Abstract
The fission yeast model system Schizosaccharomyces pombe is used to study fundamental biological processes. To continue to fill gaps in the S. pombe gene deletion collection, we constructed a set of 90 haploid gene deletion strains covering many previously uncharacterized genes. To begin to understand the function of these genes, we exposed this collection of strains to a battery of stress conditions. Using this information in combination with microscopy, proteomics and mini-chromosome loss assays, we identified genes involved in cell wall integrity, cytokinesis, chromosome segregation and DNA metabolism. This subset of nonessential gene deletions will add to the toolkits available for the study of biological processes in S. pombe.
Keywords: Gene deletions, Schizosaccharomyces pombe, fission yeast, cytokinesis, stress sensitivities, DNA metabolism
Introduction
The utility of large-scale genetic screening for deciphering functional relationships for genes of unknown or redundant function is well established (reviewed in (Costanzo et al., 2011)). Although this approach was developed in budding yeast (reviewed in (Dixon et al., 2009)), it has been successfully adapted for use in the fission yeast, Schizosaccharomyces pombe (Kennedy et al., 2008, Kelly et al., 2011, Chen et al., 2012, Navarro et al., 2012, Pan et al., 2012, Ryuko et al., 2012, Hayles et al., 2013, Zhang et al., 2013, Zhou et al., 2013, Rallis et al., 2014, Ucisik-Akkaya et al., 2014, Chen et al., 2015)(reviewed in (Costanzo et al., 2011)). The majority of S. pombe genetic screens have been performed with a haploid collection of about 3,300 protein coding gene deletions constructed by and obtained from Bioneer Corp. This valuable collection represents a significant portion of the 3,576 non-essential genes known at the time (Kim et al., 2010). At the time this ambitious deletion project began, it was estimated that 4914 protein coding genes existed in the S. pombe genome. Since then, additional protein-coding genes have been identified or predicted (Bitton et al., 2011, Rhind et al., 2011, Hayles et al., 2013), and the current annotated S. pombe genome sequence predicts 5124 protein coding genes (http://www.pombase.org/) (Wood et al., 2012, McDowall et al., 2015). Thus, the particular collection utilized for many genetic screens did not include all possible viable S. pombe gene deletions.
We have constructed an additional 90 haploid, isogenic gene deletion mutants that were not available in the version 3 collection from Bioneer Corp or in a complementary collection that added 281 strains (Chen et al., 2015). We assayed cell growth for each of these gene deletions under a variety of different environmental stresses and found new players in the processes of cell wall integrity, mitosis, and chromosome segregation.
Materials and Methods
Yeast strains, media, and materials
The parent strain used for constructing the gene deletions was ade6-M210 ura4-D18 leu1-32 h+ (KGY247) unless otherwise noted below. All chemicals were from Sigma-Aldrich (St. Louis, MO) unless indicated otherwise.
The following gene deletion strains were generated previously: SPAC23C4.04c, SPAC27E2.12, SPAPB1A10.16, SPCC1393.14, SPAC2F7.16c, SPBC16G5.19, SPBC17G9.06c, SPBC685.08, SPBC6B1.12c, SPBC800.14c, SPBP23A10.11c, SPBPB8B6.06c, SPAC15E1.03, SPBC215.15, SPAC23D3.14c, SPAC9.09, SPAPB15E9.01c, SPBC21D10.06c, SPBPB8B6.03 as kanMX4 deletions and graciously provided to us (Spirek et al., 2010). We attempted to confirm each of these deletions using the dual flank and gene PCR reactions, as described below. Genes underlined indicate gene deletions we were able to confirm. The others were not confirmed bky our PCR methods and were therefore not included in our analysis.
PCR mediated gene deletions
Gene deletions were accomplished using a two-step PCR method we previously described (Chen et al., 2015), with the 2nd round PCR being performed as described (Krawchuk et al., 1999) with the following modifications. Briefly, in addition to the components added as described (Chen et al., 2015), 0.4 μM (final concentration) of each of the two outer most primers (5F and 3R, as shown in Figure 1 in Chen et al., 2015) was also included in the 2nd round PCR reaction, and the amount of Easy A Hi-Fi polymerase (Agilent Technologies, Santa Clara, CA) was increased to 1 μl per 50-μl reaction. The program used by the 2nd round PCR was modified as following: 94°C, 1 min; 7 cycles of (94°C for 30 sec; 45°C for 5 min; and 72°C for 4 min); 20 cycles of (94°C for 1 min; 55°C for 1 min; and 72°C for 4 min); 72°C for 10 min. 5 μl of PCR product was checked on 0.8% agarose gel. In cases where no good 2nd round PCR products were produced, Pfu Turbo (Agilent Technologies, Santa Clara, CA) or PrimeStar GXL (Clontech, Mountain View, CA) polymerases were also used.
Generation of kanMX6 resistant strains by marker exchange
Existing deletion strains containing the ura4+ marker (kindly provided by Paul Russell, Takashi Toda, Jian-Qiu Wu, Sergio Moreno, and Peter Baumann) were used to generate kanMX6 deletion strains as follows. Homologous sequences shared between the ura4 and kanMX6 cassettes were exploited to generate a PCR product for marker exchange from ura4 to kanMX6. Primers containing the homologous region were used to amplify the kanMX6 cassette. Sequences were as follows: 5′-GCTACAAATCCCACTGGCTATATGTATGCATTTGTGTTAAAAAAGTTTGTATAGATT ATTTAATCTACTCAGCATTCTTTCGGATCCCCGGGTTAATTAA-3′ and 5′-GTGATATTGACGAAACTTTTTGACATCTAATTTATTCTGTTCCAACACCAATGTTTAT AACCAAGTTTTATCTTGTTTGTGAATTCGAGCTCGTTTAAAC-3′. The kanMX6 cassette was amplified from the same construct as described for gene deletion (pFA6-kanMX6 [Bahler, 1998]) and the resulting PCR product was transformed into ura4+ deletion strains using lithium acetate transformations (Keeney et al., 1994) and single colonies were selected on YE-G418 plates at 29°C for 3–5 days and colonies were checked by PCR.
Yeast transformations
Transformations were performed as described previously (Chen et al., 2015). Briefly, S. pombe strain leu1-32 ura4-D18 ade6-M210 h+ was grown in YE media at 32°C overnight to mid-log phase, washed in TE/LiAc (10 mM Tris, pH 7.6, 1 mM EDTA, 100 mM lithium acetate) twice, and re-suspended in TE/LiAc. 100 μl of the suspension was incubated with transforming DNA at room temperature for 10 min., mixed with 260 μl of TE/LiAc/PEG (40% PEG in TE/LiAc), vortexed gently and then incubated at 32°C for 30–45 min. 43 μl of DMSO was then added to each tube followed by incubation at 42°C for 6 min. Cells were then washed with sterile H2O before being re-suspended in 5 ml of YE media and incubated at 29°C overnight. 500 μl of the cell suspension was then plated on YE-G418 plates and incubated at 29°C for 3–5 days. Each plate was replica-plated 3 times to fresh YE-G418 plates before colonies were checked by PCR.
Whole cell PCR checking of yeast transformants
Whole cell PCR was used to check and select the colony with the correct deletion. A pointed toothpick was used to add a small amount of cells to each reaction. For each PCR reaction, 5 μl of 4 μM of both forward and reverse oligos were mixed with 10 μl GoTaq green. The following PCR program was used: 95°C, 1 min; 12 cycles of (95°C for 45 seconds, 60°C for 1 min, and 72°C for 1 min); 25 cycles of (95°C for 45 seconds, 52°C for 45 seconds, and 72°C for 1 min). 10 μl of each PCR product was checked on an 0.8% agarose gel.
Sensitivity/resistance assay
Wildtype and deletion strains were grown in yeast extract media (YE) in 96-well plates to saturation. 10-fold serial dilutions of each strain were made and spotted onto minimal medium agar plates or YE agar plates in the absence or presence of the following additions: brefeldin A (BFA, 25 μM); bleomycin (1 μg/ml, Bleo); calcofluor (Calc, 0.5 mg/ml); cycloheximide (CHX, 10 μg/ml); EGTA (5 mM); hydroxyurea (HU, 7.5 mM); KCl (1 M); latrunculin A (Cayman chemical, Ann Arbor, Michigan) (LatA, 0.20 μM); Methyl 1-(butylcarbamoyl)-2-benzimidazolecarbamate (Benomyl, Ben, 10 μg/ml) (Roy et al., 1982); methyl methanesulfonate (MMS, 0.01%); sodium dodecyl sulfate (SDS, 0.005%); sorbitol (Sorb, 1.2 M); thiabendazole (TBZ, 12.5 μg/ml). Plates were incubated at 29°C or other temperatures as indicated for 2–8 days and then colonies were imaged with a scanner. The following criteria were used to define not sensitive (NS), sensitive (S), very sensitive (VS) and resistant (R) strains: deletion strains that grew similarly or within 1 dilution factor of wildtype cells were scored as NS, deletion strains that grew at 2 or more dilutions less than wildtype under same conditions were classified as S, deletion strains that did not grow when wildtype cells grew were classified as VS, and deletion strains that grew better than wild-type cells were scored R.
Microscopy methods
For protein localization studies, live cells were imaged. For visualization of DNA, cells were fixed with ice cold 70% ethanol. After washing with PBS the fixed cells were incubated with PBS containing 5 mg/ml methyl blue (MB) for 15 min on ice. MB-stained cells were then pelleted by centrifugation and re-suspended in PBS prior to imaging. Cells were mixed with 4′,6-diamidino-2-phenylindole (DAPI) before imaging. One of the two following microscopes were used to collect images: a spinning-disk confocal microscope (Ultraview LCI; PerkinElmer) with a 100× NA 1.40 Plan-Apochromat oil immersion objective and a 488-nm argon ion laser (GFP) or a 594-nm helium neon laser (mCherry and RFP) or a personal microscope system (DeltaVision; Applied Precision) including a microscope (IX71; Olympus), 60× NA 1.42 PlanApo and 100× NA 1.40 UPlanSApo objectives, fixed- and live-cell filter wheels, a camera (CoolSnap HQ2; Photometrics), and softWoRx imaging software. Images on the confocal were captured on a charge-coupled device camera (Orca-ER; Hamamatsu Photonics. Images were manipulated in Image J and imported into Adobe Illustrator for construction of final figures.
GFP-Trap purification
A 2 L culture of rng10-GFP was grown in 4X YE media (meaning 4 x the concentration), generating a pellet of cells (approximately 20 mL packed volume). Cells were lysed under native conditions in a glass bead beater with NP-40 buffer (10 mM sodium phosphate, pH 7.0, 0.15 M NaCl, 1% Nonidet P40, 2 mM EDTA, 50 mM sodium fluoride, 100 μM sodium orthovanadate, 5 ug/ml leupeptin) plus Protease Inhibitors (Roche). The lysate was cleared at low speed (ca. 3000 rpm) on a tabletop centrifuge. The supernatant was transferred to a clean Falcon tube and 30 μl of GFP-TRAP magnetic agarose beads (GFP-Trap®_MA, ChromoTek) pre-washed with NP-40 buffer were added and nutated at 4°C for 1 – 1.5 hr. The beads were then washed with NP-40 buffer two times (5 mL) and then once with 5ml Low-NP-40 buffer (0.02% NP-40) prior to elution from beads with 100 μl of 200 mM glycine twice. The eluate was trichloroacetic acid (TCA) precipitated (25% TCA on ice) and proteins were identified using LC-mass spectrometry (MS) as detailed below. A control purification was also performed from untagged yeast and proteins identified by MS were subtracted from the list of potential interactors.
MS analysis
Protein pellets were resuspended, digested and analyzed by LC-MS as previously described (Chen et al., 2013) with the following modifications: Peptide identifications were filtered and assembled using Scaffold (version 4.4.8; Proteome Software, Portland, OR) using the following filters: minimum of 99.0% protein identification probability, minimum of 2 unique peptides, minimum of 95% peptide identification probability. Protein hits were further filtered in Excel to define proteins localized to the division site (TSC ≥ 5) according to Pombase.org (Wood et al., 2012).
Results and Discussion
To expand the toolkits available for S. pombe genetic screens (Gregan et al., 2006, Kim et al., 2010, Rumpf et al., 2010, Spirek et al., 2010, Hayles et al., 2013) we have assembled a collection of 90 gene deletion strains, many of which were missing from previously available deletion collections. We analyzed the morphology of each deletion strain and assayed the growth of this cohort of strains using a battery of stress tests, identifying new players in cell wall integrity, mitosis and chromosome segregation.
Gene deletion construction
We used a PCR-based strategy to make 71 new S. pombe gene deletions, replacing ORFs with the kanMX6 cassette (Wach et al., 1994, Bahler et al., 1998) as we described previously (Chen et al., 2015). Briefly, gene flanks were amplified using PCR and then using overlap extension, fused to the kanMX6 cassette. This product was then transformed into cells and deletions were selected on YE-G418 plates at 29°C. To confirm replacement of the ORFs with kanMX6, PCR reactions were performed on G418-resistant colonies (1–2 per targeted gene). Two PCR reactions targeted the gene flanks from the kanMX6 cassette (3′ and 5′) and a third reaction targeted the gene sequence to make sure the ORF had been removed (ORF check).
Deletion marker and genetic background exchange
A number of additional deletion strains were acquired from other labs as ura4+ or kanR knockout strains to add to the new deletion strain cohort. For ura4+ strains (kindly provided by Paul Russell, Takashi Toda, Jian-Qiu Wu, Sergio Moreno, and Peter Baumann), we used a PCR-based strategy to swap kanMX6 cassettes into the deletion locus. Briefly, the kanMX6 cassette was amplified with flanks corresponding to sequences inside the ura4 cassette to allow for homologous recombination of the marker cassette without using gene-specific sequences. We successfully applied this strategy, swapping ura4+ cassettes out for kanMX6 in seven strains. About a dozen kanR deletion strains constructed previously (Spirek et al., 2010) were confirmed using our PCR methods outlined in experimental methods and we crossed these strains to acquire the appropriate auxotrophic markers and mating type such that they were isogenic with other gene deletion strains in common use (Kim et al., 2010) prior to stress testing. Using these two approaches, a collection of 90 gene deletion strains were constructed (Table S1) and have been deposited in the yeast NBRP, Japan (http://yeast.lab.nig.ac.jp/nig/index_en.html).
Morphology of gene deletion strains
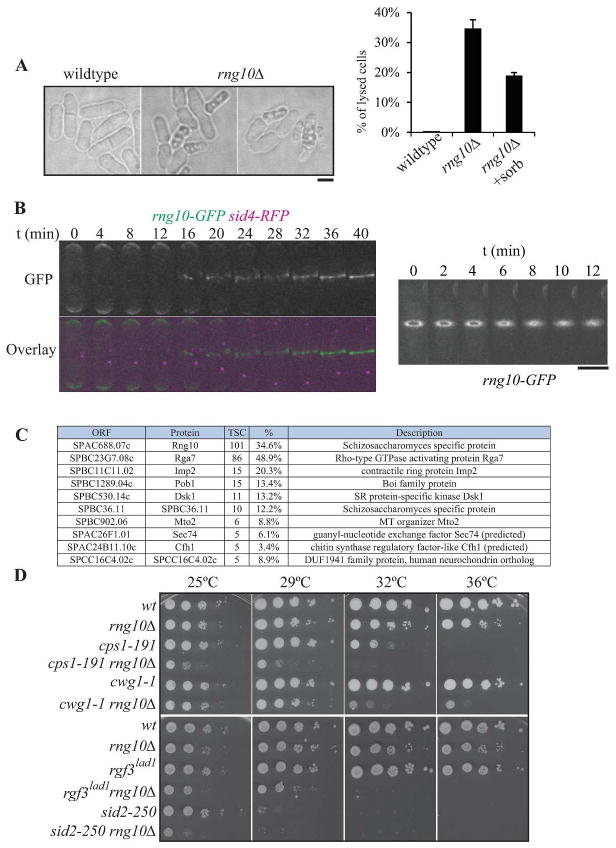
To identify strains with aberrant phenotypes, cells of each deletion strain were examined by light microscopy. One strain exhibited a cell lysis phenotype in over 30% of cells (SPAC688.07c, Fig. 1A). Lysis was partially but not completely suppressed by the addition of sorbitol to the medium (Fig. 1A, right panel). To define the cellular localization of the protein encoded by SPAC688.07c, sequences encoding GFP were endogenously integrated at the 3′ end of SPAC688.07c. The GFP fusion protein was visualized in a strain also containing the spindle pole body (SPB) protein Sid4 fused to RFP (Fig. 1B). Imaging revealed localization of the protein at cell tips in interphase and the cell division site during mitosis and cytokinesis. Because of this localization, this gene product has been named Rng10 for ring protein 10. Real time imaging of live cells indicates that Rng10 arrives at the division site as cells enter anaphase B (Fig. 1B, left panel) and constricts with the contractile ring (CR), but a portion of the protein remains at the cell division site during septation (Fig. 1B, right panel). Purification of Rng10-GFP using GFP-TRAP® beads followed by LC-MS analysis revealed that Rng10’s top protein interactor was Rga7, an F-BAR protein involved in cytokinesis (Martin-Garcia et al., 2014, Arasada et al., 2015) (Fig. 1C), indicating a possible direct role for Rng10 in cytokinesis. Interestingly, rng10Δ exhibits a cell lysis phenotype reminiscent of rga7Δ (Fig. 1A) (Martin-Garcia et al., 2014), suggesting that they may act together to ensure cell wall integrity. Consistent with this idea, rng10 showed negative genetic interactions with mutations in genes encoding cell wall synthetic enzymes Cps1/Bgs1 (Liu et al., 2000) and Cwg1/Bgs4 (Ribas et al., 1991) and positive regulators of those enzymes, the RhoGEF Rgf3 (Tajadura et al., 2004, Morrell-Falvey et al., 2005, Mutoh et al., 2005) and the septation initiation network kinase, Sid2 (Balasubramanian et al., 1998) (Fig. 1D).
Figure 1. Characterization of SPAC688.07cΔ (rng10Δ).
A) Differential interference contrast (DIC) images of wildtype and rng10Δ cells, illustrating the cell lysis phenotype. Quantitation of cell lysis is included to the right (n=100 for each data point, the average from three replicates and standard deviation are plotted). Scale bar, 5 μm. B) Montages of live cell images of rng10-GFP sid4-RFP cells. Left panel emphasizes the arrival time of Rng10 to the division site; right panel shows Rng10 localization during contractile ring constriction. Scale bar for both right and left panels, 5 μm. C) List of proteins that localize to the cell division site identified by LC-MS of a Rng10-GFP GFP-trap experiment. TSC = total spectral counts. D) Negative genetic interactions of rng10Δ with mutations in glucan synthase genes (cps1/bgs1 and bgs4/cwg1) and septation regulatory genes (rgf3 and sid2). 10-fold dilutions of the indicated strains were spotted on YE plates and incubated for 2–3 days at the indicated temperatures.
Stress sensitivities of the deletion strains
Many of these deletions strains have no functional assignment (Wood et al., 2012); thus, to help define the roles of these uncharacterized genes in biological processes and expand knowledge of genes of known function, the growth of each of the 90 deletion strains was assayed under 16 different stress conditions. Specifically, the strains were tested for growth at 29°C on rich YE medium containing fungal microtubule destabilizing drugs thiabendazole (TBZ) or Methyl 1-(butylcarbamoyl)-2-benzimidazolecarbamate (Benomyl, Ben) (Roy et al., 1982), actin cytoskeleton destabilizing drug latrunculin A (LatA)(Ayscough, 1997), DNA replication inhibitor hydroxyurea (HU) (Slater, 1973, Yarbro, 1992), DNA damage inducing drugs bleomycin (Bleo) (Povirk, 1996) and methyl methanesulfonate (MMS) (Beranek, 1990), secretory pathway inhibitor brefeldin A (BFA) (Turi et al., 1994), protein synthesis inhibitor cycloheximide (CHX), cell wall/membrane compromising agents calcofluor (Calc) or sodium dodecyl sulfate (SDS), high salt (KCl), osmolyte sorbitol (Sorb), or the calcium chelator EGTA, and for growth on rich medium at low (19°C) or high temperature (36°C). Finally, the deletion strains were also assayed for growth at 29°C on minimum media plates (Min).
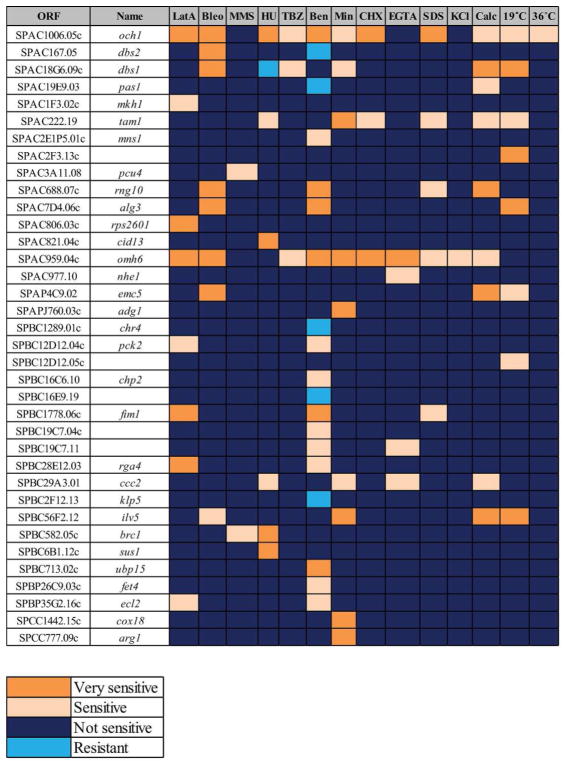
The sensitivities of the 36 deletion strains whose growth was affected by at least one stress are summarized in Figure 2. Loss of a few genes resulted in pleotropic effects (i.e. multiple sensitivities, e.g. och1 and omh6), likely because these genes are involved in core cellular functions, but most deletion strains had unique patterns of sensitivities.
Figure 2. Summary of the deletions that showed sensitivity or resistance to any stress condition.
LatA = latrunculin A, Bleo = bleomycin, MMS = methyl methanesulfonate, HU = hydroxyurea, TBZ = thiabendazole, Ben = benomyl, Min= minimal media, CHX = cycloheximide, Calc = calcofluor. The following criteria were used to define not sensitive (NS), sensitive (S), very sensitive (VS) and resistant (R) strains: deletion strains that grew similarly or within 1 dilution factor of wildtype cells were scored as NS, deletion strains that grew at 2 or more dilutions less than wildtype under same conditions were classified as S, deletion strains that did not grow when wildtype cells grew were classified as VS, and deletion strains that grew better than wild-type cells were scored R.
New players in DNA metabolism
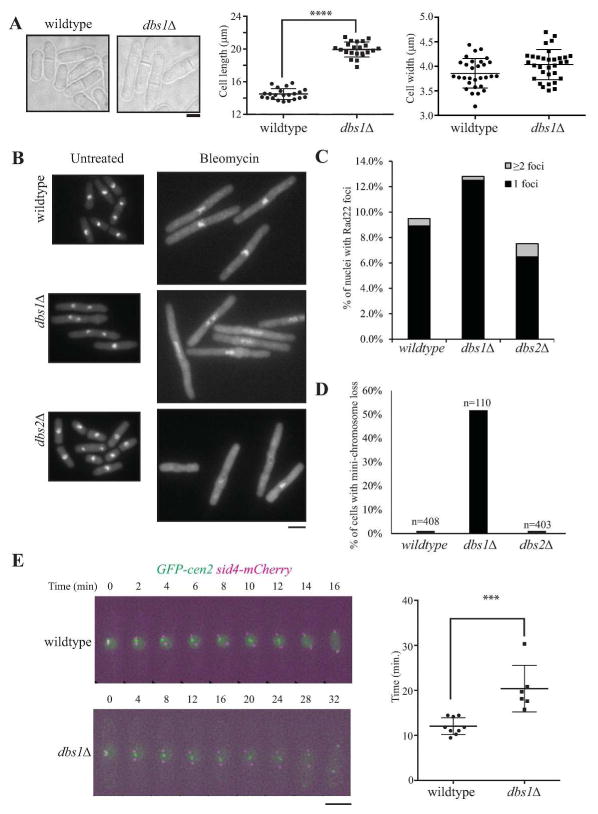
There were 14 deletion mutants sensitive to one or more drugs that interfere with DNA metabolism (Fig. 2, Table S1). The three drugs we used differ in their modes of action: bleomycin causes base loss, single and double strand breaks (Stubbe et al., 1987), MMS methylates bases, blocking the replicative polymerases and causing replication stress (Beranek, 1990, Lundin et al., 2005), and HU inhibits the enzyme ribonucleotide reductase, leading to a depletion of dNTPs and to a stalling of replication forks (Slater, 1973, Yarbro, 1992). Thus the deletion strain sensitivities varied in their response to these perturbations. Two gene deletion strains (SPAC18G6.09cΔ and SPAC167.05Δ) that showed selective sensitivity to bleomycin (they were not sensitive to HU or MMS) were named deletion bleomycin sensitive (dbs) 1 and 2, respectively and were analyzed further.
Dbs1 (SPAC18G6.09c) is only found amongst Schizosaccharomyces species and encodes a protein that has no known domain structure and localizes to the nucleus and cytoplasm (Matsuyama et al., 2006, Wood et al., 2012). We found that dbs1Δ cells are significantly longer (19.9 μm vs 14.5 μm) but not wider (4.03 μm vs 3.86 μm) relative to wildtype (Fig. 3A), possibly because they contain damaged DNA, which may trigger a G2/M checkpoint delay. SPAC167.05Δ (dbs2Δ) was previously described as having a meiotic defect (Gregan et al., 2005), localizes to the nucleus and cytoplasm (Matsuyama et al., 2006) and contains a universal stress protein A domain (Wood et al., 2012). When exposed to bleomycin, both dbs1Δ and dbs2Δ exhibited elongated phenotypes, like wildtype cells (Fig. 3B), indicating an intact DNA damage checkpoint response. However, unlike wildtype, the DNA of dbs1Δ and dbs2Δ appeared indistinct and/or fragmented in many cells, suggesting a possible defect in chromosome organization. Using Rad22-GFP-positive foci (Noguchi et al., 2009) as a marker for DNA damage, we quantitated the number of Rad22-GFP foci in nuclei of wildtype and mutant cells (Fig. 3C). There were more Rad22-GFP foci in dbs1 cells, but fewer in dbs2 cells compared to wild-type (Fig. 3C), suggesting that Dbs2 may be important for assembly or maintenance of DNA repair foci. To explore the possibility that these strains were defective in chromosome segregation, we performed mini-chromosome loss assays and found that dbs1Δ, but not dbs2Δ lost mini-chromosomes at an elevated rate, indicating that dbs1Δ has a defect in chromosome segregation (Fig. 3D). Given the very high rate of mini-chromosome loss in cells lacking Dbs1, we next defined Dbs1’s impact on mitotic timing and chromosome movement. Wildtype and dbs1Δ cells containing spindle pole (Sid4-mCherry) and kinetochore markers (GFP-cen2) were imaged and analyzed for the timing of metaphase and anaphase A, measuring the time from spindle pole body separation to spindle elongation (Fig. 3E). While wildtype cells progressed through this phase of constant spindle length with an average time of 12 min., dbs1Δ cells were delayed and on average spent much longer in metaphase and/or anaphase A (20 min.), indicating significant defects in chromosome segregation and highlighting a role for Dbs1 in mitotic progression. It will be interesting in the future to determine at what cellular locale Dbs1 impacts mitotic progression.
Figure 3. Characterization of deletion sensitive in bleomycin (dbs) strains.
A) Left, DIC images of wildtype and dbs1Δ cells, illustrating the elongated cell phenotype. Scale bar, 5 μm. Middle, quantitation of cell lengths at septation of wildtype and dbs1Δ cells measured in Image J (****p = <0.0001). Right, quantitation of cell widths at septation of wildtype and dbs1Δ cells measured in Image J. B) Images of DAPI staining of wildtype, dbs1Δ and dbs2Δ cells untreated or treated with 1.25 μg/ml bleomycin for 4h at 32°C. C) Percent of nuclei containing 1 or 2+ Rad22-GFP DNA repair foci in wildtype, dbs1Δ and dbs2Δ strains. D) Percent of colonies exhibiting mini-chromosome loss in wildtype and dbs1Δ, and dbs2Δ strains. E) Live cell microscopy of the indicated strains growing at 32°C. Scale bar, 5 μm. Right, quantitation of time from onset of SPB duplication until onset of anaphase B. (***p = 0.0006). Significance was determined by two-tailed unpaired t-test.
In bacteria, the universal stress domain containing proteins, like Dbs2, are thought to play a role in resistance to DNA damaging agents (Kvint et al., 2003), but a role for them in yeast has not been reported. Although dbs2Δ cells have no defect in mini-chromosome retention, they are very sensitive to bleomycin and have fewer Rad22-GFP DNA repair foci, suggesting a conserved role for this protein in protection from DNA damage through an unknown mechanism.
Morphogenesis factors
Eight gene deletions showed increased sensitivity only to latrunculin A (LatA) or LatA plus one additional factor (Fig. 2, Table S1). Loss of either the actin bundler Fim1 or the Rho-GAP Rga4 renders cells very sensitive to LatA treatment, likely because inhibition of actin filament formation further disrupted the actin cytoskeleton resulting in cell death. This is the first report of sensitivities of fim1Δ cells to external stressors.
In sum, we have generated 90 new S. pombe deletion strains in an effort to fill gaps in available deletion collections. In addition to the 36 deletions for which we identified sensitivities to one or more stress conditions, 54 have no obvious phenotypes. These genes may have biological functions not tested in our panel of stress conditions, or they may have redundant functions that mask their functions. Nonetheless, the deletions reported here will provide an important resource for the characterization of these gene functions and, in combination with other deletion collections (Kim et al., 2010, Spirek et al., 2010, Chen et al., 2015) for future genome-wide genetic screens.
Supplementary Material
Acknowledgments
We thank Paul Russell (Scripps Research Institute), Susan L. Forsburg (University of Southern California), Gennaro D’Urso (University of Miami), Peter Baumann (Howard Hughes Medical Institute and Stowers Institute for Medical Research), Li-Lin Du (National Institute of Biological Sciences, Beijing), Anthony M. Carr (University of Sussex), Fred Winston (Harvard Medical School), Danesh Moazed (Howard Hughes Medical Institute and Harvard Medical School), Rob Martienssen (Howard Hughes Medical Institute and Cold Spring Harbor Laboratory), Phong Tran (University of Pennsylvania, Perelman School of Medicine), Takashi Toda (The Francis Crick Institute and Hiroshima University), Jian-Qiu Wu (The Ohio State University), and Sergio Moreno (CSIC/University of Salamanca) for providing reagents and/or financial support for this project. This work was supported by NIH GM101035 to K.L.G. and GMGM098815 to N.R.
Footnotes
In preparation for YEAST March 2016 as a “Genome Screen Report”
References
- Arasada R, Pollard TD. A role for F-BAR protein Rga7p during cytokinesis in S. pombe. J Cell Sci. 2015;128:2259–2268. doi: 10.1242/jcs.162974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough KR. High Rates of Actin Filament Turnover in Budding Yeast and Roles for Actin in Establishment and Maintenance of Cell Polarity Revealed Using the Actin Inhibitor Latrunculin-A. The Journal of Cell Biology. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, McCollum D, Chang L, et al. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 1998;149:1265–1275. doi: 10.1093/genetics/149.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- Bitton DA, Wood V, Scutt PJ, et al. Augmented Annotation of the Schizosaccharomyces pombe Genome Reveals Additional Genes Required for Growth and Viability. Genetics. 2011;187:1207–1217. doi: 10.1534/genetics.110.123497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Beckley JR, McDonald NA, et al. Identification of new players in cell division, DNA damage response, and morphogenesis through construction of Schizosaccharomyces pombe deletion strains. G3 (Bethesda) 2015;5:361–370. doi: 10.1534/g3.114.015701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Broadus MR, McLean JR, et al. Comprehensive proteomics analysis reveals new substrates and regulators of the fission yeast clp1/cdc14 phosphatase. Mol Cell Proteomics. 2013;12:1074–1086. doi: 10.1074/mcp.M112.025924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, McCroskey S, Guo W, et al. A Genetic Screen to Discover Pathways Affecting Cohesin Function in Schizosaccharomyces pombe Identifies Chromatin Effectors. G3: Genes|Genomes|Genetics. 2012;2:1161–1168. doi: 10.1534/g3.112.003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Myers CL, et al. Charting the genetic interaction map of a cell. Current Opinion in Biotechnology. 2011;22:66–74. doi: 10.1016/j.copbio.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Dixon SJ, Costanzo M, Baryshnikova A, et al. Systematic Mapping of Genetic Interaction Networks. Annu Rev Genet. 2009;43:601–625. doi: 10.1146/annurev.genet.39.073003.114751. [DOI] [PubMed] [Google Scholar]
- Gregan J, Rabitsch PK, Rumpf C, et al. High-throughput knockout screen in fission yeast. Nat Protoc. 2006;1:2457–2464. doi: 10.1038/nprot.2006.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J, Rabitsch PK, Sakem B, et al. Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr Biol. 2005;15:1663–1669. doi: 10.1016/j.cub.2005.07.059. [DOI] [PubMed] [Google Scholar]
- Hayles J, Wood V, Jeffery L, et al. A genome-wide resource of cell cycle and cell shape genes of fission yeast. Open Biol. 2013;3:130053. doi: 10.1098/rsob.130053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney JB, Boeke JD. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly FD, Nurse P. Spatial control of Cdc42 activation determines cell width in fission yeast. Molecular Biology of the Cell. 2011;22:3801–3811. doi: 10.1091/mbc.E11-01-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy PJ, Vashisht AA, Hoe KL, et al. A Genome-Wide Screen of Genes Involved in Cadmium Tolerance in Schizosaccharomyces pombe. Toxicological Sciences. 2008;106:124–139. doi: 10.1093/toxsci/kfn153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-U, Hayles J, Kim D, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawchuk MD, Wahls WP. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast. 1999;15:1419–1427. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1419::AID-YEA466>3.0.CO;2-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvint K, Nachin L, Diez A, et al. The bacterial universal stress protein: function and regulation. Curr Opin Microbiol. 2003;6:140–145. doi: 10.1016/s1369-5274(03)00025-0. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang H, Balasubramanian MK. A checkpoint that monitors cytokinesis in Schizosaccharomyces pombe. J Cell Sci. 2000;113( Pt 7):1223–1230. doi: 10.1242/jcs.113.7.1223. [DOI] [PubMed] [Google Scholar]
- Lundin C, North M, Erixon K, et al. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005;33:3799–3811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Garcia R, Coll PM, Perez P. F-BAR domain protein Rga7 collaborates with Cdc15 and Imp2 to ensure proper cytokinesis in fission yeast. J Cell Sci. 2014;127:4146–4158. doi: 10.1242/jcs.146233. [DOI] [PubMed] [Google Scholar]
- Matsuyama A, Arai R, Yashiroda Y, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- McDowall MD, Harris MA, Lock A, et al. PomBase 2015: updates to the fission yeast database. Nucleic Acids Res. 2015;43:D656–661. doi: 10.1093/nar/gku1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell-Falvey JL, Ren L, Feoktistova A, et al. Cell wall remodeling at the fission yeast cell division site requires the Rho-GEF Rgf3p. J Cell Sci. 2005;118:5563–5573. doi: 10.1242/jcs.02664. [DOI] [PubMed] [Google Scholar]
- Mutoh T, Nakano K, Mabuchi I. Rho1-GEFs Rgf1 and Rgf2 are involved in formation of cell wall and septum, while Rgf3 is involved in cytokinesis in fission yeast. Genes Cells. 2005;10:1189–1202. doi: 10.1111/j.1365-2443.2005.00908.x. [DOI] [PubMed] [Google Scholar]
- Navarro FJ, Nurse P. A systematic screen reveals new elements acting at the G2/M cell cycle control. Genome Biol. 2012;13:R36. doi: 10.1186/gb-2012-13-5-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Ansbach AB, Noguchi C, et al. Assays Used to Study the DNA Replication Checkpoint in Fission Yeast. Methods in Molecular Biology, Springer Science + Business Media. 2009:493–507. doi: 10.1007/978-1-60327-815-7_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Lei B, Zhou N, et al. Identification of novel genes involved in DNA damage response by screening a genome-wide Schizosaccharomyces pombe deletion library. BMC Genomics. 2012;13:662. doi: 10.1186/1471-2164-13-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk LF. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 1996;355:71–89. doi: 10.1016/0027-5107(96)00023-1. [DOI] [PubMed] [Google Scholar]
- Rallis C, Lopez-Maury L, Georgescu T, et al. Systematic screen for mutants resistant to TORC1 inhibition in fission yeast reveals genes involved in cellular ageing and growth. Biology Open. 2014;3:161–171. doi: 10.1242/bio.20147245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas JC, Diaz M, Duran A, et al. Isolation and characterization of Schizosaccharomyces pombe mutants defective in cell wall (1-3)beta-D-glucan. J Bacteriol. 1991;173:3456–3462. doi: 10.1128/jb.173.11.3456-3462.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Fantes PA. Benomyl resistant mutants of Schizosaccharomyces pombe cold-sensitive for mitosis. Curr Genet. 1982;6:195–201. doi: 10.1007/BF00390338. [DOI] [PubMed] [Google Scholar]
- Rumpf C, Cipak L, Novatchkova M, et al. High-throughput knockout screen in Schizosaccharomyces pombe identifies a novel gene required for efficient homolog disjunction during meiosis I. Cell Cycle. 2010;9:1802–1808. doi: 10.4161/cc.9.9.11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryuko S, Ma Y, Ma N, et al. Genome-wide screen reveals novel mechanisms for regulating cobalt uptake and detoxification in fission yeast. Molecular Genetics and Genomics. 2012;287:651–662. doi: 10.1007/s00438-012-0705-9. [DOI] [PubMed] [Google Scholar]
- Slater ML. Effect of reversible inhibition of deoxyribonucleic acid synthesis on the yeast cell cycle. J Bacteriol. 1973;113:263–270. doi: 10.1128/jb.113.1.263-270.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirek M, Benko Z, Carnecka M, et al. S. pombe genome deletion project: An update. Cell Cycle. 2010;9:2399–2402. doi: 10.4161/cc.9.12.11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbe J, Kozarich JW. Mechanisms of bleomycin-induced DNA degradation. Chem Rev. 1987;87:1107–1136. [Google Scholar]
- Tajadura V, Garcia B, Garcia I, et al. Schizosaccharomyces pombe Rgf3p is a specific Rho1 GEF that regulates cell wall beta-glucan biosynthesis through the GTPase Rho1p. J Cell Sci. 2004;117:6163–6174. doi: 10.1242/jcs.01530. [DOI] [PubMed] [Google Scholar]
- Turi TG, Webster P, Rose JK. Brefeldin A sensitivity and resistance in Schizosaccharomyces pombe. Isolation of multiple genes conferring resistance. J Biol Chem. 1994;269:24229–24236. [PubMed] [Google Scholar]
- Ucisik-Akkaya E, Leatherwood JK, Neiman AM. A Genome-Wide Screen for Sporulation-Defective Mutants in Schizosaccharomyces pombe. G3: Genes|Genomes|Genetics. 2014;4:1173–1182. doi: 10.1534/g3.114.011049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pöhlmann R, et al. New heterologous modules for classical or PCR-based gene disruptions inSaccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wood V, Harris MA, McDowall MD, et al. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 2012;40:D695–699. doi: 10.1093/nar/gkr853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbro JW. Mechanism of action of hydroxyurea. Semin Oncol. 1992;19:1–10. [PubMed] [Google Scholar]
- Zhang L, Ma N, Liu Q, et al. Genome-Wide Screening for Genes Associated with Valproic Acid Sensitivity in Fission Yeast. PLoS ONE. 2013;8:e68738. doi: 10.1371/journal.pone.0068738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Ma Y, Fang Y, et al. A Genome-Wide Screening of Potential Target Genes to Enhance the Antifungal Activity of Micafungin in Schizosaccharomyces pombe. PLoS ONE. 2013;8:e65904. doi: 10.1371/journal.pone.0065904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.