater E, Bilezikjian LM, Vale W. Betaglycan binds inhibin and can mediate functional antagonism of activin signalling. Nature. 2000;404:411–414. doi: 10.1038/35006129. [DOI] [PubMed] [Google Scholar]
- Liu F, Ventura F, Doody J, Massague J. Human type II receptor for bone morphogenic proteins (BMPs): extension of the two-kinase receptor model to the BMPs. Mol Cell Biol. 1995;15:3479–3486. doi: 10.1128/mcb.15.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias-Silva M, Hoodless PA, Tang SJ, Buchwald M, Wrana JL. Specific Activation of Smad1 Signaling Pathways by the BMP7 Type I Receptor, ALK2. J. Biol. Chem. 1998;273:25628–25636. doi: 10.1074/jbc.273.40.25628. [DOI] [PubMed] [Google Scholar]
- Mishina Y, Suzuki A, Ueno N, Behringer RR. Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 1995;9:3027–3037. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- Mishina Y, Hanks MC, Miura S, Tallquist MD, Behringer RR. Generation of Bmpr/Alk3 conditional knockout mice. Genesis. 2002;32:69–72. doi: 10.1002/gene.10038. [DOI] [PubMed] [Google Scholar]
- Nicol L, Faure MO, McNeilly JR, Fontaine J, Taragnat C, McNeilly AS. Bone morphogenetic protein-4 interacts with activin and GnRH to modulate gonadotrophin secretion in LbetaT2 gonadotrophs. J Endocrinol. 2008;196:497–507. doi: 10.1677/JOE-07-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvis GD, Jamin SP, Kwan KM, Mishina Y, Kaartinen VM, Huang S, Roberts AB, Umans L, Huylebroeck D, Zwijsen A, et al. Functional redundancy of TGF-beta family type I receptors and receptor-Smads in mediating anti-Mullerian hormone-induced Mullerian duct regression in the mouse. Biol Reprod. 2008;78:994–1001. doi: 10.1095/biolreprod.107.066605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka F, Shimasaki S. A novel function of bone morphogenetic protein-15 in the pituitary: selective synthesis and secretion of FSH by gonadotropes. Endocrinology. 2002;143:4938–4941. doi: 10.1210/en.2002-220929. [DOI] [PubMed] [Google Scholar]
- Qiao S, Nordstrom K, Muijs L, Gasparoni G, Tierling S, Krause E, Walter J, Boehm U. Molecular Plasticity of Male and Female Murine Gonadotropes Revealed by mRNA Sequencing. Endocrinology. 2016;157:1082–1093. doi: 10.1210/en.2015-1836. [DOI] [PubMed] [Google Scholar]
- Safwat N, Ninomiya-Tsuji J, Gore AJ, Miller WL. Transforming growth factor beta-activated kinase 1 is a key mediator of ovine follicle-stimulating hormone beta-subunit expression. Endocrinology. 2005;146:4814–4824. doi: 10.1210/en.2005-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallon C, Faure MO, Fontaine J, Taragnat C. Dynamic regulation of pituitary mRNAs for bone morphogenetic protein (BMP) 4, BMP receptors, and activin/inhibin subunits in the ewe during the estrous cycle and in cultured pituitary cells. J Endocrinol. 2010;207:55–65. doi: 10.1677/JOE-10-0035. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suszko MI, Balkin DM, Chen Y, Woodruff TK. Smad3 mediates activin-induced transcription of follicle-stimulating hormone beta-subunit gene. Mol Endocrinol. 2005;19:1849–1858. doi: 10.1210/me.2004-0475. [DOI] [PubMed] [Google Scholar]
- Suszko MI, Lo DJ, Suh H, Camper SA, Woodruff TK. Regulation of the rat follicle-stimulating hormone beta-subunit promoter by activin. Mol Endocrinol. 2003;17:318–332. doi: 10.1210/me.2002-0081. [DOI] [PubMed] [Google Scholar]
- Takeda M, Otsuka F, Takahashi H, Inagaki K, Miyoshi T, Tsukamoto N, Makino H, Lawson MA. Interaction between gonadotropin-releasing hormone and bone morphogenetic protein-6 and-7 signaling in LbetaT2 gonadotrope cells. Mol Cell Endocrinol. 2012;348:147–154. doi: 10.1016/j.mce.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dijke P, Yamashita H, Sampath TK, Reddi AH, Estevez M, Riddle DL, Ichijo H, Heldin CH, Miyazono K. Identification of type I receptors for osteogenic protein-1 and bone morphogenetic protein-4. The Journal Of Biological Chemistry. 1994;269:16985–16988. [PubMed] [Google Scholar]
- Tran S, Lamba P, Wang Y, Bernard DJ. SMADs and FOXL2 synergistically regulate murine FSHbeta transcription via a conserved proximal promoter element. Mol Endocrinol. 2011;25:1170–1183. doi: 10.1210/me.2010-0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida K, Nakatani M, Yamakawa N, Hashimoto O, Hasegawa Y, Sugino H. Activin isoforms signal through type I receptor serine/threonine kinase ALK7. Mol Cell Endocrinol. 2004;220:59–65. doi: 10.1016/j.mce.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Vogt J, Traynor R, Sapkota GP. The specificities of small molecule inhibitors of the TGFss and BMP pathways. Cell Signal. 2011;23:1831–1842. doi: 10.1016/j.cellsig.2011.06.019. [DOI] [PubMed] [Google Scholar]
- Wang Y, Bernard DJ. Activin A induction of murine and ovine follicle-stimulating hormone beta transcription is SMAD-dependent and TAK1 (MAP3K7)/p38 MAPK-independent in gonadotrope-like cells. Cell Signal. 2012;24:1632–1640. doi: 10.1016/j.cellsig.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Weiss J, Guendner MJ, Halvorson LM, Jameson JL. Transcriptional activation of the follicle-stimulating hormone beta-subunit gene by activin. Endocrinology. 1995;136:1885–1891. doi: 10.1210/endo.136.5.7720634. [DOI] [PubMed] [Google Scholar]
- Wen S, Ai W, Alim Z, Boehm U. Embryonic gonadotropin-releasing hormone signaling is necessary for maturation of the male reproductive axis. Proc Natl Acad Sci U S A. 2010;107:16372–16377. doi: 10.1073/pnas.1000423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen S, Schwarz JR, Niculescu D, Dinu C, Bauer CK, Hirdes W, Boehm U. Functional characterization of genetically labeled gonadotropes. Endocrinology. 2008;149:2701–2711. doi: 10.1210/en.2007-1502. [DOI] [PubMed] [Google Scholar]
- Yi SE, LaPolt PS, Yoon BS, Chen JY, Lu JK, Lyons KM. The type I BMP receptor BmprIB is essential for female reproductive function. Proc Natl Acad Sci U S A. 2001;98:7994–7999. doi: 10.1073/pnas.141002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon BS, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR, Lyons KM. Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc Natl Acad Sci U S A. 2005;102:5062–5067. doi: 10.1073/pnas.0500031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Juengel JL, Dodds KG, Laird M, Dearden PK, McNeilly AS, McNatty KP, Wilson T. The activin receptor-like kinase 6 Booroola mutation enhances suppressive effects of bone morphogenetic protein 2 (BMP2), BMP4, BMP6 and growth and differentiation factor-9 on FSH release from ovine primary pituitary cell cultures. J Endocrinol. 2008;196:251–261. doi: 10.1677/JOE-07-0148. [DOI] [PubMed] [Google Scholar]
- Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, Lin HY, Bloch KD, Peterson RT. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]