Abstract
Polyprenyl phosphate-GlcNAc-1-phosphate transferase (WecA) is an essential enzyme for the growth of Mycobacterium tuberculosis (Mtb) and some other bacteria. Mtb WecA catalyzes the transformation from UDP-GlcNAc to decaprenyl-P-P-GlcNAc, the first membrane-anchored glycophospholipid that is responsible for the biosynthesis of mycolylarabinogalactan in Mtb. Inhibition of WecA will block the entire biosynthesis of essential cell wall components of Mtb in both replicating and non-replicating states, making this enzyme a target for development of novel drugs. Here, we report a fluorescence-based method for the assay of WecA using a modified UDP-GlcNAc, UDP-Glucosamine-C6-FITC (1), a membrane fraction prepared from an M. smegmatis strain, and the E. coli B21WecA. Under the optimized conditions, UDP-Glucosamine-C6-FITC (1) can be converted to the corresponding decaprenyl-P-P-Glucosamine-C6-FITC (3) in 61.5% yield. Decaprenyl-P-P-Glucosamine-C6-FITC is readily extracted with n-butanol and can be quantified by ultraviolet-visible (UV-Vis) spectrometry. Screening of the compound libraries designed for bacterial phosphotransferases resulted in the discovery of a selective WecA inhibitor, UT-01320 (12) that kills both replicating and non-replicating Mtb at low concentration. UT-01320 (12) also kills the intracellular Mtb in macrophages. We conclude that the WecA assay reported here is amenable to medium- and high-throughput screening, thus facilitating the discovery of novel WecA inhibitors.
Keywords: Prenyl-phosphate-GlcNAc-1-phosphate Transferase, WecA, Bacterial phosphotransferases, Mycobacterium tuberculosis, Fluorescence-based assay, HTS, WecA inhibitors
The eradication of tuberculosis (TB) remains a challenge for basic, translational, and clinical research scientists (1). Clinical responses of multidrug-resistant (MDR)-TB patients to the first line drugs are poor, or in some cases there is no response at all. For MDR strains of Mycobacterium tuberculosis (Mtb), treatment length of TB chemotherapy will be at least 20–28 months. The treatment of extensively drug-resistant (XDR)-TB takes substantially longer than MDR-TB (2,3). Therefore, it is very important to discover promising approaches to improve current TB treatment. Mtb can persist in host tissues for months to decades without replicating, yet with the ability to resume growth, but current TB drugs are not effective against non-replicating Mtb in vivo at therapeutic concentrations. The ability of Mtb to survive in host macrophages by entering dormant state is one factor that requires the long duration of TB chemotherapy (4–6).
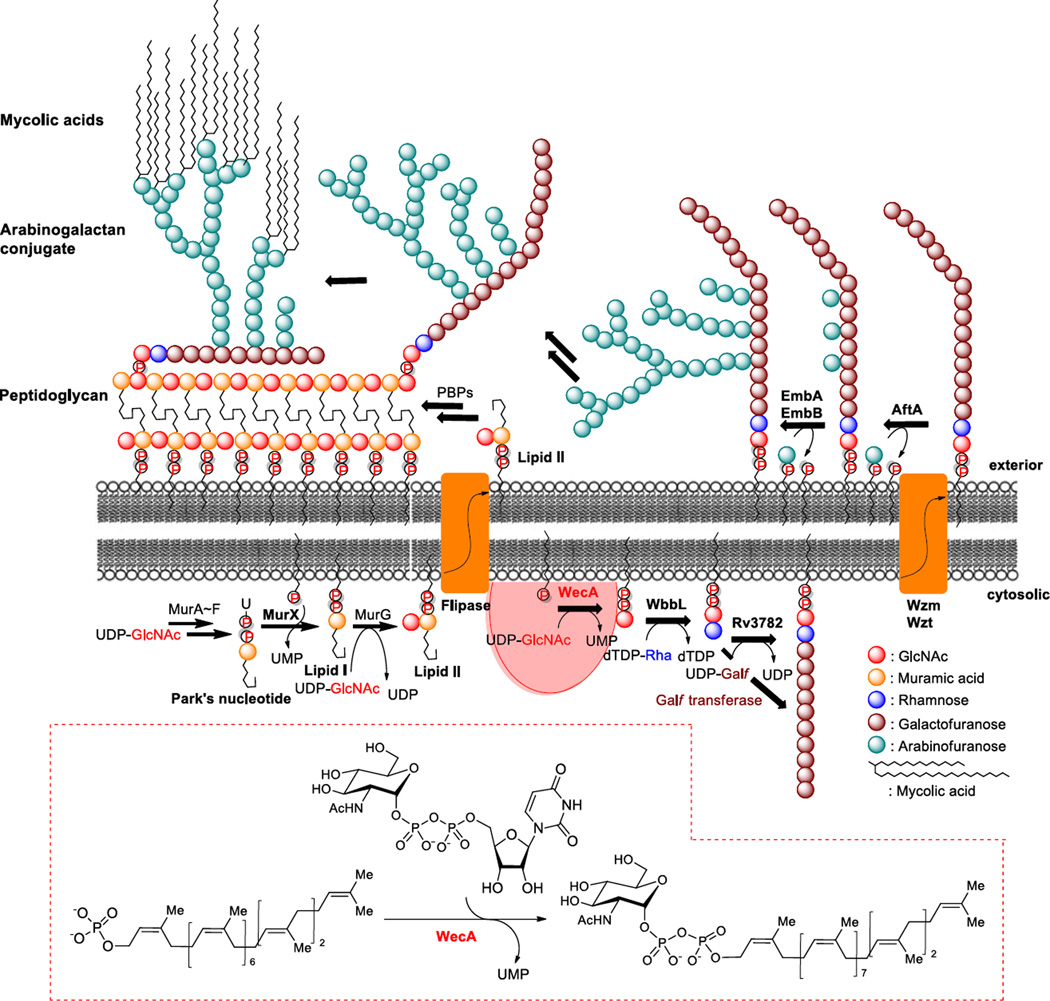
Mtb cell walls play an important role in survival of Mtb inside the macrophages (7,8). Comparisons from gene expression studies of Mtb at exponential phase and non-replicating states indicated that the genes associated with cell envelope biosynthesis and assembly (e.g. arabinosyl transferases, lipoarabinomannan biosynthesis, mycolic acid biosynthesis, and other enzymes associated with reconstructions of cell walls) are up-regulated (8,9). Therefore, inhibition of the committed step of the mycolylarabinogalactan synthesis of Mtb cell wall may enable non-replicating Mtb to become susceptible to current TB drugs, as well as blocking Mtb survival in host macrophages. Prenyl-phosphate-GlcNAc-1-phosphate transferase (WecA) is a polyprenyl-phosphate N-acetylhexosamine-1-phosphate transferase that catalyzes formation of decaprenyl-GlcNAc-pyrophosphate (decaprenyl-P-P-GlcNAc) in Mtb. As illustrated in Fig. 1, WecA is the first membrane-associated step that anchors the phospho-GlcNAc moiety of UDP-GlcNAc to decaprenyl phosphate (C50-P) [undecaprenyl phosphate (C55-P) in E. coli], leading to the formation of mycolylarabinogalactan. On the other hand, translocase I (MurX/MraY) is highly specific for UDP-N-acetylmuramyl-pentapeptide (Park’s nucleotide), forming undeca- or decaprenyl-diphosphoryl-N-acetylmuramate-pentapeptide (lipid I). WecA and MurX/MraY belong to the polyprenyl-phosphate N-acetylhexosamine 1-phosphate transferase enzyme superfamily (P2HPT). Besides these integral membrane proteins, P2HPT includes the bacterial phosphotransferases, TagO, WbcO, WbpL, and RpgG, and the eukaryotic UDP-GlcNAc:dolichyl-P GlcNAc-1-P transferase (GPT) (10).
FIG 1.
The biosynthetic pathway of peptidoglycan and mycolylarabinogalactan in M. tuberculosis.
Inhibition of WecA may prevent the biosynthesis of mycolylarabinogalactan conjugate, the essential component of the cell wall for M. tuberculosis to survive in macrophages.
Both WecA and MurX/MraY enzymes are essential for Mtb growth; however, MurX/MraY inhibitors are effective in killing only replicating Mtb under aerobic conditions (11–14). Although WecA inhibitors have the potential to be effective TB drugs that kill non-replicating Mtb under oxygen depleted conditions, only a few molecules are known to interfere with WecA and their effectiveness against non-replicating (or dormant) Mtb has been poorly characterized (15). WecA-catalyzed reactions have been performed via UDP-[14C]GlcNAc, C50-P (or C55-P), and either purified WecA or crude membranes containing WecA, the reported assays require separation of the product by chromatography (10,15–17). These assays are inadequate to systematically characterize library molecules in a high-throughput manner (18,19). We identified new UDP-GlcNAc fluorescent probes, UDP-Glucosamine-C6-FITC (1) and UDP-Glucosamine-C6-Dansyl (2), both of which can be recognized by the MurG transglycosylase, which is an essential peptidoglycan biosynthetic enzyme (20,21). Interestingly, under optimized conditions the water-insoluble decaprenyl-Glucosamine-C6-FITC (3) and decaprenyl-Glucosamine-C6-Dansyl (4) analogues could be biosynthesized with the WecA-containing membrane fractions obtained from M. tuberculosis, M. smegmatis, and E. coli. In the present work, we report a convenient and reliable enzyme assay for WecA to identify antimycobacterial WecA inhibitor molecules and validate it by detecting a selective WecA inhibitor, UT-01320 (12) against non-replicating Mtb and intracellular Mtb.
MATERIALS AND METHODS
Chemical materials and methods
Difco Middlebrook 7H10 agar, Middlebrook 7H9 broth, Tryptic soy agar, Tryptic soy broth, MOPS, tris(hydroxymethyl)aminomethane, 2-mercaptoethanol, sucrose and triton-X 100 were purchased from Sigma-Aldrich. ADC enrichment was purchased from Fisher Scientific. Magnesium chloride and potassium chloride were purchased from VWR. All reagents and solvents were commercial grade and were used as received without further purification unless otherwise noted. Flash chromatography was performed with Whatman silica gel (Purasil 60 Å, 230–400 Mesh). Analytical thin-layer chromatography was performed with 0.25 mm coated commercial silica gel plates (EMD, Silica Gel 60F254) visualizing at 254 nm, or developed with ceric ammonium molybdate or anisaldehyde solutions by heating on a hot plate. 1H-NMR spectral data were obtained using 400, and 500 MHz instruments. 13C-NMR spectral data were obtained using 100 and125 MHz instruments. For all NMR spectra, δ values are given in ppm and J values in Hz.
WecA assay substrates
UDP-Glucosamine-C6-FITC (1), decaprenyl phosphate (C50-P), undecaprenyl phosphate (C55-P), and other prenyl phosphates evaluated in this article were chemically synthesized from the corresponding starting materials.
UDP-Glucosamine-C6-FITC (1)
To a stirred solution of UDP-Glucosamine-C6-NH2 (5.9 mg, 8.3 µmol) in 0.1 M aq. NaHCO3 (0.20 mL) was added fluorescein isothiocyanate (6.4 mg, 0.017 mmol) in DMF (0.20 mL). After 5 h at room temperatures (r.t.), the reaction mixture was filtered. The filtrate was purified by reverse phase HPLC [column: HYPERSIL GOLD™ (175 Å, 12 µm, 250 × 10 mm), solvents: a gradient elution of 0:100 to 30:70 CH3CN : 0.05 M aq. NH4HCO3 over 30 min, flow rate: 2.0 mL/min, UV: 500 nm] to afford UDP-Glucosamine-C6-FITC (1, 7.1 mg, 78%, retention time: 23 min). 1H NMR (400 MHz, Deuterium Oxide) δ 7.90 (d, J = 8.1 Hz, 1H), 7.71 – 7.63 (m, 1H), 7.61 – 7.51 (m, 1H), 7.37 – 7.30 (m, 3H), 7.30 – 7.20 (m, 4H), 5.99 – 5.85 (m, 2H), 5.52 (d, J = 6.3 Hz, 1H), 4.35 – 4.27 (m, 3H), 4.26 – 4.13 (m, 2H), 4.12 – 4.00 (m, 2H), 3.92 – 3.84 (m, 1H), 3.80 (dd, J = 16.2, 3.2 Hz, 1H), 3.76 – 3.69 (m, 2H), 3.62 – 3.56 (m, 2H), 3.54 – 3.47 (m, 1H), 3.45 – 3.37 (m, 1H), 1.70 – 1.57 (m, 4H), 1.46 – 1.33 (m, 4H).
UDP-Glucosamine-C6-Dansyl (2)
UDP-Glucosamine-C6-Dansyl (2) was synthesized according to the procedure described for 1, but with dansyl chloride instead of fluorescein isothiocyanate. 1H NMR (500 MHz, Deuterium Oxide) δ 7.97 (d, J = 8.1 Hz, 1H), 5.97 (s, 1H), 5.96 (d, J = 12.2 Hz, 1H), 5.55 – 5.52 (m, 1H), 4.38 – 4.33 (m, 2H), 4.28 – 4.21 (m, 2H), 4.20 – 4.14 (m, 3H), 4.13 – 4.01 (m, 2H), 3.93 – 3.89 (m, 1H), 3.88 – 3.84 (m, 1H), 3.80 (dd, J = 12.6, 4.2 Hz, 1H), 3.77 – 3.69 (m, 2H), 3.57 – 3.50 (m, 1H), 1.70 – 1.60 (m, 4H), 1.44 – 1.36 (m, 4H).
Decaprenyl-P-P-Glucosamine-C6-FITC (3) and decaprenyl-P-P-Glucosamine-C6-Dansyl (4)
Decaprenyl-P-P-Glucosamine-C6-FITC (3) and Decaprenyl-P-P-Glucosamine-C6-Dansyl (4) were synthesized according to the procedure described previously (22–24). The product was purified by reverse-phase HPLC [column: HYPERSIL GOLD™ (175 A, 12 µm, 250 × 10 mm), solvents: a gradient elution of 85 : 15 to 100 : 0 MeOH : 0.05 M aqueous NH4HCO3 over 30 min, flow rate: 2.0 mL/min, UV: 485 nm for 3 and 350 nm for 4].
Preparation of polyprenyl phosphates
Decaprenyl phosphate (C50-P) and undecaprenyl phosphate (C55-P)
Leaves of Magnolia kobus, (Rosaceae) were collected in the Botanical Garden of Polish Academy of Sciences, Powsin, dried in open air, pulped using mortar and pestle and extracted with acetone/hexane (1:1, v/v) for 48h at r.t. After filtration and evaporation of the solvents, the crude mixture of lipids containing polyprenols was dissolved in hexane and purified by silica gel chromatography (hexanes to 1:1 hexanes/Et2O) (25,26). The crude polyprenol mixture was purified by C18-silica gel chromatography (LiChroprep RP-18, 40–63µm, Merck, solvent: MeOH to acetone:MeOH = 10/1). Fractions containing pure decaprenol and undecaprenol were pooled, evaporated, dissolved in hexane and stored at −20 °C. Polyprenyl phosphates were synthesized via the procedures reported by Danilov et al. (27). Stirred solution of decaprenol in CHCl3 was added Cl3CN and nBu4NH2PO4. After 30 min at room temperature, all volatiles were evaporated and the residue was dissolved in THF. Aqueous ammonia was added to the mixture and kept overnight at 4 °C. The supernatant was collected, evaporated and dissolved in CHCl3/MeOH (2:1, v/v) containing 3% of water and loaded onto DEAE-Sephadex A-25 column. Unreacted decaprenol was eluted with CHCl3/MeOH (2:1) and the decaprenyl phosphate was eluted with ammonium acetate (0–100 mM) in CHCl3/MeOH (2:1). Purified decaprenyl phosphate was dissolved in CHCl3/MeOH (2:1) containing 3% of NH3 and stored at −20°C. Undecaprenyl phosphate was synthesized by the same method described for decaprenyl phosphate.
Bacterial strains and growth of bacteria
Mycobacterium tuberculosis (H37Rv) was obtained through BEI Resources, NIAID/NIH. Mycobacterium smegmatis (ATCC 607), and Escherichia coli K-12 (ATCC 29425) were obtained from ATCC. E. coli B21 wecA strain was obtained from Dr. McNeil (Colorado State University). A single colony of each bacterial strain was obtained on a Difco Middlebrook 7H10 nutrient agar enriched with 10% oleic acid, albumin, dextrose and catalase (OADC for M. tuberculosis), albumin, dextrose and catalase (ADC for M. smegmatis), and on Tryptic Soy agar (for E. coli). Seed cultures were obtained in Middlebrook 7H9 broth enriched with OADC (for M. tuberculosis), ADC (for M. smegmatis), and in Tryptic Soy broth (for E. coli), respectively. Each strain was grown to mid-log phase.
Preparation of membrane fraction P-60 containing WecA
M. smegmatis cells were harvested by centrifugation (4,700 RPM) at 4 °C followed by washing with 0.9% saline solution (thrice) and ~5g of pellet (wet weight) was collected. The washed cell pellets were suspended in homogenization buffer (containing 50 mM MOPS of pH = 8, 10 mM MgCl2 and 5 mM 2-mercaptoethanol) and disrupted by probe sonication on ice (10 cycles of 60s on and 270s off). The resulting suspension was centrifuged at 1,000 ×g for 10 min at 4 °C to remove unbroken cells. The supernatant was centrifuged at 25,000 ×g for 40 min at 4 °C (3 to 4 times). All pellets in each tube were pooled and a second sonication was performed (10 cycles of 60s on and 270s off). The lysate was centrifuged once at 25,000 ×g for 1h and the supernatant was subjected to ultracentrifugation at 60,000 ×g for 1h at 4 °C. The supernatant was discarded and the membrane fraction containing WecA enzyme (P-60) was suspended in the TRIS-HCl buffer (pH 7.5, containing 2-mercaptoethanol) (28,29). Total protein concentrations are about 8~10 mg/mL (30). Aliquots were stored in eppendorf tubes at −80°C. Similarly, the membrane fractions (P-60) were prepared from M. tuberculosis, and E. coli, respectively.
WecA assay
UDP-Glucosamine-C6-FITC (1) [2 mM stock solution; 0.56 µL (22.5 µM)], MgCl2 [0.5 M; 4 µL (40 mM)], CHAPS (5%; 10 µL (1 %)), β-mercaptoethanol [50 mM; 5 µL (5 mM)], tris-buffer (pH=8.0; 50mM, 9.74 µL), decaprenyl phosphate dissolved in Tris-HCl (50mM, pH 8) : PEG : DMSO (5 : 1 : 2) [4 mM, 1.7 µL (140 µM)], and inhibitor [0–100 µM, in DMSO (1 µL)] were place in a 1.5 mL Eppendorf tube. P-60 (18 µl) was added (the total volume of the reaction mixture: 50 µL) to the reaction mixture. The reaction mixture was incubated for 2 h at 37 °C, and quenched with n-butanol (150 µL). Two phases were mixed via vortex and centrifuged at 10,000 ×g for 3 min. The upper butanol phase was assayed via reverse-phase HPLC or UV-Vis Synergy HT plate reader. For HPLC analyses, the butanol phase (25 µl) was injected into HPLC (solvent: CH3OH:0.05 M aq. NH4HCO3 = 85 : 15, UV: 485 nm, flow rate: 0.5 mL/min, Column: Kinetex 5µ C8 100Å, 150 × 4.60 mm); the area of the peak for decaprenyl-P-P-Glucosamine-C6-FITC (3) was quantified to obtain the IC50 value. For the UV-Vis-based assay, the n-butanol phase (20 µL) was transferred to a 384 well black plate and fluorescence was measured at an excitation of 485nm and emission of 528nm. The IC50 values were calculated from plots of the percent product inhibition versus the inhibitor concentration.
MurX assay
MurX assay was performed via the procedure reported previously (31). Park’s nucleotide-Nε-C6-dansyl and neryl phosphate were chemically synthesized according to the reported procedures (22–24). Park’s nucleotide-Nε-C6-dansyl [2 mM stock solution; 3.75 µL (75 µM)], MgCl2 [0.5 M; 10 µL (50 mM)], KCl [2 M, 10 µL (200 mM)], triton X100 (0.5%; 11.25 µL), tris-buffer (pH = 8; 50mM, 2.5 µL), neryl phosphate (10 mM, 45 µL), and inhibitor (0–100 µM, in DMSO (2.5 µL)) were placed in a 500 µL Eppendorf tube. P-60 (15 µl) was added (the total volume of the reaction mixture: 100 µL) to the reaction mixture. The reaction mixture was incubated for 1 h at room temperature (26 °C), and quenched with CHCl3 (200 µL). Two phases were mixed via vortex and centrifuged at 25,000 ×g for 10 min. The upper aqueous phase was assayed via reverse-phase HPLC. The water phase (10 µl) was injected into HPLC (solvent: CH3CN : 0.05 M aq. NH4HCO3 = 25 : 75, UV: 350 nm, flow rate: 0.5 mL/min, Column: Kinetex 5u C8 100Å, 150 × 4.60mm), and the area of the peak for lipid I-neryl derivative was quantified to obtain the IC50 value. The IC50 values were calculated from plots of the percent product inhibition versus the inhibitor concentration (32).
Kinetic parameter evaluation via WecA activity assay
Evaluation of kinetic parameters was performed through P-60-catalyzed WecA synthesis. Km and Vmax were determined by Michaelis-Menten enzyme kinetics. The correlation (Michaelis-Menten plot) between the concentrations of UDP-Glucosamine-C6-FITC (1,×axis) and rate (V) of decaprenyl-Glucosamine-C6-FITC (3, y axis) formation was obtained using GraphPad Prism Software (17,33,34).
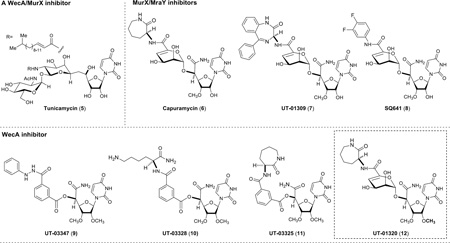
Compounds
All antimycobacterial molecules screened against WecA and MurX were synthesized in our laboratory, except for tunicamycin (sigma) and the TB drugs (sigma). All molecules were diluted with DMSO to be the concentration of 1.0 mg/100 µL (stock solution). The WecA assays developed here were tolerated to 2.5% of DMSO concentrations in total volume of the reaction solution (100 µL). The maximum tolerated concentration for DMSO in WecA-catalyzed decaprenyl-P-P-Glucosamine-C6-FITC (3) reaction was determined.
Determination of MICs
M. tuberculosis was cultured to be at an optical density of 0.4–0.5. Each compound (8 µL) stored in DMSO (1 mg/100 µL) was placed in a sterile 96 well plate and a serial dilution was conducted with the culturing broth (total volume of 100 µL). The bacterial suspension (100 µL) was added to each well (total volume of 200 µL). The bacterial culture in a plate treated or non-treated with compounds was incubated for 14 days at 37 °C in a incubator (120 rpm). Resazurin (0.01%, 20 µL) was added to each well and incubated at 37 °C for 5h. The MIC values were determined according to NCCLS method (pink = growth, blue = no visible growth). The absorbance of each well was also measured at 570 nm and 600 nm via a microplate reader.
Luminescence-based low-oxygen-recovery assay (LORA) assay
These assays were performed according to the reported procedures (35). In brief, M. tuberculosis H37Rv cells were transformed by mixing at least 1.0 µg of the purified plasmid, pFCA-luxAB and incubating at room temperature for 30 min, followed by electroporation. M. tuberculosis pFCA-luxAB strain cultured was diluted in Middlebrook 7H12 broth, and sonicated for 15s. The cultures were diluted to obtain an A570 of 0.03 to 0.05 and 3,000 to 7,000 RLUs per 100 µl. Two-fold serial dilutions of antimicrobial agents were prepared in black 96-well microtiter plates (100 µl), and 100 µl of the cell suspension was added. The microplate was placed under anaerobic conditions (oxygen concentration, less than 0.16%) by using an Anoxomat model WS-8080 (MART Microbiology) and three cycles of evacuation and filling with a mixture of 10% H2, 5% CO2, and 85% N2. Incubation was continued for 10 days, and transferred to an ambient gaseous condition (5% CO2-enriched air) incubator for a 28 h “recovery.” Each culture (100 µl) was transferred to white 96-well microtiter plates for determination of luminescence.
Killing effect against intracellular M. tuberculosis
J774A.1 cells were seeded at 2.5 × 105 cells/well in 24-well dishes or 1 × 105 cells/well in 8-well chamber slides and incubated overnight at 37 °C in DMEM. A transformant Mtb CDC1551 expressing tdTomato was grown in 7H9 Middlebrook medium supplemented with OADC. The Mtb cells were harvested at an optical density of 0.5, washed and re-suspended in saline. J774A.1 cells were maintained in cell culture medium and were infected by Mtb (106 bacteria in 0.2 mL of media): a multiplicity of infection (MOI) of≈10 (bacteria/cell). The extracellular bacteria were removed by washing with PBS. The infected macrophages were treated with antibacterial agents at 2x and x4 MIC concentrations and the relative intensity of the fluorescence was measured [emission wavelength (581 nm)] via UV–Vis spectroscopy in 24, 48, and 72h for inhibition of intracellular bacterial growth. Surviving Mtb cells were confirmed by CFU method (36).
Resynthesis of UT-01320 (12)
UT-01320 (12) was resynthesized according to the procedures reported previously (37–39). Purity of 12 was determined to be over 98% via HPLC analysis (column: Kinetex 5u C18 100A, 250 × 4.60 mm, solvent: MeOH/H2O = 25/75, flow rate: 0.5 mL/min., UV: 254 nm).
RESULTS
Development of a convenient WecA transferase assay
Although extensive studies on assay developments of MurX/MraY-type phosphotransferases have been reported (31,32,40–42), there are a limited number of studies reported on practical WecA assays (15,17,18,43). The reported WecA assay protocols are not amenable to HTS and they require radiolabeled UDP-GlcNAc and chromatographic separation of the product for quantitation. To date, WecA-catalyzed reactions have been carried out under a variety of biochemical conditions in which the effects of pH, concentrations of divalent cation and detergent, and effects of additives (ATP, EDTA, etc) were evaluated. Moreover, specificity of WecA against the structure of polyprenyl phosphate was reported (44–50). After extensive optimization efforts, we could achieve the high-yielding WecA-catalyzed reactions with UDP-Glucosamine-C6-FITC (1) or UDP-Glucosamine-C6-Dansyl (2) and exogenous decaprenyl phosphate (C50-P) that significantly increases reliability of WecA assays.
Specificity of WecA against nucleotide substrate and optimization of reaction conditions
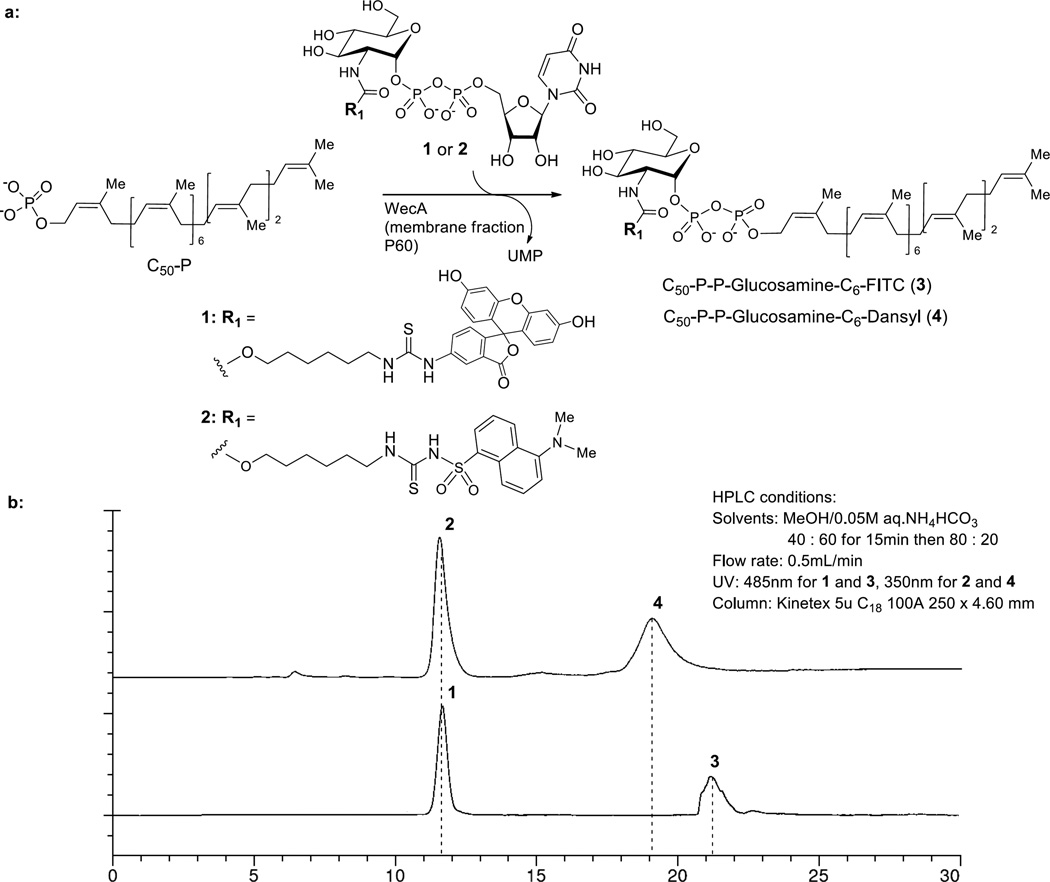
While developing high-throughput screens for GlcNAc transferase such as MurG, we identified that UDP-Glucosamine-C6-FITC (1) is recognized by Mtb MurG to form the decaprenyl-P-P-GlcNAc-MurNAc-(pentapeptide) (lipid II) analogue. We sought to explore whether these modified UDP-GlcNAc derivatives could be applied for WecA assays. To facilitate the screening for WecA inhibitors, specificity of WecA against UDP-Glucosamine-C6-FITC (1) and UDP-Glucosamine-C6-Dansyl (2) was examined using the membrane fraction (P-60) prepared from M. smegmatis and exogenous decaprenylphosphate (C50-P). Because WecA and MurX/MraY use different UDP-2-deoxy-2-N-acetyl-D-hexosamine donor substrates, we first applied the optimized reaction conditions established for MurX/MraY (50 mM MgCl2, 0.5% Triton-X, 50 mM Tris-HCl buffer at pH 8.0) to the WecA-catalyzed transformation from 1 (or 2) to C50-P-P-Glucosamine-C6-FITC (3) (or C50-P-P-Glucosamine-C6-Dansyl (4)) (31). The results show that P-60-catalyzed phosphotransfer reactions of 1 and 2 with C50-P resulted in the formation of 3 and 4 with 7.5–10% yields in 2 h. The generated WecA products 3 and 4 could be separated by partition with n-butanol (n-BuOH). In this process, majority of 1 and 2 remained in the water phase. The consumption of 1 or 2 (water phase) and generation of 3 or 4 (n-BuOH phase) could be monitored via reverse-phase HPLC (solvent: CH3OH:0.05 M aq. NH4HCO3 = 40 : 60 to 80 : 20, UV: 485 or 350 nm, flow rate: 0.5 mL/min). Under these conditions, the retention times of UDP-Glucosamine-C6-FITC (1) and C50-P-P-Glucosamine-C6-FITC (3) were 11.6 min. and 21.2 min., respectively (Fig. 2). The HPLC retention times of 3 and 4 were identical to those of the chemically synthesized molecules. Because of wider applicability of fluorescein (FITC) for bioanalysis, we decided to utilize UDP-Glucosamine-C6-FITC (1) towards the development of a WecA assay.
FIG 2.
WecA-catalyzed biosynthesis of C50-P-P-Glucosamine-C6-fluorescent analogues 3 and 4.
a: UDP-GlcNAc donor analogues 1 and 2 can be recognized by WecA. b: Fluorescence detection-based HPLC at the endpoint of the reaction.
Although the synthesis of C50-P-P-Glucosamine-C6-FITC (3) could be achieved with 7.5–10% yield in 2 h using the UDP-GlcNAc fluorescent probe 1, C50-P, and P-60, optimization of reaction condition is important to improve assay efficiency and to reduce false-positive and false-negative errors in high-throughput screening (HTS) assays. The effects of adding ATP, EDTA, detergent, and co-factors have been previously reported for WecA-catalyzed reactions by using radiolabeled UDP-GlcNAc and undecaprenyl phosphate (C55-P). In general, conversion of over 20% or more of reactions provides reliable analyses in fluorescence-based assays for screening; enzyme inhibitory assays of conversion of 10% or below produce high standard errors by false-positive and false-negative assay data.
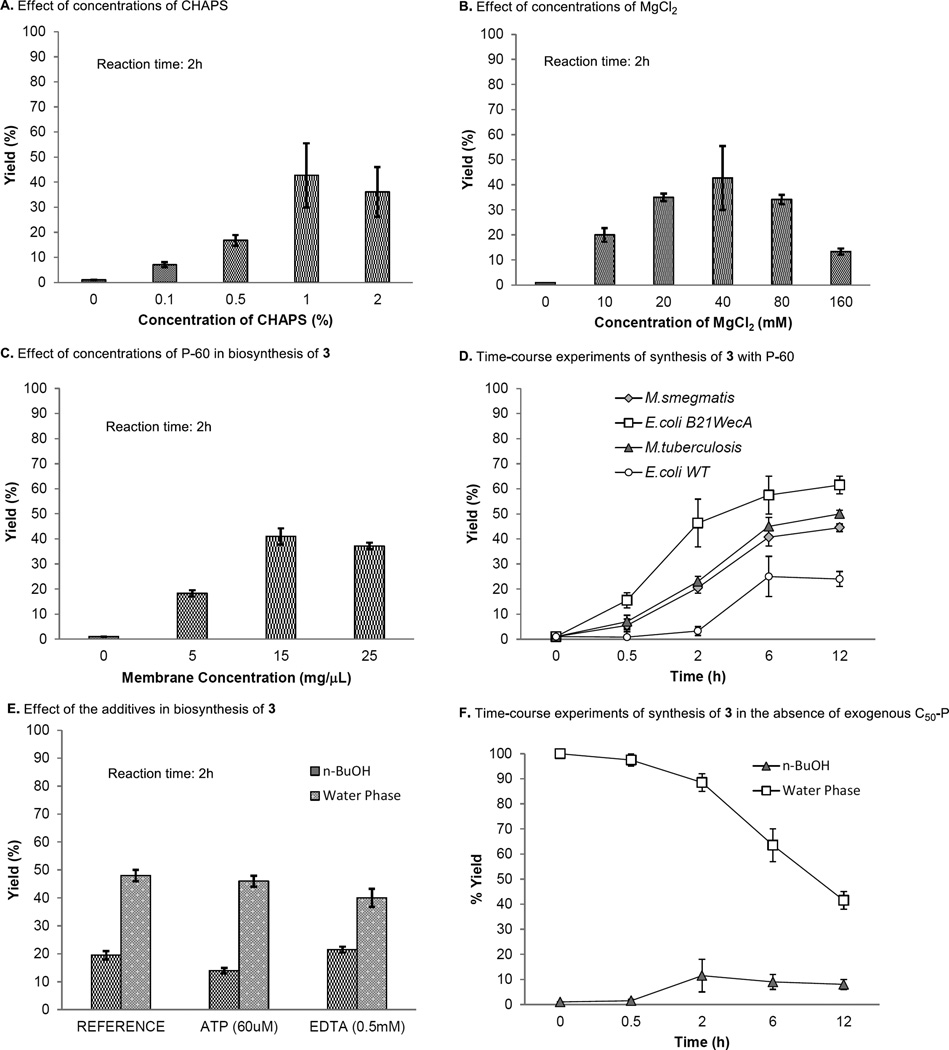
WecA-catalyzed synthesis of C50-P-P-Glucosamine-C6-FITC (3) with UDP-Glucosamine-C6-FITC (1) and decaprenyl phosphate (3 equivalents against 1) in the presence of 0.5% of triton X (optimized concentrations for MurX/MraY) furnished 3 in 7.5–10% yields after 2h (31). As summarized in Fig 3A, the effect of surfactant, CHAPS was observed in the transformation from 1 to 2 (50); it was determined that 1% of CHAPS is the ultimate concentration of the surfactant. Under the conditions with 1% of CHAPS, the optimum MgCl2 concentration was 40 mM (17) (Fig 3B); the same reaction furnished 3 in 46% yield at 37 °C (pH 8.0) after 2 h. In the time-course experiments, application of higher concentrations of WecA enzyme (P-60) increased the product yield of 3. However, saturation of the product formation was observed at the concentration of P-60 at 15 µL (1mg P-60/µL) (Fig 3C). Under the optimized conditions, the synthesis of 3 was increased in a time-dependent manner; WecA-catalyzed transformation of 1 with decaprenyl phosphate (3 equivalents) furnished 3 in 57.5% yield after 6 h (Fig 3D). Among the other reaction parameters examined for M. smegmatis WecA-catalyzed synthesis of 3, the reaction temperatures (30 vs. 37 °C) and pH (8.0 vs. 8.3) did not noticeably affect the reaction rate. Although the other groups reported effects of ATP or EDTA in WecA-catalyzed GlcNAc-P-P-C55 formation (15,17,18), we concluded that these additives show no effect in the transformation from 1 to 3 with the P-60 obtained from M. smegmatis (Fig 3E). It is worth mentioning that the active P-60 membrane consumed UDP-Glucosamine-C6-FITC (1) in the absence of exogenous C50-P but formed 3 by using endogenous polyprenyl phosphate(s). As illustrated in Fig 3F, the time-course study demonstrated that the consumption of 1 was well correlated to the generation of 3; in this reaction, 3 was formed in 5–10% yield after 2–12h. The optimized conditions established with the P-60 membrane fraction from M. smegmatis were effective in the same reaction with the other sources of P-60 prepared from Mtb and E. coli. At the concentration of P-60 [15 µL (1mg P-60/µL)] optimized in Fig 3C, the P-60 prepared from a WecA overexpressed E. coli strain (E. coli B21wecA) dramatically improved the transformation from 1 to 3 compared to that with the P-60 from a wild type E. coli. (Fig 3D). Quantitations of the remaining 1 and generated 3 in each reaction mixture were performed via reverse-HPLC. Signal-to-noise ratio of HPLC analyses using UV (485 nm) is excellent for quantitation of 25 µL of the assay mixtures. The range of linearity was established by injections (via an auto sampler) of six concentrations of chemically synthesized 3 and 1 (r ≥ 0.9), and limit of detection was determined to be much lower than 0.1 µM concentrations. The Km value for UDP-Glucosamine-C6-FITC (1) was 194.7 µM at concentrations of 140 µM of exogenous decaprenyl phosphate; this was similar to the Km values obtained with undecaprenyl phosphate (Km: 195.5 µM). The Vmax for C50-P-P-Glucosamine-C6-FITC (3) synthesis by the crude membrane containing M. smegmatis WecA (P-60) was determined to be 0.404 µM/min. through the Michaelis-Menten plot. Significant difference in WecA-catalyzed phosphoryltransfer reactions is that the transformation between UDP-Glucosamine-C6-FITC (1) and C50-P-P-Glucosamine-C6-FITC (3) is not a reverse process, whereas the recent studies have concluded that Thermotoga maritima WecA-catalyzed reaction shows equilibrium between UDP-GlcNAc and C55-P-P-GlcNAc (51). Similarly, MraY/MurX-catalyzed phosphoryltransfer of polyprenyl phosphate to Park’s nucleotide is known to be an equilibrium reaction (31).
FIG 3.
WecA-catalyzed biosynthesis of C50-P-P-Glucosamine-C6-FITC (3).
A: Effect of the phase transfer catalyst CHAPS. B: Effect of concentrations of MgCl2. C: Effect of concentrations of P-60 obtained from M. smegmatis. D: Time-course experiments of biosynthesis of 3 with different sources of membrane fractions. E: Effect of additives (ATP and EDTA). F: Time-course experiments of biosynthesis of 3 in the absence of exogenous C50-P.
Development of UV/Vis spectroscopy-based assay for WecA
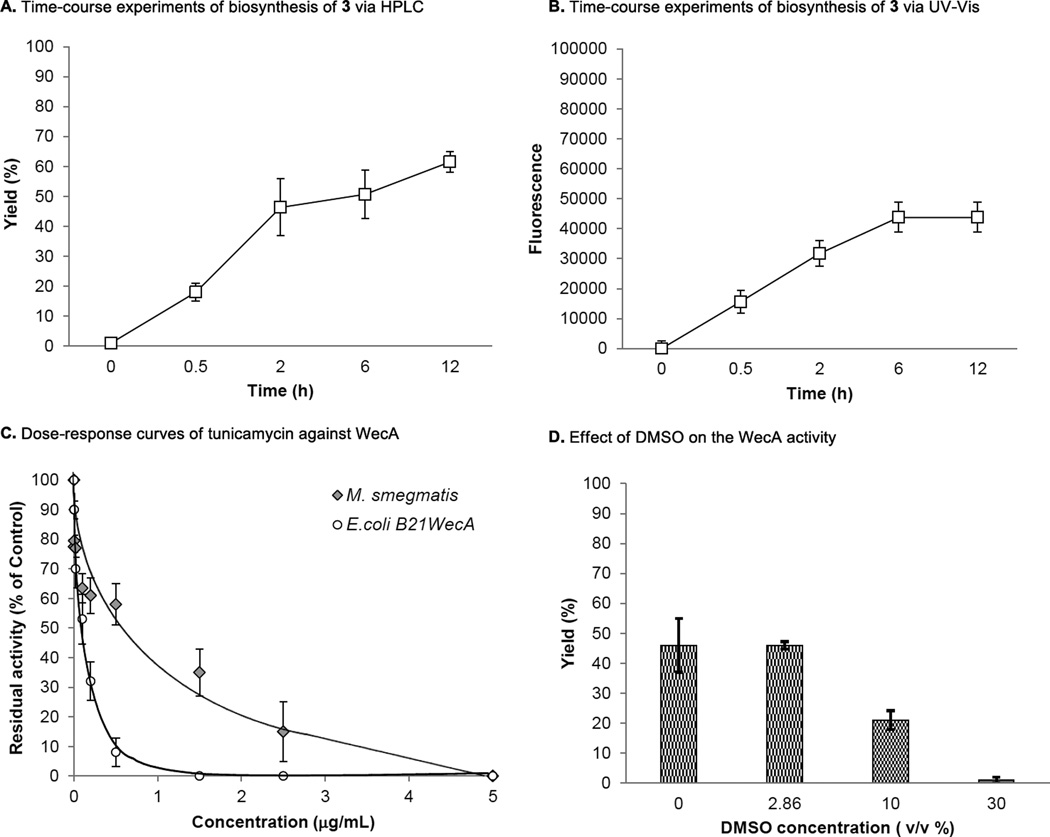
C50-P-P-Glucosamine-C6-FITC (3) is readily extracted with n-BuOH and UDP-Glucosamine-C6-FITC (1) remains in water media. Taking advantage of the hydrophobic character of 3, the fluorescence in n-BuOH extract of WecA reaction was monitored via ultraviolet-visible (UV-Vis) spectrometry (excitation of 485nm, emission of 528nm); the UV-Vis-based assay was performed at a sufficiently high concentration of 1 (22.5 µM) for UV-Vis spectrometry and enough concentrations of C50-P that fulfill the Km value. Progress of the WecA-catalyzed reaction of 1 was determined periodically and monitored for 12h. As shown in Fig 4B, an increase in fluorescence signal was observed in a time-dependent manner that was well-correlated to the yield curve obtained via the HPLC method (Fig 4A). A new assay method developed here was validated by demonstrating the inhibition of WecA activity by a known MurX/WecA inhibitor, tunicamycin (5); generation of 3 was inhibited by tunicamycin in a dose-dependent manner (Fig 4C). The IC50 value of tunicamycin against M. smegmatis WecA and E.coli WecA was determined to be 0.59 µg/mL and 0.12 µg/mL by constructing dose-response curves via UV-Vis spectrometry (Fig 4B). The data summarized in Table 1 were obtained from at least three independent experiments and the standard deviation was less than 10%. It was determined that DMSO did not inhibit the WecA assays at 5% (v/v) concentrations. However, inhibition of the reactions was started at 10% (v/v) of DMSO; approximately 50% of the enzyme activity was reduced at this concentration (Fig 4D).
FIG 4.
Validation of WecA assays via UV-Vis.
A: Time-course experiments of biosynthesis of 3 monitored via HPLC. B: Time-course experiments of biosynthesis of 3 monitored via UV-Vis. C: Dose-response curves of tunicamycin against E.coli and M. smegmatis WecA. D: Effect of DMSO on the WecA activity.
Table 1.
Assays against a library of capuramycin analogues against the WecA enzyme (P-60 from E. coli B21WecA).
 | |||||
|---|---|---|---|---|---|
| entry | Compound | WecA inhibition (%)a | IC50 (µg/mL)b | ||
| 0.02 µg/mL | 2 µg/mL | 20 µg/mL | |||
| 1 | Tunicamycin (5) | 36.0 (38.5)c | 97.0 (100) | 100 (100) | 0.120 ± 7.80 |
| 2 | Capuramycin (6) | - | - | - | - |
| 3 | UT-01309 (7) | - | - | - | - |
| 4 | SQ641 (8) | - | - | - | - |
| 5 | UT-03347 (9) | 11.0 (15.5) | 23.0 (29.0) | 80.5 (84.5) | ND |
| 6 | UT-03328 (10) | 0 (0) | 23.0 (28.0) | 60.5 (75.5) | ND |
| 7 | UT-03325 (11) | 30.0 (35.0) | 35.0 (45.0) | 55.0 (60.5) | ND |
| 8 | UT-01320 (12) | 42.0 (45.0) | 100 (100) | 100 (100) | 0.0354 ± 9.14 |
| 9 | DMSO | 0 (0) | 0 (0) | 0 (0) | - |
1) Reaction conditions: UDP-Glucosamine-C6-FITC (1, 0.56 µL), MgCl2 (0.5 M; 4 µL); CHAPS (5%; 10 µl), β-mercaptoethanol (50 mM, 5µL) Tris-buffer (pH = 8.0; 50 mM, 9.74 µL), C50-P (4mM, 1.7 µL), P-60 (E. coli B21WecA) (18 µL), 37 °C, 2h.; The Km :194.7 µM (for 1).
Each experiment was performed three times, and the average IC50 values were summarized.
WecA inhibitory activity in parentheses were obtained via HPLC.
Specificity of WecA against the structures of prenyl phosphate
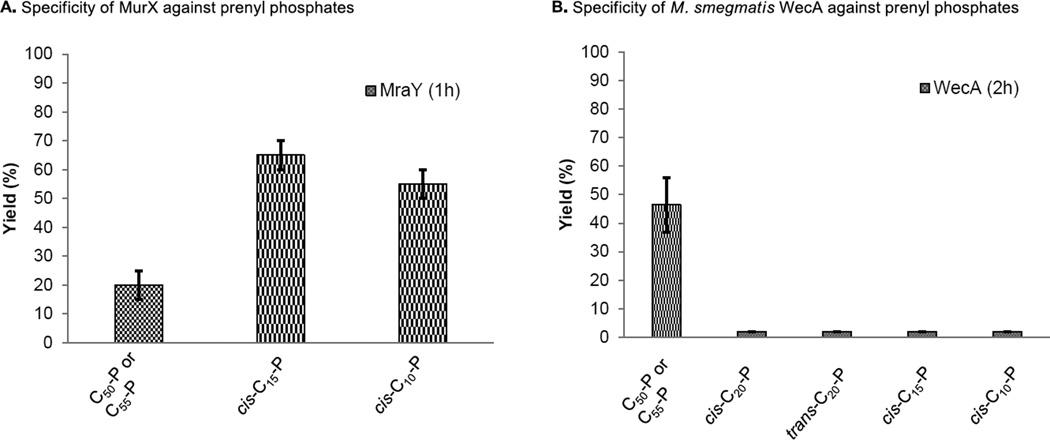
We identified that neryl phosphate (cis-C10-P) and (2Z,6E)-farnesyl phosphate (cis-C15-P) could be recognized by the MurX/MraY-type phosphotransferases to form the corresponding lipid I analogues, whereas, trans-C10-P and -C15-P, and cis- and trans-phytols (C20-P) were not recognized by MurX/MraY enzymes (24,31). To investigate the specificity of WecA against truncated prenyl phosphates, we evaluated WecA-catalyzed transformations of UDP-Glucosamine-C6-FITC (1) with a series of synthetic prenyl phosphates. Due to the fact that endogenous decaprenyl phosphate (C50-P) exist in the M. smegmatis P-60 membrane could be utilized in the WecA-catalyzed transformation of 3, generations of prenyl-P-P-Glucosamine-C6-FITC analogues with the truncated lipid-P were determined by comparison of the retention times of the chemically synthesized analogues via HPLC. The generated products were also quantitated by HPLC. Unlike MurX/MraY enzymes, C10-P, C15-P, and C20-P were not incorporated by WecA; the corresponding prenyl-P-P-Glucosamine-C6-FITC derivatives were not formed (Fig 5). In these experiments, C50-P-P-Glucosamine-C6-FITC was generated in 5–10% yield. Undecaprenyl phosphate (C55-P) was equally effective to C50-P in WecA catalyzed transformation from 1 to 3 using P-60 prepared from M. smegmatis.
FIG 5.
Comparison of specificity of MurX and WecA against prenyl phosphates.
A: Specificity of MurX against C55-P, C50-P, cis-C15-P and cis-C10-P. B: Specificity of M. smegmatis WecA against C55-P, C50-P, cis- and trans-C20-P, cis-C15-P and cis-C10-P.
Identification of new WecA inhibitors by screening a library of capuramycin-based analogues
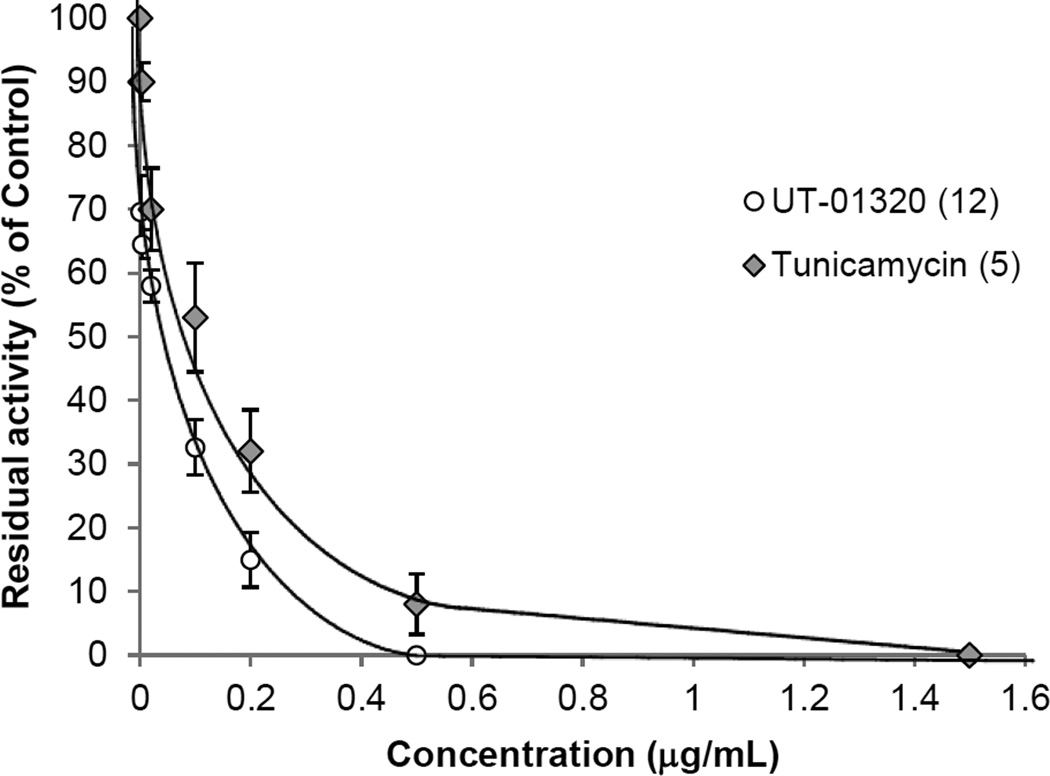
A natural product, capuramycin is unique in the development of MurX/MraY inhibitors for new TB drugs (52–54). We have generated a series of capuramycin analogues towards improving its in vivo efficacy (37,38). To identify WecA inhibitors, we applied the UV-Vis-based WecA assay to screen an optimized library of 50 capuramycin analogues in duplicate with 384-well plates. In these screening, the P-60 membrane fraction isolated form E. coli B21WecA was applied. All compounds were screened at three different concentrations (0.02, 2.0, and 20 µg/mL). Each plate contained four control wells: the first one with the denatured P-60 (heated at 100 °C for 5 min.), the second one without P-60, the third one addition of 30% (v/v) of DMSO and the forth one with tunicamycin at 20 µg/mL. Under the assay conditions, four molecules were identified as WecA inhibitors. The identified WecA inhibitors were confirmed by the HPLC-based assays at 0.02 2.0, and 20 µg/mL concentrations. A dose-response curve of 12 against WecA obtained via UV-Vis spectrometry determined the IC50 value of 12 to be 35.4 ng/mL (Fig 6). The observed inhibition of the WecA enzyme by 12 is more potent than that of tunicamycin (IC50 0.12 µg/mL). It is important to note that the inhibitory activities of WecA inhibitors identified using P-60 from E. coli B21wecA was well-correlated to those against the WecA containing membranes from M. smegmatis ATCC607 (Fig 4C).
FIG 6.
Dose-response curves for the WecA inhibitory activities of UT-01320 (12) and tunicamycin (5).
WecA containing membrane fraction (P-60) was prepared from E. coli B21WecA and applied to the inhibition studies.
Mycobactericidal activity of UT-01320
We reported that UT-01320 (12) killed non-replicating (dormant) Mtb at low concentrations under low-oxygen conditions, whereas selective MurX inhibitors killed only replicating Mtb under aerobic conditions (11). We resynthesized enough amounts of 12 to characterize its in vitro efficacy against WecA enzymes and to assay against Mtb under different growth conditions. Efficacy of newly synthesized 12 was confirmed by assay against replicating and non-replicating Mtb. As previously reported, UT-01320 did not exhibit MurX enzyme inhibitory activity even at 400 µg/mL concentrations; however, it was noticed that 12 inhibited WecA with the IC50 values of 35.4 ng/mL (E.coli wecA) and 50.7 ng/mL (M. smegmatis wecA), respectively. Effectiveness of 12 against non-replicating Mtb was first determined by using an Mtb H37Rv (pFCA-luxAB) under low-oxygen conditions (LORA). UT-01320 is a selective WecA inhibitor that kills both replicating and non-replicating Mtb at 1.5 (MABA) and 2.58 µg/mL (LORA), respectively (35); the MICLORA/MICMABA value was determined to be 1.72 (MICLORA/MICMABA = 7.35 for rifampicin). Effectiveness of 12 against non-replicating Mtb was confirmed by a modified Wayne model and nutrient starving conditions.
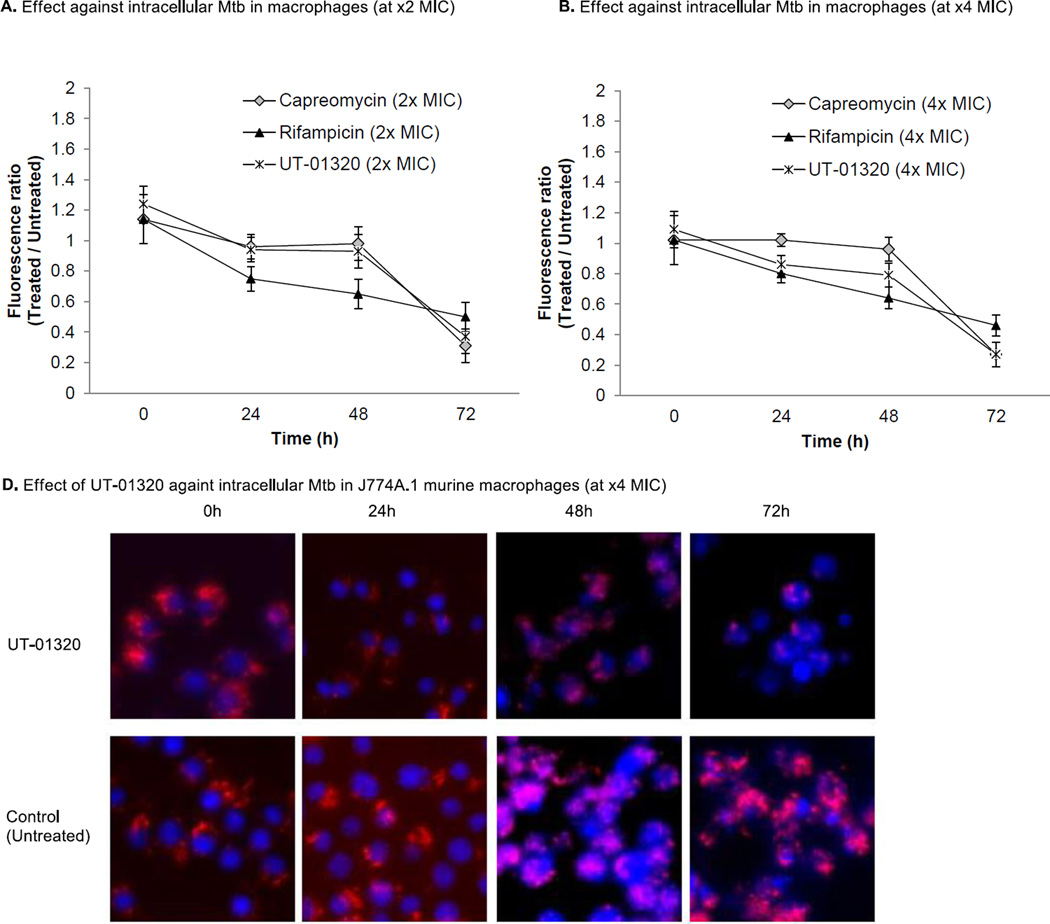
Effectiveness of 12 in non-replicating Mtb was also confirmed via assays using a Wayne model under low-oxygen conditions at the MICLORA concentrations (2.58 µg/mL) (11). Activity of 12 against intracellular Mtb was evaluated. Murine macrophage cell J774A.1 infected by a transformant Mtb CDC1551 containing tdTomato (MOI = 10) were treated with 12 (at x2 and x4 MICLORA). After 24, 48, and 72h of incubation, the relative intensity of the fluorescence was measured (emission wavelength (581 nm)) via UV–Vis spectroscopy (Fig. 7A and 7B). Alternatively, the lysates were tenfold serially diluted in 7H10-S broth and inoculated on 7H11-S plates to determine the number of viable cell-associated Mtb to confirm the bactericidal effect of 12 against intracellular Mtb in 72h. UT-01320 killed Mtb in infected macrophages at x2 MICLORA concentrations within 72h. Fig 7A and 7B showed clearly that 12 kills intracellular Mtb as effectively as capreomycin (a 2nd line TB drug). Bactericidal effect of 12 against the intracellular Mtb was distinguished from that of rifampicin in 72h. The intracellular activity of 12 was visualized via fluorescence confocal microscopy; significant reduction of intracellular Mtb was observed in 48–72h compared to untreated control cultures (Fig 7C).
FIG 7.
Effect of UT-01320 (12) and representative TB drugs against intracellular Mtb CDC1551-tdTomato (a transformant Mtb CDC1551 containing tdTomato) in macrophages (J774A.1 cells).
A: Time-kill curve for intracellular Mtb at 2xMIC concentration. B: Time-kill curve for intracellular Mtb at 4xMIC concentration. C: Fluorescence confocal microscopy of J774A.1 murine macrophages infected with tdTomato (red) expressing Mtb after the treatment of UT-01320 (at 4xMIC). Four shows images were obtained for the treated and untreated cells for 72h. Fixed cells were stained with a blue fluorescent stain, DAPI (4’,6-diamidino-2-phenylindole) to visualize nuclei.
DISCUSSION
The bacterial translocase I (MraY/MurX) and polyprenyl phosphate-GlcNAc-1-phosphate transferase (WecA/TagO) utilize undecaprenyl or decaprenyl phosphate as the donors, but use different UDP-2-deoxy-2-N-acetyl-D-hexosamine acceptors (55–60). Function of MurX/MraY in peptidoglycan biosynthesis has been studied extensively and MurX is considered a promising drug target for developing new TB drugs (11,54,61). Some MurX inhibitors are known to kill Mtb much faster than the FDA-approved TB drugs; capuramycin analogues kill 99% of Mtb in 2–7 days, suggesting that targeting MurX will help ease the burden caused by tuberculosis (11,52). MurX inhibitors are effective only against replicating Mtb under aerobic conditions. Although essentiality of WecA in growth of Mycobacterium spp. was reported (8), WecA has never been characterized thoroughly with a selective inhibitor molecule. Recently, the MurX/MraY inhibitor, CPZEN-45 was reported to inhibit WecA at low concentrations (15). However, efficacy of CPZEN-45 against non-replicating (dormant) Mtb has not been characterized. WecA catalyzes the first step in the biosynthesis of mycolyl-arabinogalactan conjugates (Fig. 1). These exclusive cell wall lipids play an important role in survival of Mtb in macrophages. Thus, it is particularly important to discover a selective WecA inhibitor in order to study the function of WecA and cell wall biosynthesis in intracellular Mtb.
The reported assays for WecA enzymes with radiolabeled UDP-GlcNAc and polyprenyl phosphate use gel-based separation system to obtain the WecA enzyme product for quantitation. Enzyme inhibitory assays that do not depend on radioisotopes are ideal for medium- to high-throughput screenings, for which fluorescence-based bioanalytical assays are better suited. However, no fluorescent probe of UDP-GlcNAc has been developed for phosphotransferase-catalyzed reactions, although the fluorescence-incorporated UDP-GlcNAc analogue was applied in a fluorescence polarization (FP) assay to identify active-site inhibitors of MurG (62). In this study, we have demonstrated, for the first time, that UDP-Glucosamine-C6-FITC (1) can be a substrate for the WecA enzymes. In the WecA assay protocols reported previously, efficacy of addition of ATP or EDTA and inactivation of WecA enzyme by choosing the detergent, Tritox-X-100 (Hyland et al. 2003) were described (15,18). We examined efficacy of these factors thoroughly in the formation of 3 with M. smegmatis WecA. 0.5% Triton X-100 is the ultimate concentrations for MurX/MraY-catalyzed lipid I biosynthesis (31). Under the same concentration of Triton X-100, M. smegmatis WecA catalyzed transformation from 1 to 3 with 7.5–10% yields in 2 h. Under our optimized conditions using 1% CHAPS, the yield was improved to over 46% yield (pH 8.0, 37 °C). The Km value for 1 was 194.7 µM at the concentrations of 140 µM of exogenous decaprenyl phosphate (C50-P). Under the same conditions, the Km of UDP-GlcNAc was 110 µM, suggesting that the C6-linkered FITC group does not affect the catalytic activity of WecA. The Vmax for C50-P-P-Glucosamine-C6-FITC synthesis by M. smegmatis WecA (P-60) was determined to be 0.4041 µM/min. Despite that E. coli and Mtb WecA sequences have only 29% identity and 44% similarity (63), enzyme kinetics and response inhibition of tunicamycin against Mtb WecA and M. smegmatis WecA are similar to that of E. coli WecA (Fig 3D). Expression level of WecA in E. coli is significantly low and transformation of 1 with C50-P furnished 3 in less than 5% yields in 2 h, whereas over 20.5% of 3 was generated with M. smegmatis WecA. P-60 prepared from a WecA overexpressed E. coli strain (E. coli B21wecA) improved the transformation from 1 to 3; C50-P-P-Glucosamine-C6-FITC (3) was generated in 46% yield in 2 h. MurX/MraY are relatively tolerated in the structure of prenylphosphate (lipid-P); in MurX-catalyzed lipid I analogue syntheses with UDP-MurNAc-pentapeptide (Park’s nucleotide), the reactions were proceeded with cis-neryl phosphate (cis-C10-P) and (2Z,6E)-farnesyl phosphate (cis-C15-P) to form the corresponding lipid I analogues with over 60% yields in 1 h (24,31). In these reactions, generation of C50-lipid I was not observed. In contrast, WecA-catalyzed transformation of 1 with cis-C10-P or cis-C15-P or α-cis-geranylgeranyl phosphate (cis-C20-P) did not provide the corresponding prenyl-P-P-Glucosamine-C6-FITC derivatives, but generated C50-P-P-Glucosamine-C6-FITC in 5–10% yield, whose reactions were achieved by using endogenous decaprenyl phosphate (C50-P) (Fig 5). Thus, it was concluded that the WecA enzymes are specific to the prenyl substrate; in our experiments, all truncated prenyl phosphates including α-trans prenyl phosphates failed to react with 1 (43,51).
The fluorescent-based assay developed here is based on the hydrophobic property of C50-P-P-Glucosamine-C6-FITC (3) that can readily be extracted with n-BuOH and the aqueous solubility of UDP-P-P-Glucosamine-C6-FITC (1). These chemical characteristics make the WecA assays very convenient. The UV-Vis-based WecA assays show good correlation with the conversions and inhibitions by the inhibitor molecules quantitated via reverse-phase HPLC. Because WecA-catalyzed reactions optimized here yield the FITC-conjugated product in 45–50% yields (approximately 11.25 mM), quantitation of such high concentrations of the fluorescent improves the assay sensitivity significantly and diminishes assay errors caused by low conversions. To validate the fluorescent-based WecA assay developed here, we examined this assay protocol by screening a series of capuramycin analogues (a library of 50 analogues) with positive- and negative-control molecules. These assay data are summarized in Table 1. A known MraY/WecA inhibitor tunicamycin (5) exhibited the enzyme inhibitory activity in a dose-dependent manner. MraY/MurX activity of 5 was thoroughly characterized in vitro (64). The IC50 value of 5 against MurX is 2.31 µg/mL (31). Thus, we concluded that tunicamycin is a rather strong WecA inhibitor. A strong MurX inhibitor 7 (IC50 0.065 µg/mL) did not inhibit WecA at 20 µg/mL concentrations. The known MurX inhibitors, capuramycin (6) and SQ641 (8) also showed no WecA enzyme inhibitory activity at 20 µg/mL concentrations. In screening of a library of 50 capuramycin analogues via the fluorescence-based method, four molecules, 9–12 were identified to interfere with E. coli WecA at 2.0 µg/mL concentrations; the hit rate in these assays was 0.08%. The identified molecules in the first screening were confirmed by the HPLC analyses of the same n-BuOH extracts of the reaction mixtures. Thus, the UV-Vis-based assay of WecA examined here can be performed with standard analytical devises and will be adapted to medium- to high-throughput formats with close to ideal Z’-factors (0.5–1.0) (65). The Z’-factor was estimated from the data summarized in Table 1; an estimated Z’-factor was 0.60, and thus, the new WecA assay method described here is considered to be an excellent assay.
The mechanism of mycobactericidal activity of a non-MurX inhibitor, UT-01320 (12) has not been identified since its discovery, although interaction of 12 with a bacterial RNA polymerase was studied (11). However, we demonstrate here that 12 exhibited a strong WecA inhibitory activity; the reaction was inhibited completely at 2.0 µg/mL concentration. A capuramycin analogue UT-01320 (12) exhibited a strong WecA enzyme inhibitory activity with the IC50 value of 35.4 ng/mL. We also show that UT-01320 (12) kills both replicating and non-replicating Mtb at relatively low concentrations. Further, 12 could kill intracellular Mtb in 72h; the observed intracellular bactericidal activities was equal to those of capreomycin, a second line TB drug, and showed faster killing effect than rifampicin, a first line TB drug.
In conclusion, we have developed convenient methods for preparation of WecA enzyme substrates and an assay amenable to high- and medium throughput screens. We have validated the assay by screening a library of 50 capuramycin analogues and identifying UT-01320 (12) as a selective WecA inhibitor with bactericidal activity against replicating and non-replicating Mtb, as well as Mtb in macrophages. The efficacy of UT-01320 (12) against non-replicating bacteria is especially significant, suggesting that the WecA enzyme is necessary to maintain viability of Mtb in a latent state (66,67).
Acknowledgments
The National Institutes of Health is greatly acknowledged for financial support of this work (AI084411 and GM114611). We also thank University of Tennessee for generous financial support. NMR data were obtained on instruments supported by the NIH Shared Instrumentation Grant. The following reagent was obtained through BEI Resources, NIAID, NIH: Mycobacterium tuberculosis, Strain H37Rv and Gamma-Irradiated Mycobacterium tuberculosis, NR-14819. The authors gratefully acknowledge Drs. William Clemons (California Institute of Technology), Yoshimasa Ishizaki and Katsuhisa Yamazaki (The Institute of Microbial Chemistry), and Michael McNeil (Colorado State University) for useful discussions. We thank Mr. Shou M. Kurosu for proofreading this manuscript.
REFERENCES
- 1.Stover CK, Warrener P, VanDevater DR, Sherman DR, Arain TM, Langhorne MH, Anderson SW, Towell JA, Yuan Y, McMurray DN, Kreiswirth BN, Barry CE, Baker WR. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature. 2000;405:962–966. doi: 10.1038/35016103. [DOI] [PubMed] [Google Scholar]
- 2.Connolly LE, Edelstein PH, Ramakrishnan L. Why is long-term therapy required to cure tuberculosis? PLoS Med. 2007;4:435–442. doi: 10.1371/journal.pmed.0040120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miranda MS, Breiman A, Allain S, Deknuydt F, Altare F. The tuberculous granuloma: An unsuccessful host defense mechanism providing a safety shelter for the bacteria? Clin. Dev. Immunol. 2012;2012:1–14. doi: 10.1155/2012/139127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Debnath J, Siricilla S, Wan B, Crick DC, Lenaerts AJ, Franzblau SG, Kurosu M. Discovery of selective menaquinone biosynthesis inhibitors against Mycobacterium tuberculosis. J Med. Chem. 2012;55:3739–3755. doi: 10.1021/jm201608g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurosu M, Mahapatra S, Narayanasamy P, Crick DC. Chemoenzymatic synthesis of park’s nucleotide: toward the development of high-throughput screening for MraY inhibitors. Tetrahedron Lett. 2007;48:799–803. [Google Scholar]
- 6.Timothy DH, Lloyd AJ, Roper DI. Phospho-MurNAc-pentapeptide translocase (MraY) as a target for antibacterial agents and antibacterial proteins. Infect. Disord. Drug Targets. 2006;6:85–106. doi: 10.2174/187152606784112128. [DOI] [PubMed] [Google Scholar]
- 7.Grzegorzewicz AE, Ma YF, Jones V, Crick DC, Liav A, McNeil MR. Development of a microtitre plate-based assay for lipid-linked glycosyltransferase products using the mycobacterial cell wall rhamnosyltransferase WbbL. Microbiology. 2008;154:3724–3730. doi: 10.1099/mic.0.2008/023366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin Y, Xin Y, Zhang W, Ma Y. Mycobacterium tuberculosis Rv1302 and Mycobacterium smegmatis MSMEG_4947 have WecA function and MSMEG_4947 is required for the growth of M smegmatis. FEMS Microbiol. Lett. 2010;310:54–61. doi: 10.1111/j.1574-6968.2010.02045.x. [DOI] [PubMed] [Google Scholar]
- 9.Bacon J, Alderwick LJ, Allnutt JA, Gabasova E, Watson R, Hatch KA, Clark SO, Jeeves RE, Marriott A, Rayner E, Tolley H, Pearson G, Hall G, Besra GS, Wernisch L, Williams A, Marsh PD. Non-replicating Mycobacterium tuberculosis elicits a reduced infectivity profile with corresponding modifications to the cell wall and extracellular matrix. PLoS ONE. 2014;9:e87329. doi: 10.1371/journal.pone.0087329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Dabbagh B, Mengin-Lecreulx D, Bouhss A. Purification and characterization of the bacterial UDP-GlcNAc:Undecaprenyl-phosphate GlcNAc-1-phosphate transferase WecA. J Bacteriol. 2008;190:7141–7146. doi: 10.1128/JB.00676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siricilla S, Mitachi K, Wan B, Franzblau SG, Kurosu M. Discovery of a capuramycin analog that kills non-replicating Mycobacterium tuberculosis and its synergistic effects with translocase I Inhibitors. J. Antibiot. 2014;68:271–278. doi: 10.1038/ja.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandish PE, Kimura KI, Inukai M, Southgate R, Lonsdale JT, Bugg TD. Modes of action of tunicamycin, liposidomycin B, and mureidomycin A: inhibition of phospho-N-acetylmuramyl-pentapeptide translocase from Escherichia coli. Antimicrob. Agents. Chemother. 1996;40:1640–1644. doi: 10.1128/aac.40.7.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bogatcheva E, Dubuisson T, Protopopova M, Einck L, Nacy CA, Reddy VM. Chemical modification of capuramycins to enhance antibacterial activity. J. Antimicrob. Chemother. 2011;66:578–587. doi: 10.1093/jac/dkq495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koga T, Fukuoka T, Doi N, Harasaki T, Inoue H, Hotoda H, Kakuta M, Muramatsu Y, Yamamura N, Hoshi M, Hirota T. Activity of capuramycin analogs against Mycobacterium tuberculosis, Mycobacterium avium and Mycobacterium intracellular in vitro and in vivo. J. Antimicrob. Chemother. 2004;54:755–760. doi: 10.1093/jac/dkh417. [DOI] [PubMed] [Google Scholar]
- 15.Ishizaki Y, Hayashi C, Inoue K, Igarashi M, Takahashi Y, Pujari V, Crick DC, Brennan PJ, Nomoto A. Inhibition of the first step in synthesis of the mycobacterial cell wall core, catalyzed by the GlcNAc-1-phosphate transferase WecA, by the novel caprazamycin derivative CPZEN-45. J. Biol. Chem. 2013;288:30309–30319. doi: 10.1074/jbc.M113.492173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barr K, Rick PD. Biosynthesis of enterobacterial common antigen in Escherichia coli In Vitro synthesis of lipid linked intermediates. J. Biol. Chem. 1987;262:7142–7150. [PubMed] [Google Scholar]
- 17.Alexander DC, Valvano MA. Role of the rfe gene in the biosynthesis of the Escherichia coli 07-specific lipopolysaccharide and other O-specific polysaccharides containing N-Acetylglucosamine. J. Bacteriol. 1994;176:7079–7084. doi: 10.1128/jb.176.22.7079-7084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyland SA, Anderson MS. A high-throughput solid-phase extraction assay capable of measuring diverse polyprenyl phosphate: sugar-1-phosphate transferases as exemplified by the WecA, MraY, and MurG proteins. Anal. Biochem. 2003;317:156–165. doi: 10.1016/s0003-2697(03)00088-5. [DOI] [PubMed] [Google Scholar]
- 19.Berg S, Kaur D, Jackson M, Brennan PJ. The glycosyltransferases of Mycobacterium tuberculosis - roles in the synthesis of arabinogalactan, lipoarabinomannan, and other glycoconjugates. Glycobiology. 2007;17:35R–56R. doi: 10.1093/glycob/cwm010. [DOI] [PubMed] [Google Scholar]
- 20.Pasquina LW, Santa Maria JPS, Walker S. Teichoic acid biosynthesis as an antibiotic target. Curr. Opin. Microbiol. 2013;16:531–537. doi: 10.1016/j.mib.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ha S, Chang E, Lo MC, Men H, Park P, Ge M, Walker S. The kinetic characterization of Escherichia coli MurG using synthetic substrate analogues. J. Am. Chem. Soc. 1999;121:8415–8426. [Google Scholar]
- 22.Li K, Kurosu M. Synthetic studies on Mycobacterium tuberculosis specific fluorescent park’s nucleotide probe. Heterocycles. 2008;76:455–469. [Google Scholar]
- 23.Mitachi K, Mohan P, Siricilla S, Kurosu M. One-pot protection-glycosylation reactions for synthesis of lipid II analogues. Chem. Eur. J. 2014;20:1–6. doi: 10.1002/chem.201400307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitachi K, Siricilla S, Klaić L, Clemons WM, Kurosu M M. Chemoenzymatic syntheses of water-soluble lipid I fluorescent probes. Tetrahedron Lett. 2015;56:3441–3446. doi: 10.1016/j.tetlet.2015.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chojnacki T, Jankowski W, Mańkowski T, Sasak W. Preparative separation of naturally occurring mixtures of polyprenols on hydroxyalkoxypropyl-Sephadex. Anal. Biochem. 1975;69:114–119. doi: 10.1016/0003-2697(75)90572-2. [DOI] [PubMed] [Google Scholar]
- 26.Skorupińska-Tudek K, Bieńkowski T, Olszowska O, Furmanowa M, Chojnacki T, Danikiewicz W, Swiezewska E. Divergent pattern of polyisoprenoid alcohols in the tissues of Coluria geoides: A new electrospray lonization MS approach. Lipids. 2003;38:981–990. doi: 10.1007/s11745-003-1152-3. [DOI] [PubMed] [Google Scholar]
- 27.Danilov LL, Druzhinina TN, Kalinchuk NA, Maltsev SD, Shibaev VN. Polyprenyl phosphates: synthesis and structure-activity relationship for a biosynthetic system of Salmonella anatum O-specific polysaccharide. Chem. Phys. Lipids. 1989;51:191–203. doi: 10.1016/0009-3084(89)90006-6. [DOI] [PubMed] [Google Scholar]
- 28.Rezwan M, Laneelle MA, Sander P, Daffe M. Breaking down the wall: Fractionation of mycobacteria. J. Microbiol. Methods. 2007;68:2–39. doi: 10.1016/j.mimet.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 29.Wolfe LM, Mahaffey SB, Kruh NA, Dobos KM. Proteomic definition of the cell wall of Mycobacterium tuberculosis. J. Proteome. Res. 2010;9:5816–5826. doi: 10.1021/pr1005873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bouhss A, Crouvoisier M, Blanot D, Mengin-Lecreulx D. Purification and characterization of the bacterial MraY translocase catalyzing the first membrane step of peptidoglycan biosynthesis. J. Biol. Chem. 2004;279:29974–29980. doi: 10.1074/jbc.M314165200. [DOI] [PubMed] [Google Scholar]
- 31.Siricilla S, Mitachi K, Skorupinska-Tudek K, Swiezewska E, Kurosu M. Biosynthesis of a water-soluble lipid I analogue and a convenient assay for translocase I. Anal. Biochem. 2014;461:36–45. doi: 10.1016/j.ab.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapiro AB, Jahic H, Gao N, Hajec L, Rivin O. A high-throughput, homogeneous, fluorescence resonance energy transfer-based assay for phospho-N-acetylmuramoyl-pentapeptide translocase (MraY) J. Biomol. Screening. 2012;17:662–672. doi: 10.1177/1087057112436885. [DOI] [PubMed] [Google Scholar]
- 33.Ha S, Chang E, Lo MC, Men H, Park P, Ge M, Walker S. The kinetic characterization of Escherichia coli MurG using synthetic substrate analogues. J. Am. Chem. Soc. 1999;37:8415–8426. [Google Scholar]
- 34.Ma Y, Münch D, Schneider T, Sahl HG, Bouhss A, Ghoshdastider U, Wang J, Dötsch V, Wang X, Bernhard F. Preparative scale cell-free production and quality optimization of MraY homologues in different expression modes. J. Biol. Chem. 2011;286:38844–38853. doi: 10.1074/jbc.M111.301085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho SH, Warit S, Wan B, Wang CH, Pauli GF, Franzblau SG. Low-oxygen-recovery assay for high-throughput screening of compounds against nonreplicating Mycobacterium tuberculosis. Antimicrob. Agents. Chemother. 2007;51:1380–1358. doi: 10.1128/AAC.00055-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong Y, Yao H, Ren H, Subbian S, Cirillo SLG, Sacchettini JC, Rao J, Cirillo JD. Imaging tuberculosis with endogenous β-lactamase reporter enzyme fluorescence in live mice. PNAS. 2010;107:12239–12244. doi: 10.1073/pnas.1000643107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Siricilla S, Aleiwi BA, Kurosu M. Improved synthesis of capuramycin and its analogues. Chemistry. Europ. J. 2013;19:13847–13858. doi: 10.1002/chem.201302389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurosu M, Li K. Synthetic studies towards the identification of novel capuramycin analogs with antimycobacterial activity. Heterocycles. 2009;77:217–225. [Google Scholar]
- 39.Kurosu M, Li K, Crick DC. Concise synthesis of capuramycin. Org. Lett. 2009;11:2393–2396. doi: 10.1021/ol900458w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Branstorm AA, Midha S, Longley CB, Han K, Baizman ER, Axelrod HR. Assay for identification of inhibitors for bacterial MraY translocase and MurG transferase. Anal. Biochem. 2000;280:315–319. doi: 10.1006/abio.2000.4530. [DOI] [PubMed] [Google Scholar]
- 41.Solapure SM, Raphael P, Gayathri CN, Barde SP, Chandrakala B, Das KS, deSousa SM. Development of a microplate-based scintillation proximity assay for MraY using a modified substrate. J. Biomol. Screen. 2005;10:149–156. doi: 10.1177/1087057104272007. [DOI] [PubMed] [Google Scholar]
- 42.Ravishankar S, Prasanna Kumar V, Chandrakala B, Jha RK, Solapure SM, deSousa SM. Scintillation proximity assay for inhibitors of Escherichia coli MurG and, optionally, MraY. Antimicrob. Agents. Chemother. 2005;49:1410–1418. doi: 10.1128/AAC.49.4.1410-1418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Dabbagh B, Blanot D, Mengin-Lecreulx D, Bouhss A. Preparative enzymatic synthesis of polyprenyl-pyrophosphoryl-N-acetylglucosamine, an essential lipid intermediate for the biosynthesis of various bacterial cell envelope polymers. Anal. Biochem. 2009;391:163–165. doi: 10.1016/j.ab.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 44.Price NP, Momany FA. Modeling bacterial UDP-HexNAc: polyprenol-P HexNAc-1-P transferases. Glycobiology. 2005;15:29R–42R. doi: 10.1093/glycob/cwi065. [DOI] [PubMed] [Google Scholar]
- 45.Amer AO, Valvano MA. Conserved amino acid residues found in a predicted cytosolic domain of the lipopolysaccharide biosynthetic protein WecA are implicated in the recognition of UDP-N-acetylglucosamine. Microbiology. 2001;147:3015–3025. doi: 10.1099/00221287-147-11-3015. [DOI] [PubMed] [Google Scholar]
- 46.Anderson MS, Eveland SS, Price NP. Conserved cytoplasmic motifs that distinguish sub-groups of the polyprenol phosphate:N-acetylhexosamine-1-phosphate transferase family. FEMS Microbiol. Lett. 2000;191:169–175. doi: 10.1111/j.1574-6968.2000.tb09335.x. [DOI] [PubMed] [Google Scholar]
- 47.Merino S, Jimenez N, Molero R, Bouamama L, Regué M, Tomás JM. A UDP-HexNAc:Polyprenol-P GalNAc-1-P Transferase (WecP) representing a new subgroup of the enzyme family. J. Bacteriol. 2011;193:1943–1952. doi: 10.1128/JB.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patel KB, Toh E, Fernandez XB, Hanuszkiewicz A, Hardy GG, Brun YV, Bernards MA, Valvano MA. Functional characterization of UDP-Glucose:Undecaprenyl-Phosphate Glucose-1-Phosphate transferases of Escherichia coli and Caulobacter crescentus. J. Bacteriol. 2012;194:2646–2657. doi: 10.1128/JB.06052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lehrman MA. A family of UDP-GlcNAc/MurNAc: polyisoprenol-P GlcNAc/MurNAc-1-P transferases. Glycobiology. 1994;4:768–771. doi: 10.1093/glycob/4.6.768. [DOI] [PubMed] [Google Scholar]
- 50.Rush JS, Rick PD, Waechter CJ. Polyisoprenyl phosphate specificity of UDP-GlcNAc:undecaprenyl phosphate N-acetylglucosaminyl 1-P transferase from E. coli. Glycobiology. 1997;7:315–322. doi: 10.1093/glycob/7.2.315. [DOI] [PubMed] [Google Scholar]
- 51.Al-Dabbagh, Olatunji S, Crouvoisier M, El Ghachi M, Blanot D, Mengin-Lecreulx D, Bouhss A. Catalytic mechanism of MraY and WecA, two paralogues of the polyprenyl-phosphate N-acetylhexosamine 1-phosphate transferase superfamily. Biochimie. 2016;127:249–257. doi: 10.1016/j.biochi.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 52.Nikonenko BV, Reddy VM, Protopopova M, Bogatcheva E, Einck L, Nacy CA. Activity of SQ641, a capuramycin analog, in a murine model of tuberculosis. Antimicrob. Agents. Chemother. 2009;53:3138–3139. doi: 10.1128/AAC.00366-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winn M, Goss RJM, Kimura K, Bugg TDH. Antimicrobial nucleoside antibiotics targeting cell wall assembly: Recent advances in structure–function studies and nucleoside biosynthesis. Nat. Prod. Rep. 2009;27:279–304. doi: 10.1039/b816215h. [DOI] [PubMed] [Google Scholar]
- 54.Yamashita A, Norton E, Petersen PJ, Rasmussen BA, Singh G, Yang Y, Mansour TS, Ho DM. Muraymycins, novel peptidoglycan biosynthesis inhibitors: synthesis and SAR of their analogues. Bioorg. Med. Chem. Lett. 2003;13:3345–3350. doi: 10.1016/s0960-894x(03)00671-1. [DOI] [PubMed] [Google Scholar]
- 55.Dini C. MraY inhibitors as novel antibacterial agents. Curr. Top. Med. Chem. 2005;5:1221–1236. doi: 10.2174/156802605774463042. [DOI] [PubMed] [Google Scholar]
- 56.Lehrer J, Vigeant KA, Tatar LD, Valvano MA. Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-Antigen lipopolysaccharide. J. Bacteriol. 2007;189:2618–2628. doi: 10.1128/JB.01905-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clarke BR, Bronner D, Keenleyside WJ, Severn WB, Richards JC, Whitfield C. Role of Rfe and RfbF in the initiation of biosynthesis of D-galactan I, the lipopolysaccharide O antigen from Klebsiella pneumoniae serotype O1. J. Bacteriol. 1995;177:5411–5418. doi: 10.1128/jb.177.19.5411-5418.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schäffer C, Wugeditsch T, Messner P, Whitfield C. Functional expression of enterobacterial O-polysaccharide biosynthesis enzymes in Bacillus subtilis. Appl. Environ. Microbiol. 2002;68:4722–4730. doi: 10.1128/AEM.68.10.4722-4730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D'Elia MA, Pereira MP, Chung YS, Zhao W, Chau A, Kenney TJ, Sulavik MC, Black TA, Brown ED. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J. Bacteriol. 2006;188:4183–4189. doi: 10.1128/JB.00197-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amer AO, Valvano MA. The N-Terminal region of the Escherichia coli WecA (Rfe) protein, containing three predicted transmembrane helices, is required for function but not for membrane insertion. J. Bacteriol. 2000;182:498–503. doi: 10.1128/jb.182.2.498-503.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chung BC, Mashalidis EH, Tanino T, Kim M, Matsuda A, Hong J, Ichikawa S, Lee SY. Structural insights into inhibition of lipid I production in bacterial cell wall synthesis. Nature. 2016;533:557–560. doi: 10.1038/nature17636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Helm JS, Hu Y, Chen L, Gross B, Walker S. Identification of active-site inhibitors of MurG using a generalizable, High-throughput glycosyltransferase screen. J. Am. Chem. Soc. 2003;125:11168–11169. doi: 10.1021/ja036494s. [DOI] [PubMed] [Google Scholar]
- 63.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinform. App. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 64.Brandish PE, Kimura KI, Inukai M, Southgate R, Lonsdale JT, Bugg TDH. Modes of action of tunicamycin, liposidomycin B, and mureidomycin A: Inhibition of phospho-N-acetylmuramyl-pentapeptide translocase from Escherichia coli. Antimicrob. Agents and Chemother. 1996;40:1640–1644. doi: 10.1128/aac.40.7.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang JH, Chung TDY, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 66.Griffin JE, Gawronski JD, DeJesus MA, Ioerger TR, Akerley BJ, Sassetti CM. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PloS Pathog. 2011;7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bacon J, Alderwick LJ, Allnutt JA, Gabasova E, Watson R, Hatch KA, Clark SO, Jeeves RE, Marriott A, Rayner E, Tolley H, Pearson G, Hall G, Besra GS, Wernisch L, Williams A, Marsh PD. Non-replicating Mycobacterium tuberculosis elicits a reduced infectivity profile with corresponding modifications to the cell wall and extracellular matrix. PLoS ONE. 2014;9:e87329. doi: 10.1371/journal.pone.0087329. [DOI] [PMC free article] [PubMed] [Google Scholar]