Abstract
Background
Clostridium difficile infection (CDI) may not respond to initial therapy and frequently recurs but predictors of response and recurrence are inconsistent. The impact of specific alterations in the gut microbiota determining treatment response and recurrence in patients with CDI is unknown.
Aim
To assess microbial signatures as predictors of treatment response and recurrence in CDI.
Methods
Pre-treatment stool samples and clinical metadata including outcomes were collected prospectively from patients with their first CDI episode. Next generation 16s rRNA sequencing using MiSeq Illumina platform was performed and changes in microbial community structure were correlated with CDI outcomes.
Results
Eighty-eight patients (median age 52.7 years, 60.2% female) were included. Treatment failure occurred in 12.5% and recurrence after response in 28.5%. Patients who responded to treatment had an increase in Ruminococcaceae, Rikenellaceae, Clostridiaceae, Bacteroides, Faecalibacterium and Rothia compared to non-responders. A risk-index built from this panel of microbes differentiated responders (mean 0.07±0.24) from non-responders (0.52±0.42; p=0.0002). Receiver operating characteristic (ROC) curve demonstrated that risk-index was a strong predictor of treatment response with an area under the curve (AUC) of 0.85. Among clinical parameters tested, only proton-pump inhibitor use predicted recurrent CDI (OR 3.75, 95%CI 1.27-11.1, p=0.01). Patients with recurrent CDI had statistically significant increases in Veillonella, Enterobacteriaceae, Streptococci, Parabacteroides and Lachnospiraceae compared to patients without recurrence and a risk index was able to predict recurrence (AUC=0.78).
Conclusions
Gut microbiota signatures predict treatment response and recurrence potentially allowing identification of CDI patients that may benefit from early institution of alternate therapies.
Keywords: Clostridium difficile infection, recurrence, treatment response, microbiome, prediction models
Introduction
Clostridium difficile infection (CDI) is a common infection in the United States with 450,000 infections and 29,000 deaths per year 1-3, with a tremendous economic impact (attributable costs ranging from $8,911 to $30,049 per hospitalized patient) 4. The pathophysiology of CDI is complex with alterations in gut microbiota playing an important role in susceptibility to CDI 5. A recent study reported an increase in Firmicutes, Proteobacteria, and Actinobacteria and decreases in Bacteroidetes in CDI patients and an altered ratio of Bacteroidetes to Firmicutes was found to be significant after controlling for confounding factors 6. Another study demonstrated that several bacterial species within the Ruminococcaceae, Lachnospiraceae, Bacteroides, and Porphyromonadaceae were largely absent in CDI cases and highly associated with non-diarrheal controls 7. Similarly alterations in Ruminococcus gnavus, certain Enterobacteriaceae, Verrucomicrobia and Enterococcus have been related to development of CDI 7-9. Risk factors for CDI include increasing age, antibiotic exposure, proton pump inhibitor use, hospitalization, immunosuppression, and comorbidities 10-15. Among these gut microbiota changes associated with antibiotic use, have been best studied in animal models and provide strong evidence for alterations in gut microbiota composition and function in susceptibility to primary and recurrent C. difficile 5, 16.
The management of CDI includes use of oral antibiotics but the rate of treatment failure (up to 35%) and recurrence (60% or higher after 3 episodes) is concerning 17, 18. Alternate treatment strategies such as fecal microbiota transplant are highly effective in patients with recurrent CDI 19, 20. Studies have assessed clinical features which may predict CDI response, but there is a lack of robust clinical or microbial biomarkers predictive of response to primary therapy 21-23. Severe CDI and hospital admission are clinical predictors of metronidazole failure but these have not been validated. Increasing age, concomitant antibiotic use, decreased anti-toxin IgG levels, the presence of comorbidities and potentially the use of gastric acid suppression medications have been associated with recurrent CDI. Anti-toxin IgG levels are not clinically available 24, 25.
Despite the identification of these risk factors, clinical models to predict the risk of recurrent CDI are not robust enough for routine clinical use and there are no models to predict response to initial antibiotic therapy 24, 26. Hence, there is a need for biomarkers and better models to predict these treatment outcomes. While the role gut microbiota alterations play in CDI susceptibility has been established 27, their role in determining outcomes of CDI treatment has not been investigated. We used next generation sequencing to characterize microbial communities in patients with primary CDI to identify differences in microbial community structure and key taxa that can predict treatment response and the risk of recurrence after successful treatment.
Materials and Methods
Study design
We prospectively recruited 88 patients (median age 52.7 years, interquartile range 36.9 – 65.1; 60.2% female) with their first CDI episode (from 3/2012 – 9/2013) as identified from the Clinical Microbiology Laboratory at Mayo Clinic, Rochester, Minnesota and collected an aliquot from the stool samples that led to the diagnosis. Details on clinical data acquisition and analysis are outlined in supplement 1.
Sequencing and analytic methods
16S rRNA gene sequencing and data analyses
After fecal DNA isolation (MoBio, Carlsbad, CA fecal DNA kit), amplicons spanning the variable region 4 of bacterial 16S rRNA were generated and sequenced using MiSeq Illumina platform at the Mayo Clinic Medical Genome Facility, Rochester, MN. We used 515F TATGGTAATTGTGTGCCAGCMGCCGCGGTAA and barcoded 806R primers AGTCAGTCAGCCGGACTACHVGGGTWTCTAAT. The amplicons were ~400 base pairs. The sequencing was bidirectional but only forward reads were used in the study due to low quality scores of reverse sequences. We applied built-in functions of the Quantitative Insights Into Microbial Ecology (QIIME) pipeline for data analysis 28. The 16S rRNA sequencing data from the Illumina runs were trimmed, demultiplexed, chimera filtered and assigned to operational taxonomic units (OTUs) using packages implemented in QIIME 1.8.0 software 28. Further details of 16S rRNA data analysis are outlined in supplement 1.
In order to study differences in alpha diversity between our cohort of patients and healthy subjects, we included data from healthy subjects from a previously published study (global gut dataset) 29. The differences in gut microbiota composition between healthy controls and patients with CDI have been previously described 7. We included adult US individuals and compared the following alpha diversity metrics with our cohort of CDI patients: phylogenetic diversity whole tree, observed species and Chao 1 index. The methodology applied in both the studies was similar as samples were stored at −80C in both studies. DNA extraction was done using MO BIO power soil DNA isolation kit in both studies and the same protocol (Earth Microbiome Protocol) was followed including bead beating the fecal samples and the same region of 16S was amplified using the same primers and the same PCR protocol 30.
In order to identify significant associations between microbial community abundances and clinical metadata, we applied a linear multivariate regression model specifically developed for microbiome data (MaAsLin, Multivariate microbial Association by Linear models 31. We used default parameters within MaAsLin as described by Morgan et al 31. Clinical data were selected by boosting, as microbial communities are known to be high dimensional, to identify those most associated with each microbial feature over potential covariates. Selected clinical data were then used in a general linear model including clinical data as predictor and taxonomic relative abundances as response.
Comparisons of relative abundance of taxa between responders and non-responders, or between patients with and without recurrence after initial response, was performed using Linear discriminant analysis Effect Size (LEfSe), a non-parametric Mann-Whitney U test applied to detect features with significant differential abundance with respect to the groups compared, followed by a Linear Discriminant Analysis (LDA) to estimate the effect size of each differentially abundant feature 32. As proposed, an LDA score (log 10) > 2 was considered significant.
A risk index was built to differentiate responders from non-responders to initial treatment or patients with and without recurrence after initial response, based on taxonomy on a non-collapsed OTU table. All the taxa with a LDA score (log 10) > 2 were included in the calculation of the risk index. In order to build the risk index to predict the patients who would respond or not respond to treatment, the relative abundances (arcsine square root transformed) of the taxa associated with the responders to treatment (based on the LEfSe output, all taxa with a LDA score (log 10) > 2) were summed and the relative abundances of the taxa associated with nonresponders to treatment (based on the LEfSe output, all taxa with a LDA score (log 10) > 2) were summed. Then the difference between these two sums (relative abundance of the taxa associated with no response to treatment minus relative abundance of the taxa associated with response to treatment) was calculated, thereby obtaining a risk index. This procedure was repeated n (overall sample size) times to obtain a risk index for each patient in the cohort. Therefore, a risk index was calculated for each patient. We had 11 risk indexes in the non-responders and 77 risk indexes in the responders, 22 risk indexes in patients who had a recurrence and 55 risk indexes in patients who did not have a recurrence of CDI. The utility of a risk index to predict a clinical outcome was recently reported by our group 33.
A leave-one-out cross-validation procedure was also conducted. This procedure calculates the risk index on the n - 1 patients (taxa that differentiated patients based on the LEfSe output) and then tested the risk index in the held-out patient that is the risk index values are predicted for each patient using a panel of microbes retrained from the other patients. A detailed description of the leave-one-out cross validation and receiver operating characteristic (ROC) curve analysis are outlined in supplement 1.
Network analyses were carried out with Cytoscape using an edge-weighted spring embedded layout 34. Importantly, we collapsed OTUs to the genus level and eliminated OTUs present in fewer than 25% of samples, to reduce the very large number of multiple hypotheses tested in correlation network analysis. We then performed Spearman correlation of taxon-taxon relative abundance and included only those links with absolute value of correlation > 0.5 and false discovery rate (FDR)-corrected p-value < 0.05.
In order to determine if competition for nutritional niches in the gut may play a role in determining response to treatment by providing C. difficile with a competitive disadvantage, we used Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) to predict Carbohydrate-Active Enzymes database - glycoside hydrolase (CAZY GH) assignments. Details on PICRUSt analysis are outlined in supplement 1.
Results
Clinical characteristics fail to predict response to standard therapy
The rate of primary non-response following recommended treatment was 12.5%. Patients were treated with either metronidazole or vancomycin (Table 1). Among those who had an initial successful response, 28.5% had recurrent CDI. Prior antibiotic exposure was noted in 59.1% of patients, and 78.8% of those with antibiotic exposure (or 36.1% of all 88 patients) were exposed to a single agent, 9.7% (or 5.7% of all 88 patients) to a combination of antibiotics and exact antibiotic name was unknown in the remaining 6 patients. Details on antibiotic use are summarized in Supplementary Table 1. Chi-square univariate analyses demonstrated no significant differences in any clinical variables including initial treatment in primary nonresponders compared to responders (Table 1).
Table 1.
Clinical characteristics of all patients
| Overall (n=88) | Treatment responder (n=77) | Treatment failure (n=11) | p-value* | Treatment success with no recurrence (n=55) | Treatment success with recurrence (n=22) | p-value** | |
|---|---|---|---|---|---|---|---|
| Age (median) | 52.7 | 53.8 | 49.9 | 0.67 | 55.6 | 49.0 | 0.62 |
| Sex (% female) | 60.2 | 59.7 | 63.6 | 0.8 | 58.2 | 63.6 | 0.65 |
| BMI, mean (Kg/M2) | 27.5 | 27.4 | 28.1 | 0.75 | 27.7 | 26.6 | 0.44 |
| Charlson comorbidity index | 1.32 | 1.32 | 1.27 | 0.53 | 1.34 | 1.27 | 0.49 |
| Prior antibiotic exposure (%) | 59.1 | 55.8 | 81.8 | 0.1 | 56.4 | 54.5 | 0.88 |
| Community-acquired (%) | 59.1 | 62.3 | 36.4 | 0.13 | 63.6 | 59.1 | 0.9 |
| Severe CDI (%) | 7 | 5.2 | 18.2 | 0.1 | 5.5 | 4.5 | 0.86 |
| Concomitant antibiotic exposure (%) | 28.4 | 27.2 | 36.4 | 0.5 | 27.3 | 27.3 | 1.0 |
| Concomitant PPI exposure (%) | 27.3 | 25.9 | 36.3 | 0.48 | 18.2 | 45.5 | 0.01 |
| Treatment with metronidazole (%) | 70.5 | 70.1 | 72.7 | 0.8 | 72.7 | 63.6 | 0.1 |
| Treatment with vancomycin (%) | 23.9 | 24.7 | 18.2 | 0.8 | 20.0 | 36.4 | 0.1 |
| Treatment with vancomycin and metronidazole | 5.6 | 5.2 | 9.1 | 0.8 | 7.3 | 0 | 0.1 |
Denotes p-value for comparison of treatment responders versus treatment failures
Denotes p-value for comparison of recurrent infections versus non-recurrent infection
CDI = Clostridium difficile infection, BMI = Body mass index, PPI = Proton pump inhibitor
Gut microbiota signatures prior to treatment predict response to therapy in CDI
Of the 88 fecal samples collected, a total of 1,449,211 high-quality 16S rRNA gene-encoding sequences were identified, representing 7,470 OTUs. The mean number of sequences obtained per sample was 16,468 ± 4,674. Importantly, since samples contained between 6,987 and 35,494 sequences, diversity analyses were rarefied at 6,987 sequences per sample to avoid bias.
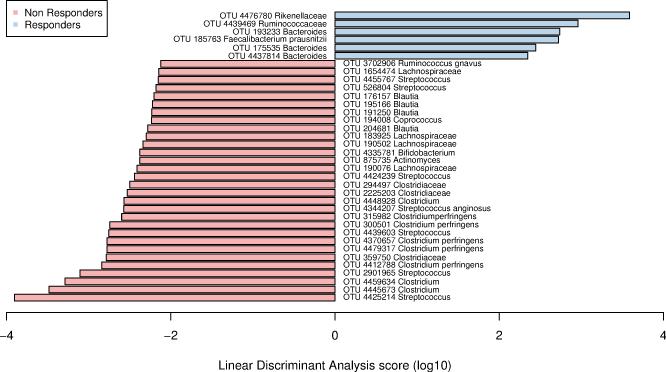
We assessed the relationship between clinical metadata and microbial measurements by identifying associations using MaAsLin (see methods above). This model investigated the relationship between microbial taxa collapsed at genus level with metadata of interest while accounting for other covariates. In our cohort of patients, we did not find significant associations between gut microbiota and the clinical characteristics (for example, Age, Sex, BMI, Charlson comorbidity Index) (Supplementary Table 2). A panel of 36 OTUs that were significantly different between primary non-responders and responders using LEfSe (corresponding to an LDA (log 10) > 2) were identified. Responders had a significant increase in relative abundance of OTUs within Ruminococcaceae, Rikenellaceae, Bacteroides, and Faecalibacterium, while nonresponders had a significant increase in Clostridiaceae, Lachnospiraceae, Blautia, Coprococcus, Streptococcus, Bifidobacterium, Ruminococcus and Actinomyces (Figure 1).
Figure 1.
Summary of the Operational Taxonomic Units associated with response or non-response to treatment using Linear discriminant analysis Effect Size analysis (LEfSe). Representative bacteria with relative abundance of at least 1% with significant differences are represented.
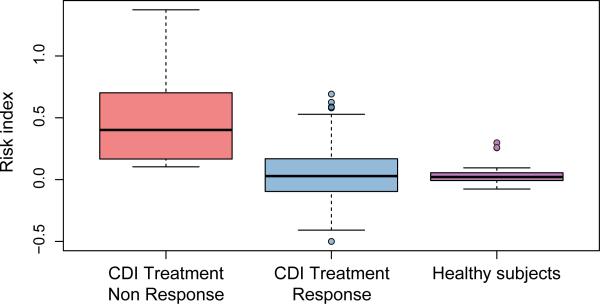
Furthermore, the ability of this panel of microbes to discriminate between responders and nonresponders using ROC curve analysis demonstrated several individual OTUs to be strong predictors of response (Supplementary Figure 1). A risk index of response to treatment was built from this panel of OTUs. The index, calculated in each patient (responders and nonresponders), corresponds to the difference between the sum of the relative abundance of all the OTUs associated with non-response to treatment (i.e. with an LDA (log 10) > 2) and the sum of the relative abundance of all the OTUs associated with response to treatment (i.e. with an LDA (log 10) > 2). The mean risk index score was significantly different [p < 0.001, one-way ANOVA (Analysis Of Variance) with post-hoc Tukey HSD (Honestly Significant Difference)] between the responders (mean score 0.07 ± 0.2) and non-responders (0.5 ± 0.4) as well as healthy subjects (0.03 ± 0.1), adult US subjects from global gut study 29, with BMI under 25, n= 60) and non-responders (Figure 2). The risk index was not significantly different between healthy subjects and responders (p = 0.57, one-way ANOVA with post-hoc Tukey HSD) suggesting that the difference in risk index in responders and non-responders is not due to variability in gut microbiome among individuals.
Figure 2.
A) Clostridium difficile Infection (CDI) Risk Index based on the differentiated Operational Taxonomic Units in non-responders, responders and healthy controls calculated using the Mann–Whitney U test: ***: p < 0.001. The boxplots denote top quartile, median and bottom quartile and individual dots represent individual patient samples.
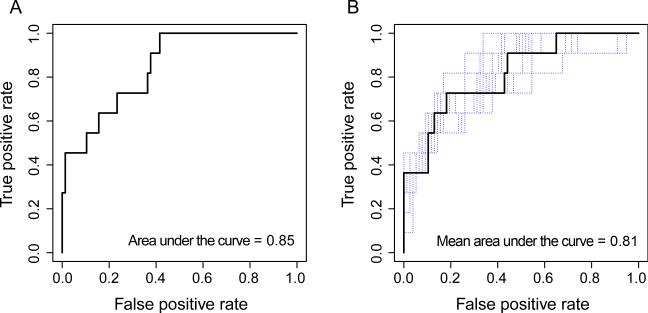
Further, ROC curve analysis showed that this risk index was a strong predictor of treatment response, with an area under the curve (AUC) of 0.85. A cut-off of 0.21 was associated with a sensitivity of 77% and a specificity of 73% (Figure 3A). Importantly, we did not find a correlation between the risk index and previous antibiotic treatment in terms of response to treatment (Pearson's product-moment correlation= 0.09, p value= 0.37). Thus, non-response was not associated with prior antibiotic administration.
Figure 3.
A) Receiver operating characteristic (ROC) curve analysis of the CDI risk index in pre-treatment fecal samples collected following 10-fold cross-validation. B) ROC curve analysis of the CDI risk index in pre-treatment fecal samples collected following 10-fold cross-validation of all the held-out samples from the leave-one-out cross-validation analysis. Following the leave-one-out procedure, the ROC curves of each leave-one-out procedure are in blue and the mean ROC curve of all the procedure is in black.
In order to assess our risk index, a leave-one-out cross-validation was performed, where the risk index was built 88 times using n-1 samples each time and then tested on the held-out sample. Thus, each held-out patient was treated as a new patient, independently from the initial cohort, on whom we tested and subsequently refined the optimal index cut-off to separate responders and non-responders. We showed that the risk index was a strong predictor of response vs. non-response (permutation test performed on the difference between the mean of those without response and the mean of those with response to treatment with 999 random permutations, p < 0.0001; Figure 3B). We also determined with this leave-one-out procedure that a CDI risk index threshold of 0.11 best predicts response to treatment, yielding a sensitivity of 70% at a specificity of 73% (mean AUC = 0.81). Thus, we found that our risk index, based on a panel of 36 OTUs that were significantly different between primary non-responders and responders using LEfSe, accurately identified patients with CDI, likely to not respond to conventional treatment.
OTU networks are disrupted in primary non-responders
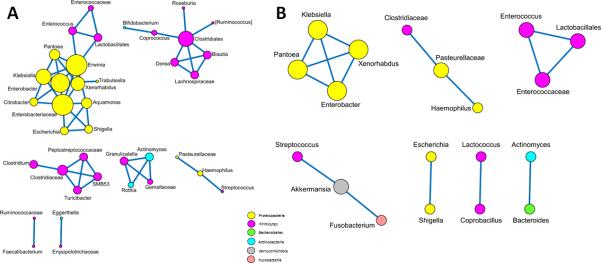
Correlations between OTU networks at the genus level were computed to differentiate responders and non-responders and demonstrated a three-fold (56 versus 16, ratio = 3.35) decrease in the number of strong taxon-taxon correlations (absolute value of Spearman correlation > 0.5 and False Discovery Rate corrected p-value < 0.05) in non-responders compared to responders, and most of the decreased nodes between responders and non-responders were members of the phylum Firmicutes (50 versus 10, ratio = 5) and Actinobacteria (7 versus 1, ratio = 7) (Figure 4).
Figure 4.
Spearman correlation demonstrating taxon-taxon relative abundance including links with absolute value of correlation > 0.5 and False Discovery Rate-corrected p-value < 0.05. Network analyses displayed with Cytoscape using an edge-weighted spring embedded layout in responders (A) and non-responders (B). Positive correlations are shown as blue links between nodes and only significant correlations are depicted. The size of the node was based on the number of correlations associated with the corresponding taxon. The color of the node is based on the phylum level: Actinobacteria (blue), Bacteroidetes (green), Firmicutes (purple), Proteobacteria (yellow), Fusobacteria (pink) and Verrucomicrobia (grey). There is a three-fold decrease in the number of taxon correlations in the non-responders and most of the decreased nodes were Firmicutes and Actinobacteria.
Gut microbial diversity is not significantly different in primary non-responders compared to responders
Unweighted and weighted UniFrac based principal coordinate analysis (PCoA) did not show significant differences in beta diversity between responders and non-responders (unweighted UniFrac distance metric: R = −0.09, p = 0.9; weighted UniFrac distance metric: R = −0.004, p = 0.5) (Supplementary Figure 2). Patient with CDI (responders and non-responders) had significantly decreased alpha diversity (p<0.001 for the 3 measures phylogenetic diversity whole tree, observed species and Chao 1 index; one-way ANOVA with post-hoc Tukey HSD) compared to healthy subjects (adult US subjects from global gut study 29). Among patients with CDI, although there was a trend towards decreased alpha diversity in non-responders compared to responders, this difference was not significant for the 3 measures phylogenetic diversity whole tree, observed species and Chao 1 index; one-way ANOVA with post-hoc Tukey HSD) (Supplementary Figure 3).
Gut microbiota functional repertoire is significantly different in responders and non-responders
We imputed functional aspects of the microbiota from 16S rRNA data using PICRUSt to predict CAZY-GH assignments. Based on LEfSe, we found that GH70 (dextransucrase) and GH38 (α-mannosidase) were increased whereas GH59 (β-galactosidase) and two carbohydrate-binding modules, CBM16 and CBM42 were significantly decreased in non-responders compared to responders with LDA score (log 10) > 2 (Supplementary Figure 4).
Gut microbiota signatures predict CDI recurrence
Among the patients who initially responded to treatment, 28.5% had recurrent CDI. The median time to recurrence was 23 days (range 15 - 56 days). There were no significant differences in patients with and without recurrent CDI (Table 1) among the clinical variables analyzed except PPI use which predicted recurrent CDI on univariate analysis (odds ratio 3.75, 95% confidence interval 1.27- 11.1, p=0.01) and multivariable analysis after controlling for age (p=0.0007) and comorbidities (p=0.0009) in separate multivariable models. However, PPI use was not associated with alterations in the gut microbiota (Supplementary Table 2).
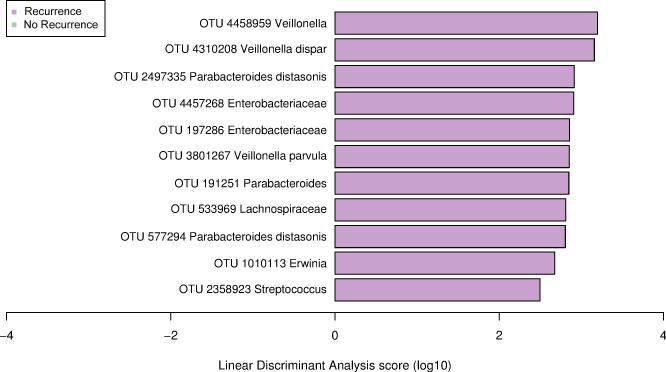
Relative abundance of eleven OTUs was significantly different between those with and without recurrence. Patients with recurrence had a significant increase in Veillonella, Enterobacteriaceae (Erwinia), Streptococcus, Parabacteroides and Lachnospiraceae using LEfSe (Figure 5). Several individual microbes were strong predictors of recurrence (Supplementary Figure 5).
Figure 5.
Summary of the Operational Taxonomic Units associated with recurrence (n=22) versus non-recurrence (n=55) after successful treatment response using Linear discriminant analysis Effect Size analysis (LEfSe).
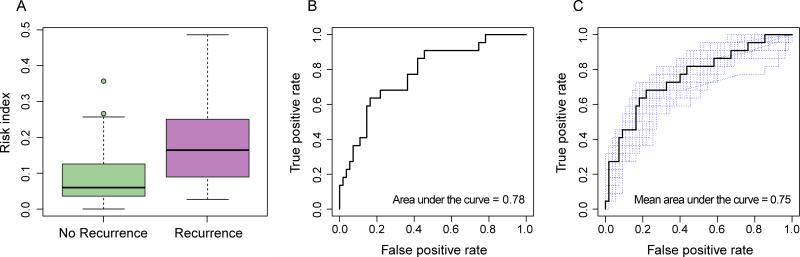
A risk index of recurrence built from this panel of microbes differentiated between patients with and without recurrence. This index included all OTUs that had a LDA score (log 10) > 2, as previously described. This index was significantly different in patients who did not have a recurrence (mean score 0.09 ±0.08) and those who did (0.19 ± 0.12) (Mann-Whitney U test, p-value = 0.0001) (Figure 6A). The ROC curve analysis showed that this risk index was a strong predictor of recurrence, with an AUC of 0.78 (Figure 6B). Moreover, a cutoff of 0.13 had a sensitivity of 78% and a specificity of 68%. As described previously, a leave-one-out cross-validation was performed, where the risk index was built 77 times using n-1 samples each time and then tested on the held-out sample. This procedure showed that the gut microbiota was a strong predictor of recurrence (permutation test performed on the difference between the mean of those with recurrence and the mean of those without recurrence with 999 random permutations, p < 0.0001; Figure 6C). This procedure also determined that a CDI risk index classification threshold of 0.11 best predicts recurrence with a sensitivity of 75% at a specificity of 69% (mean AUC = 0.75). Thus, we found that our risk index can identify patients with CDI who are likely to have recurrence after initial response. Importantly, a supervised learning method using a Random Forest model was also applied but was unable to accurately assign samples to their source population based on taxonomic profiles at the OTU level and was outperformed by the above risk index approach.
Figure 6.
A) Clostridium difficile Infection (CDI) Risk Index based on the differentiated Operational taxonomic units in patients with success versus recurrence. Mann–Whitney U test: ***: p < 0.001. Boxplots denote top quartile, median and bottom quartile. B) Receiver operating characteristic (ROC) curve analysis of the CDI Risk Index in fecal samples collected prior to treatment predicting recurrence. C) ROC curve analysis of the CDI Risk Index in fecal samples collected prior to treatment. Following the leave-one-out procedure, the ROC curves of each leave-one-out procedure are in blue and the mean ROC curve of all the procedure is in black.
Pre-treatment Gut microbial diversity is not significantly different in patients with and without recurrent CDI
Unweighted and weighted UniFrac based PCoA did not show significant differences in beta diversity between patients with and without recurrence (unweighted UniFrac distance metric: R =−0.1, p = 0.9; weighted UniFrac distance metric: R =0.02, p =0.3) (Supplementary Figure 6). Moreover, there was no significant difference in alpha diversity based on three different metrics between CDI patients with and without recurrence (Supplementary Figure 7).
Gut microbiota functional repertoire is significantly different in those with and without recurrence
As described above, we again imputed functional aspects of the microbiota from 16S rRNA data using PICRUSt to predict CAZY-GH assignments. Based on the LEfSe tool, GT30 (β-fucosidase) and a carbohydrate-binding module (CBM20) were increased in patients who had a recurrence compared to patients who did not with LDA score (log10) >2 (Supplementary Figure 8).
Discussion
In this study, we report specific gut microbiota signatures associated with the initial response to treatment and recurrence after successful treatment in patients with primary CDI. We have developed a risk index based on compositional differences among patients with CDI, which can help predict response to treatment and recurrence in patients with CDI. This is a step towards identifying CDI patients who would be candidates for early alternative therapies such as fecal microbiota transplantation (FMT). Although a significant percentage of patients either fail primary treatment or recur after successful treatment, there are no robust models to predict these outcomes. Gut microbiota play an important role in resistance to colonization by pathogens such as C. difficile, and this resistance can be altered following disruption of normal gut microbial community structure following antibiotic use or other alterations in host physiology. Several mechanisms that promote C. difficile colonization have been attributed to alteration of gut microbiota including increase in primary bile acids, increased availability of nutrients such as succinate and sialic acid, and decrease in butyrate producers in the gut 35, 36. Given the impact of gut microbiota in the pathogenesis of CDI, we profiled the microbial community in pre-treatment stool samples and identified biologically relevant microbial signatures that predict treatment response and risk of recurrent CDI.
Data on clinical predictors of response to treatment in CDI are sparse. Clinical predictors of metronidazole failure in CDI include severe CDI, recent cephalosporin use, CDI on hospital admission and intra-hospital transfer. In recurrent CDI, studies showing increasing age, concomitant antibiotic exposure and proton pump inhibitor use as risk factors are balanced by other studies with a lack of correlation between these variables and recurrent CDI 24, 25, 37. In our study, PPI use was the only clinical variable significantly associated with recurrent CDI. However, most patients had community-acquired CDI and were younger than patients with hospital-acquired CDI, which likely explains the lack of significant differences in some previously described clinical predictors of recurrent CDI such as age and comorbidities.
In our cohort, there was a significant decrease in overall microbial diversity in patients with CDI as compared to healthy individuals, but amongst patients with CDI, there was no significant decrease in the microbial diversity amongst those who failed primary therapy or had recurrence after initial success compared to CDI patients who did not have these adverse outcomes. We found an increase in taxa within Faecalibacterium, Ruminococcaceae, Bacteroides and Rikenellaceae, among others in primary responders. Interestingly, these taxa have been associated with colonization resistance against C. difficile in adult patients 27. Faecalibacterium prausnitzii is largely considered a beneficial bacterium as significant reductions in this taxa have been reported in patients with inflammatory bowel disease compared to healthy individuals 38. F. prausnitzii and members of Ruminococcaceae are capable of producing butyrate, similar to C. difficile 39, 40. Several studies indicate that CDI is accompanied by a depletion of butyrogenic bacteria 9, suggesting either an inhibitory effect of butyrate or the loss of a nutrient niche, which can be occupied by C. difficile. Thus, the presence of F. prausnitzii and Ruminococcaceae may improve the response to treatment by providing niche competition or direct inhibition. An increase in Bacteriodetes represented by greater abundance of the families Bacteroidaceae, Rikenellaceae and Porphyromonadaceae has been reported following successful FMT for treatment of CDI 41. Interestingly a higher relative abundance of Bacteroides and Ricknellaceae was seen in primary responders in our study, suggesting they may synergize with conventional treatment for exclusion of C. difficile. While the mechanism of action remains unclear, recent studies have shown that certain Bacteroides spp. may play a role in preventing infection with C. difficile 42, 43. B. fragilis influences the development of the immune response and B. thetaiotaomicron stimulates Paneth cells to produce antibacterial peptides, which may prevent pathogens from colonizing 44, 45. While indirect, these data suggest a synergistic role for members of the gut microbial community in augmenting treatment response.
Interestingly, even though overall gut microbial diversity is not significantly different in responders compared to non-responders, there is a three-fold increase in the number of strong taxon-taxon correlations in responders as compared to non-responders, and most of the decreased nodes in non-responders were members of the phylum Firmicutes. This suggests that in addition to differences in individual taxa, the overall microbial community structure plays an important role in augmenting response to primary therapy by more effectively excluding C. difficile.
In patients with recurrent CDI we found increases in taxa that were different than in primary non-responders. Several of these have been previously associated with recurrent CDI or potentially promote the growth of C. difficile. There was an increase in Enterobacteriaceae, Veillonella and Parabacteroides among others in patients with recurrent CDI compared to patients without recurrence. Several studies both in mice and in humans demonstrate an increase of Enterobacteriaceae in CDI 7, 27, 46. A persistent expansion of Enterobacteriaceae has also been shown after treatment with clindamycin suggesting it may play a role in susceptibility to colonization with C. difficile. Parabacteroides diastonis, a succinate producer, has been previously associated with CDI 47, 48 perhaps since increased succinate availability has been associated with expansion of C. difficile. The low concentration of succinate present in the microbiota of conventional mice is transiently elevated upon antibiotic treatment or chemically induced intestinal motility disturbance, and C. difficile exploits this succinate spike to expand in the perturbed intestine 35. Further studies are needed to identify mechanisms by which these bacteria may contribute to failure of treatment or recurrence after successful treatment.
The primary strength of our study is the ability to predict the clinically important outcomes of treatment failure and recurrence in patients with primary CDI by characterizing the gut microbiota of pre-treatment stool samples. It is possible that strain level differences in C. difficile contribute to the outcome but these were not assessed in the current study. This study helps define a new diagnostic paradigm, but as with any initial finding, there are limitations. The risk index developed in our study likely needs to be prospectively validated in a larger cohort of CDI patients given the relatively small sample size. We did not have information on the patients’ dietary histories at the time of diagnosis so we were unable to assess how diet may influences microbial composition and CDI outcomes. Additionally, detailed analyses comparing microbiome changes due to prior antibiotic exposure would need to be considered in future studies. We also used healthy controls from a previously published study and there could be a study effect given the samples were run at different times as has been pointed out in previous studies. This is usually attributed to differences in DNA extraction protocol, primers and data analysis 49. However, as these were not different between our study and the healthy controls, we do not expect a study effect. Prior studies, which have investigated study effects have looked at healthy controls from different studies where study specific effects can be seen. However as we are investigating differences in healthy controls and patients with CDI, we expect the true biological effect to overcome inter-individual study differences. While this was not a primary aim of our study we do note that our findings are similar to those previously reported 7. Both these studies used Illumina platform for sequencing, we used Miseq and the healthy controls were sequenced on HiseqA cross validation has been previously published showing that the data from these two platforms do not introduce a bias in the results 30. The risk index determined in our study and applied to the healthy controls was similar in responders and healthy controls (from different studies) and different from non-responders, further supporting the validity of our finding.
In conclusion, gut microbiota signatures can be used to predict response to and recurrence after initial treatment in patients with CDI. This finding will need to be validated in a larger cohort and future work will focus on understanding interaction of individual taxa with C. difficile to understand mechanisms by which they may determine outcome of treatment. Nevertheless, these biomarkers potentially allow identification of subsets patients that may be initially treated with more effective therapies such as newer antibiotics, fecal microbiota transplantation or defined microbial consortia instead of a prolonged therapeutic trial.
Supplementary Material
Acknowledgments
Financial support:
This research was made possible by support from NIH K08 DK100638 (PK), Global Probiotic Council (PK), Minnesota Partnership for Biotechnology and Genomics (PK), and Center for Individualized Medicine, Mayo Clinic, Rochester and CTSA Grant Number UL1 TR000135 from the National Center for Advancing Translational Sciences (NCATS), a component of the National Institutes of Health (NIH) and the National Institute Of Diabetes And Digestive And Kidney Diseases of the National Institutes of Health under Award Number P30 DK084567 (Clinical Core). Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH. EM was supported by the Robert Tournut award from the French National Society of Gastroenterology.
Footnotes
Guarantor of the article: Purna Kashyap
Specific author contributions:
S.K.: Study concept, data acquisition, data analysis, data interpretation, drafting and revision of manuscript; E.M.: Data acquisition, data analysis, data interpretation, drafting and revision of manuscript; B.S.: Data acquisition, data analysis and data interpretation; R.P.: Data analysis, data interpretation, drafting and revision of manuscript; D.K.: Study concept, data analysis, data interpretation, drafting and revision of manuscript D.S.P.: Study concept, data analysis, data interpretation, drafting and revision of manuscript; P.K.: Study concept, data acquisition, data analysis, data interpretation, drafting and revision of manuscript. All authors have reviewed and approved the final draft submitted.
Conflict of interest: None
References
- 1.Kelly CP, LaMont JT. Clostridium difficile--more difficult than ever. N. Engl. J. Med. 2008;359(18):1932–40. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 2.Magill SS, Edwards JR, Bamberg W, et al. Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 2014;370(13):1198–208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lessa FC, Mu Y, Bamberg WM, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372(9):825–34. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nanwa N, Kendzerska T, Krahn M, et al. The economic impact of Clostridium difficile infection: a systematic review. Am J Gastroenterol. 2015;110(4):511–9. doi: 10.1038/ajg.2015.48. [DOI] [PubMed] [Google Scholar]
- 5.Buffie CG, Bucci V, Stein RR, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517(7533):205–8. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manges AR, Labbe A, Loo VG, et al. Comparative metagenomic study of alterations to the intestinal microbiota and risk of nosocomial Clostridum difficile-associated disease. J. Infect. Dis. 2010;202(12):1877–84. doi: 10.1086/657319. [DOI] [PubMed] [Google Scholar]
- 7.Schubert AM, Rogers MA, Ring C, et al. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. mBio. 2014;5(3):e01021–14. doi: 10.1128/mBio.01021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rousseau C, Poilane I, De Pontual L, Maherault AC, Le Monnier A, Collignon A. Clostridium difficile carriage in healthy infants in the community: a potential reservoir for pathogenic strains. Clin. Infect. Dis. 2012;55(9):1209–15. doi: 10.1093/cid/cis637. [DOI] [PubMed] [Google Scholar]
- 9.Antharam VC, Li EC, Ishmael A, et al. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol. 2013;51(9):2884–92. doi: 10.1128/JCM.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khanna S, Pardi DS. Clostridium difficile Infection: New Insights Into Management. Mayo Clin. Proc. 2012;87(11):1106–17. doi: 10.1016/j.mayocp.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leffler DA, Lamont JT. Clostridium difficile Infection. N. Engl. J. Med. 2015;373(3):287–8. doi: 10.1056/NEJMc1506004. [DOI] [PubMed] [Google Scholar]
- 12.Press A, Ku B, McCullagh L, Rosen L, Richardson S, McGinn T. Developing a Clinical Prediction Rule for First Hospital-Onset Clostridium difficile Infections: A Retrospective Observational Study. Infect. Control Hosp. Epidemiol. 2016:1–5. doi: 10.1017/ice.2016.97. [DOI] [PubMed] [Google Scholar]
- 13.Settle CD, Wilcox MH, Fawley WN, Corrado OJ, Hawkey PM. Prospective study of the risk of Clostridium difficile diarrhoea in elderly patients following treatment with cefotaxime or piperacillin-tazobactam. Aliment. Pharmacol. Ther. 1998;12(12):1217–23. doi: 10.1046/j.1365-2036.1998.00428.x. [DOI] [PubMed] [Google Scholar]
- 14.Turco R, Martinelli M, Miele E, et al. Proton pump inhibitors as a risk factor for paediatric Clostridium difficile infection. Aliment. Pharmacol. Ther. 2010;31(7):754–9. doi: 10.1111/j.1365-2036.2009.04229.x. [DOI] [PubMed] [Google Scholar]
- 15.Dalton BR, Lye-Maccannell T, Henderson EA, Maccannell DR, Louie TJ. Proton pump inhibitors increase significantly the risk of Clostridium difficile infection in a low-endemicity, non-outbreak hospital setting. Aliment. Pharmacol. Ther. 2009;29(6):626–34. doi: 10.1111/j.1365-2036.2008.03924.x. [DOI] [PubMed] [Google Scholar]
- 16.Allegretti JR, Kearney S, Li N, et al. Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Aliment. Pharmacol. Ther. 2016 doi: 10.1111/apt.13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leffler DA, Lamont JT. Clostridium difficile infection. N Engl J Med. 2015;372(16):1539–48. doi: 10.1056/NEJMra1403772. [DOI] [PubMed] [Google Scholar]
- 18.Khanna S, Pardi DS. Clostridium difficile infection: management strategies for a difficult disease. Therapeutic advances in gastroenterology. 2014;7(2):72–86. doi: 10.1177/1756283X13508519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cammarota G, Masucci L, Ianiro G, et al. Randomised clinical trial: faecal microbiota transplantation by colonoscopy vs. vancomycin for the treatment of recurrent Clostridium difficile infection. Aliment. Pharmacol. Ther. 2015 doi: 10.1111/apt.13144. [DOI] [PubMed] [Google Scholar]
- 20.Guo B, Harstall C, Louie T, Veldhuyzen van Zanten S, Dieleman LA. Systematic review: faecal transplantation for the treatment of Clostridium difficile-associated disease. Aliment. Pharmacol. Ther. 2012 doi: 10.1111/j.1365-2036.2012.05033.x. [DOI] [PubMed] [Google Scholar]
- 21.Louie TJ, Miller MA, Mullane KM, et al. Fidaxomicin versus vancomycin for Clostridium difficile infection. N. Engl. J. Med. 2011;364(5):422–31. doi: 10.1056/NEJMoa0910812. [DOI] [PubMed] [Google Scholar]
- 22.Zar FA, Bakkanagari SR, Moorthi KM, Davis MB. A comparison of vancomycin and metronidazole for the treatment of Clostridium difficile-associated diarrhea, stratified by disease severity. Clin. Infect. Dis. 2007;45(3):302–7. doi: 10.1086/519265. [DOI] [PubMed] [Google Scholar]
- 23.Johnson S, Louie TJ, Gerding DN, et al. Vancomycin, metronidazole, or tolevamer for Clostridium difficile infection: results from two multinational, randomized, controlled trials. Clin. Infect. Dis. 2014;59(3):345–54. doi: 10.1093/cid/ciu313. [DOI] [PubMed] [Google Scholar]
- 24.Hu MY, Katchar K, Kyne L, et al. Prospective derivation and validation of a clinical prediction rule for recurrent Clostridium difficile infection. Gastroenterology. 2009;136(4):1206–14. doi: 10.1053/j.gastro.2008.12.038. [DOI] [PubMed] [Google Scholar]
- 25.Shivashankar R, Khanna S, Kammer PP, et al. Clinical predictors of recurrent Clostridium difficile infection in out-patients. Aliment. Pharmacol. Ther. 2014;40(5):518–22. doi: 10.1111/apt.12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eyre DW, Walker AS, Wyllie D, et al. Predictors of first recurrence of Clostridium difficile infection: implications for initial management. Clin Infect Dis. 2012;55(Suppl 2):S77–87. doi: 10.1093/cid/cis356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seekatz AM, Young VB. Clostridium difficile and the microbiota. J. Clin. Invest. 2014;124(10):4182–9. doi: 10.1172/JCI72336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–4. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13(9):R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montassier E, Al-Ghalith GA, Ward T, et al. Pretreatment gut microbiome predicts chemotherapy-related bloodstream infection. Genome Med. 2016;8(1):49. doi: 10.1186/s13073-016-0301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe. 2014;16(6):770–7. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng KM, Ferreyra JA, Higginbottom SK, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502(7469):96–9. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonald EG, Milligan J, Frenette C, Lee TC. Continuous Proton Pump Inhibitor Therapy and the Associated Risk of Recurrent Clostridium difficile Infection. JAMA internal medicine. 2015;175(5):784–91. doi: 10.1001/jamainternmed.2015.42. [DOI] [PubMed] [Google Scholar]
- 38.Cao Y, Shen J, Ran ZH. Association between Faecalibacterium prausnitzii Reduction and Inflammatory Bowel Disease: A Meta-Analysis and Systematic Review of the Literature. Gastroenterol Res Pract. 2014;2014:872725. doi: 10.1155/2014/872725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 2002;217(2):133–9. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 40.Rios-Covian D, Gueimonde M, Duncan SH, Flint HJ, de Los Reyes-Gavilan CG. Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol. Lett. 2015;362(21) doi: 10.1093/femsle/fnv176. [DOI] [PubMed] [Google Scholar]
- 41.Hamilton MJ, Weingarden AR, Unno T, Khoruts A, Sadowsky MJ. High-throughput DNA sequence analysis reveals stable engraftment of gut microbiota following transplantation of previously frozen fecal bacteria. Gut Microbes. 2013;4(2):125–35. doi: 10.4161/gmic.23571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hopkins MJ, Macfarlane GT. Changes in predominant bacterial populations in human faeces with age and with Clostridium difficile infection. J. Med. Microbiol. 2002;51(5):448–54. doi: 10.1099/0022-1317-51-5-448. [DOI] [PubMed] [Google Scholar]
- 43.Hopkins MJ, Macfarlane GT. Nondigestible oligosaccharides enhance bacterial colonization resistance against Clostridium difficile in vitro. Appl. Environ. Microbiol. 2003;69(4):1920–7. doi: 10.1128/AEM.69.4.1920-1927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4(3):269–73. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 45.Mazmanian SK, Kasper DL. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat Rev Immunol. 2006;6(11):849–58. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- 46.Rea MC, O'Sullivan O, Shanahan F, et al. Clostridium Difficile Carriage in Elderly Subjects and Associated Changes in the Intestinal Microbiota. J. Clin. Microbiol. 2011 doi: 10.1128/JCM.05176-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang JY, Antonopoulos DA, Kalra A, et al. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 2008;197(3):435–8. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 48.Lawley TD, Clare S, Walker AW, et al. Targeted Restoration of the Intestinal Microbiota with a Simple, Defined Bacteriotherapy Resolves Relapsing Clostridium difficile Disease in Mice. PLoS pathogens. 2012;8(10):e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lozupone CA, Stombaugh J, Gonzalez A, et al. Meta-analyses of studies of the human microbiota. Genome Res. 2013;23(10):1704–14. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.