Abstract
Fluorescence lifetime imaging (FLIm) and Raman spectroscopy are two promising methods to support morphological intravascular imaging techniques with chemical contrast. Both approaches are complementary and may also be used in combination with OCT / IVUS to add chemical specificity to these morphologic intravascular imaging modalities. In this contribution, both modalities were simultaneously acquired from two human coronary specimens using a bimodal probe. A previously trained SVM model was used to interpret the fluorescence lifetime data; integrated band intensities displayed in RGB false color images were used to interpret the Raman data. Both modalities demonstrate unique strengths and weaknesses and these will be discussed in comparison to histologic analyses from the two coronary arteries imaged.
Keywords: fluorescence lifetime, Raman spectroscopy, bimodal, imaging, fiber probes, atherosclerosis
1. Introduction
The rupture of vulnerable atherosclerotic plaques is responsible for both heart attack and stroke. Currently, the morphological structure of vulnerable plaques (a fibrous cap and underlying necrotic core) can be imaged in-vivo in coronary arteries with intravascular optical coherence tomography (OCT). The size of the lipid or necrotic core can be determined with intravascular ultrasound (IVUS). However, both techniques have significant drawbacks. OCT accurately visualizes fibrous caps, but has a very high false positive rate for identification of lipid [1]. IVUS can clearly visualize calcium but has very limited resolution [2]. Either technique would greatly benefit from the addition of an imaging modality that could detect plaque components with high chemical specificity.
Both fluorescence lifetime imaging (FLIm) and Raman spectroscopy can provide specific information about plaque components [3-5] and have been implemented in intravascular catheters [4-7]. In particular, FLIm is useful for quickly measuring lipid to collagen ratios in the fibrous cap[8], which can provide information about plaque stability. Raman spectroscopy can provide further chemical specificity, and is able to, for instance, distinguish between calcifications, cholesterol or carotenoids [9-14]. FLIm can create maps of the surface composition at comparable speed with IVUS or OCT whereas Raman spectroscopy is slower and more suitable for point measurements rather than mapping. Thus the two modalities are complementary as FLIm provides real-time compositional information whereas Raman spectroscopy is more suitable to further investigate regions of interest with high specificity. A FLIm and Raman spectroscopy probe was recently reported that allows for detailed biochemical characterization acquired from tissue [15]. Here we report a comparison of FLIm and Raman spectroscopy with a bi-modal probe for characterization of plaque composition in ex-vivo human coronary arteries.
2. Materials and methods
2.1 Instrumental Setup
The combined FLIm and Raman probe used in this study to image the coronary specimens was described previously[15]. Briefly, the probe consists of one central and nine surrounding 300 μm UV Silica/Silica fibers. The central fiber is used for Raman excitation (785 nm) and equipped with appropriate filters to suppress background fiber fluorescence. Two of the off-centered fibers are used as unfiltered FLIm excitation and collection fibers. The other fibers are used as collection fibers for the Raman signals and are equipped with appropriate filters to block the 785 nm excitation light. The whole fiber package contains a MgF2 lens to focus the excitation on a central spot (450 μm diameter) providing a lateral resolution of 250 μm for the two modalities, resulting in a total outer diameter of 2.2 mm. The working distance of the probe was determined to be less than 1 mm for both channels.
The central excitation fiber was connected to a 785 nm multimode laser (B&W Tec) and the output power at the probe tip was set to 100 mW. All collection fibers of the probe were arranged as a line in an SMA plug, which was connected and aligned to a Holospec spectrometer (Kaiser optical systems) with a low wavenumber transmission grating (LGP785, Kaiser optical systems). The spectrometer was coupled to a Pixis 400B CCD (Princeton Instruments), which was thermoelectrically cooled to −70 °C. The readout of the CCD was triggered externally by a computer to start each line of the lateral scan.
The FLIm component of the probe involved a bifurcated setup for excitation and collection of fluorescence. The excitation was generated using a 355 nm fiber laser (Fianium HE, 355 nm, 80 ps, 10 kHz – 1 MHz, 0.5 μJ). The resulting fluorescence emission was collected using a custom-built wavelength selection module (WSM) as explained previously[16]. This module resolves emission wavelengths with certain bandwidths [CH1 - 390/40 dominated by collagen, CH2 - 452/45 – dominated by elastin and lipids, CH3 - 542/50 dominated by extracellular lipids) using a series of dichroic mirrors and band-pass filters and couples each waveband into delay fibers with different lengths (1, 10 and 19) to generate optical delays between the fluorescence signals from the four wavelength channels. Signals from all channels are detected by a non-gated microchannel plate (MCP) photomultiplier tube (PMT) (R3809U-50, Hamamatsu, 45 ps FWHM) and the signals are amplified (C5594, Hamamatsu, 36 dB) and digitized (PXIe-5185, National Instrument, 12.5 GS/s sampling rate, 3 GHz analog bandwidth). The pulse energy was set to 200 nJ/pulse for all experiments.
Both modalities will penetrate into the samples with different depths based on excitation wavelength. While FLIm has a rather low penetration depth (355 nm excitation wavelength) of about 100 – 200 μm, Raman-spectroscopy (785 nm excitation wavelength) has a higher penetration depth between 400 and 600 μm. This will result in different information acquired from each modality since the imaged volumes differ.
2.2 Sample treatment and imaging
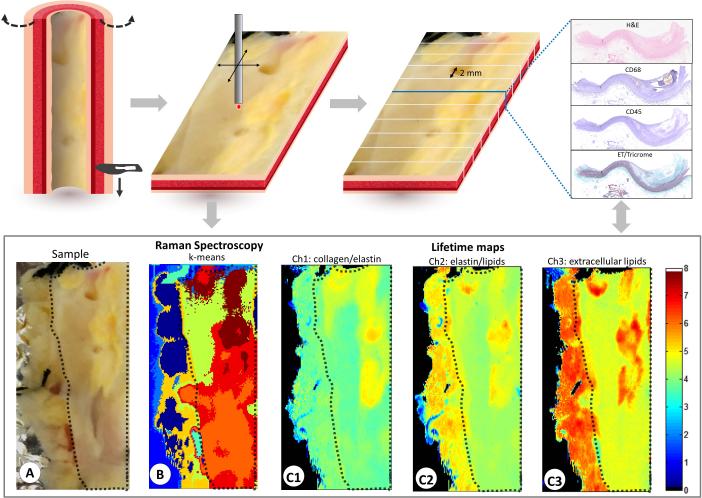
Human left anterior descending (LAD) coronary specimens were obtained from two different donors from the University of Pennsylvania with approval from the institutional review board. Specimens were obtained from a heart transplantation recipient and from a brain-dead organ donor, and prospective informed consent for research use of heart tissue was obtained from the subject and next-of-kin, respectively. Epicardial coronary artery segments were immediately chilled in isopentane, rapidly frozen in liquid nitrogen, shipped overnight and stored at −80 °C. Specimens were thawed in a 37°C phosphate-buffered saline bath and cut open parallel to the direction of blood flow to expose the lumen surface. The cut specimen was sutured to a non-fluorescent rubber mat for imaging (see Fig. 1). The surface of the samples was adjusted to the scanning plane with a 6 axis stage. Both FLIm and Raman images were recorded simultaneously as lateral raster scans with a resolution of 250 μm in the x-direction and a step size of 125 μm in the y-direction with an acquisition time of 22 ms per pixel for both modalities with the excitation powers as mentioned previously (FLIm: 355 nm, 80 ps, 10 kHz – 1 MHz, 0.5 μJ / Raman: 785 nm, 100 mW). This resulted in a bimodal image with 108 × 120 pixels (27 mm × 15 mm) and a total acquisition time of 285 seconds. The total acquisition time is limited by the maximum read out rate of the spectrometer CCD. As shown in overview Fig. 1, following FLIm-Raman bimodal imaging, the specimens were fixed in formalin, decalcified and cut at 2 mm intervals along the entire length of the vessel. Four tissue sections were cut from each 2 mm interval and stained with H&E, CD68, CD45, and Elastin-Masson's Trichrome. The lumen surface of the vessel is delineated by the dashed black boundary on the white light image of the vessel in Fig. 1A. Areas outside this boundary can be disregarded as the signal is from the outer adventitial/fatty tissue.
Figure 1.
Visualization of the datasets generated in this study. The samples were cut along the aorta in the direction of blood flow (long axis) and flattened before the bimodal scan. The white light image of the LAD (A), the k-means Raman spectroscopy data (B), the lifetime maps for the 3 channels (C1 – C3) are shown at the bottom. Histopathology sections were obtained at 2 mm intervals along the entire length of the specimen and H&E, CD68, CD45 and elastic trichrome were done on sequential cuts at each interval.
The fluorescence lifetime values from CH1 (dominated by collagen/elastin), CH2 (dominated by elastin/lipids) and CH3 (dominated by extracellular lipids like cholesterol and lipoproteins) are shown in Fig.1. The acquired Raman map was clustered by a k-means algorithm to determine the most prominent compounds in the area of interest.
2.3 Data evaluation
2.3.1 Histopathology
H&E, Elastin-Masson's Trichrome, CD68 and CD45 stained histopathology sections were obtained at 2 mm intervals along the entire length of the specimen. All histology slides were digitized at 20X magnification (Aperio Scanscope, Aperio Technologies) for analysis. With the help of an expert pathologist, regions of interest on the sections were identified as one of seven pathological groups (also investigated in our previous studies [17])- namely, diffuse intimal thickening (DIT), pathologic intimal thickening (PIT), thin-cap fibroatheroma (TCFA), thick-cap fibroatheroma (ThCFA), ThCFA with macrophage/lymphocyte infiltration in the fibrous cap (ThCFAM), fibrotic plaque (FT) and fibrocalcific plaque (FC). These groups are in accordance with the American Heart Association's modified definitions [18]. The histologic sections were co-registered every 2 mm with an accuracy of about ± 0.25 mm and compared to FLIm classification and Raman false-color maps.
2.3.2 Fluorescence lifetime imaging
The measured fluorescence decay intensity F (t) is a convolution of the actual fluorescence decay p(t) and the instrument impulse response function x(t). In discrete time, for N equal sampling time points ti = iδt , i = 0....., N −1 and sampling interval δt gives:
where k = 0, ..., N − 1. p(k) was expanded on to a set of discrete ordered Laguerre basis functions, bl. such that:
where L and α are the basis parameters and cl is the lth expansion coefficient. Using this formula, p(k) was computed in Matlab using a fast constrained least-square deconvolution technique[19]. The average lifetime was calculated from p(k) as follows:
Consequently, for every measurement in a given region of interest (ROI), the absolute fluorescence intensity, fluorescence lifetime and Laguerre expansion coefficients are computed to generate 2-D intensity and average lifetime maps. The fluorescence intensity is computed as the numerical integration of the decay curve p(k) for each wavelength sub-band.
The fluorescence lifetime values from the three channels were used to classify each pixel into one of seven possible pathological groups. A hierarchical classification scheme was implemented using a previously trained Support Vector Machine (SVM) binary classifier [17]. The SVM was initially developed from data recorded from ex vivo human LAD specimens using an intravascular FLIm catheter. A color-coded classification map was generated based on the pixel by pixel classification by the SVM. (See Fig. 3)
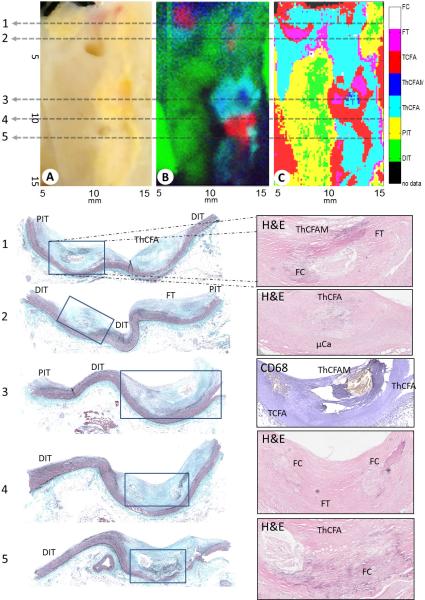
Figure 3.
VIS image of the diseased coronary (A), Raman false color image (B), FLIm classification (C). The false color map generated by identifying the 3 most prominent Raman components (triglycerides and cholesterol, carotenoids and calcium salts) (B) and the SVM classification that was generated using the lifetime maps (C). At the bottom the histology sections from the indicated positions (1-5) are shown. For visualization, the Elastin-Masson's Trichrome stained sections are shown on the left and one complementary stain diagnostically conclusive stains in the overview and one contrary stain from the zoomed areas as indicated are is shown on the right.
2.3.3 Raman imaging
Since the recorded Raman spectra include a fluorescence background as well as zero order residuals from the transmission grating due to grating imperfections, extensive Raman data preprocessing [20] in R [21] was done. A Gaussian kernel smoothing with a bandwidth of 3cm−1 was applied and a background estimation based on the statistic-sensitive, non-linear iterative peak-clipping (SNIP) algorithm [22] was carried out. Subsequently, the spectra were truncated to the wavenumber region between 600 and 1800 cm−1 followed by a vector normalization. Principal components analysis was used to reduce the dimension to 20 scores and to eliminate noise. Next, a k-means cluster analysis in the region of interest with k=6 was performed. The result with the corresponding spectra is shown in Fig. 2.
Figure 2.
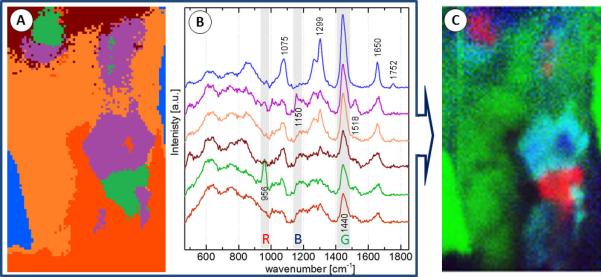
Raman processing summary: The k-means result from the region of interest with k=6 (A) and corresponding mean cluster spectra (B). Several bands of interest were selected (see text) and their integrated band intensities are displayed in RGB false color image to the left (C).
Three of the most prominent bands were selected and their integrated band intensities were displayed in RGB-false colors for visualization (Fig. 2 C). The region from 942 to 967 cm−1 (calcium salts) is displayed in red, from 1130 – 1170 cm−1 (carotenoids that are likely associated with the inflammation processes) is shown as blue and from 1425 to 1465 (triglycerides and cholesterols) is shown as green. The following discussion will only refer to the false color image.
3. Results
Fig. 3 summarizes the FLIm classification and Raman false color images from one diseased human coronary specimen imaged in this study. The bright green area of the Raman false color image (Fig 3B) is dominated by triglycerides associated with the adventitial fat of the artery. The dark green regions correspond to mostly healthy artery and may show intimal thickening or low grade pathologic intimal thickening (DIT / PIT) [5, 9, 11]. The red areas indicate either microcalcifications or calcified plaques. The blue regions show the distribution of carotenoids. When the blue and green regions overlap, they form an intermediate cyan region that indicates the presence of carotenoids and, in this case, extracellular lipids like cholesterol. Carotenoids are usually associated with inflammation and thus it can be assumed that the cyan colored regions represent inflamed and lipid-rich plaques. The tissue associated with the cyan regions imaged in this study is thicker than the penetration depth of the Raman excitation (about 700 μm) and therefore no adventitial fat contributes to the intensity in these areas, whereas the green regions alone were from adventitial fat, which is in agreement with histology.
Five histological sections are shown in Fig. 3 that present a variety of plaque types. These sections are co-registered with the gray dashed lines labeled 1 through 5.
Histology Sections 1 and 2
From left to right, PIT, ThCFAM with some calcification, fibrous tissue, ThCFA and finally DIT can be seen in the trichrome stain (Fig. 1). The FLIm classification map shows PIT, ThCFAM, fibrous tissue and TCFA. The TCFA classification from the FLIm data is likely due to extracellular lipids in the fibrous cap that lengthen the fluorescence lifetime due to the long lifetime of lipid in Ch3. Thus the signal from this region may match that of a lesion with a thinner cap. The Raman false color image shows green from the DIT/PIT areas due to the signal penetrating through the relatively normal vessel into the adventitial fat. The microcalcifications can be seen on the left side of the histology section (red in the Raman false color map), and the carotenoids and cholesterol in the ThCFA in the two lesions in this section (cyan and dark blue in the Raman false color map). Within the same section, FLIm identified the collagenous regions in the ThCFA, ThCFAM and FT while the Raman data identified microcalcifications that were not identified by FLIm as well as the cholesterol deeper in the lesions. The green signal in the Raman color map confirms the PIT/DIT classification of FLIm since the lumen and the media of the artery are relatively thin so that the Raman signal penetrated through the adventitia of the artery. Histology section 2 shows another ThCFA with microcalcifications, DIT, fibrous tissue and PIT. It is similar to histology section 1 as well as the FLIm and Raman results.
Histology Section 3
This histology section shows relatively normal PIT/DIT areas and a TCFA, ThCFAM as well as a ThCFA (all part of the same lesion) on the right of the trichrome stain. Macrophages can be observed in the corresponding zoomed in CD68 section. In the PIT/DIT areas, the Raman signal shows triglycerides (green) indicating that the signal penetrates through to the adventitia, which is in agreement with histology as this part of the artery is very thin. The ThCFAM / TCFA lesion can be easily distinguished in the Raman false color map as a ring shaped intermediate cyan cluster (cholesterol and carotenoids). In the center (blue) spot, there is much less cholesterol compared to carotenoids, which is in agreement with the CD68 stain and indicates a necrotic core. The FLIm classification also accurately identifies this ThCFAM / TCFA lesion.
Histology Section 4
In this section calcium is strongly present as well as DIT. Again the Raman signal shows that in the DIT region the signal is coming from the adventitia of the artery and the FLIm classification is DIT and PIT. The very left edge of the FLIm classification is red and demonstrates TCFA. However, this signal originates from a region of fatty tissue on the edge of the sliced open vessel, thus the signal is similar to TCFA signal although it is associated with adventitial fat. The green area from the Raman signal aligns well with the DIT/PIT FLIm classification. FLIm shows ThCFA and TCFA in the calcified region because crystalline calcium does not fluoresce and thus is difficult to be distinguished by FLIm. However, FLIm is sensitive to the extracellular lipid observed in the fibrous cap, which lengthens the fluorescence lifetime and causes the signal to match that of a lesion with a thinner fibrous cap. This explains the FLIm classification of TCFA when ThCFAM is seen in histology. Raman spectroscopy clearly identifies the presence of calcium as is demonstrated by the strong red signal. There is some lipid content as seen by the small cyan area in the Raman color map and the ThCFA/TCFA classification by FLIm.
Histology section 5
This section is very similar to section 4, but has less crystalline calcium, as is seen in histology and by the smaller red region in the Raman color map. The calcium is present in deeper regions and hence not detected in the field of view of FLIm or Raman in comparison to histology (section 4). FLIm shows ThCFA regions correctly as observed on histology. However, the FLIm map also shows regions classified incorrectly as TCFA, most likely due to the lipid content in the fibrous cap of the ThCFA that is present
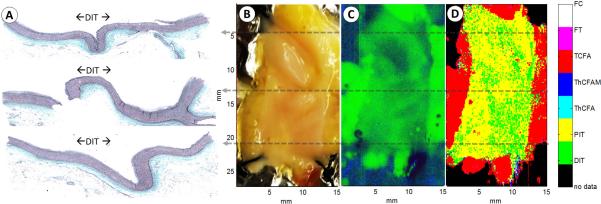
Fig. 4 summarizes the results from the second specimen that is predominantly healthy. The vessel presents DIT over the entire lumen surface and is observed as DIT/PIT in the FLIm map. The Raman false color image shows green for triglycerides in the adventitial fat—demonstrating again that in thin artery specimens (as DIT/PIT arteries often are), the Raman signal can be misleading as it appears to indicate the presence of lipid although the artery is actually healthy. In the future a morphologic imaging modality such as IVUS or OCT would be able to complement the Raman signal to identify a thin lumen, which would indicate that the triglyceride Raman signal is from the outside of the vessel or to show a thickened artery in which case the Raman signal would be indicative of triglycerides from within the artery wall.
Figure 4.
The histology section (A) on the left is in accordance with the visible image (B) as well as the false color Raman map (C) and classified FLIm image (D).
4. Discussion and Conclusion
In this work, we demonstrate for the first time, co-registered FLIm and Raman spectroscopy data from human coronary specimens. Images were acquired using a bimodal probe for simultaneous acquisition of FLIm and Raman signals. Classified image maps from FLIm and Raman false color maps were compared to histological findings with promising results. The premise for combining both imaging modalities is to complement an intravascularly compatible surface imaging technique (FLIm[7, 23]) with high molecular specificity data from Raman. The combination of FLIm and Raman may allow better understanding of plaque compositional changes as a function of disease progression in in vitro or in vivo models of disease.
The excitation wavelengths for FLIm and Raman allow probing different depths in tissue due to the variation of penetration depth of light as a function of wavelength. While the penetration depth for FLIm excitation light (355 nm) is limited to approximately 250 - 300 μm in tissue, it enables rapid compositional assessment of the vessel lumen surface. These compositional assessments particularly include tracking variations in the collagen, elastin, lipid as well as macrophage content. This enables FLIm to probe the composition of the intima and potentially determine the presence of thin fibrous-cap with an underlying necrotic core via changes in lifetime values due to signal arising from the core or the cap, depending on its thickness. Raman imaging employs a longer wavelength excitation light (785 nm) resulting in a penetration depth of at least 700 μm in tissue. These signals allow highly specific detection of molecules of interest for plaque characterization namely triglycerides, cholesterol, cholesterol esters, crystalline calcium and carotenoids. These molecules can be detected from deeper lesions as opposed to the surface. As a result, Raman adds complementary compositional information from deeper tissue to what is observed with FLIm. Additionally, Raman also provides information that is not available from FLIm such as the presence of calcium. The detection of cholesterol and carotenoids by Raman suggests the presence of a necrotic core. This information combined with FLIm features can highlight the presence of fibrous caps with or without macrophages (TCFA or ThCFA vs ThCFAM). As a result, FLIm and Raman can allow delineating features relevant for plaque characterization, namely macrophage infiltration in fibrous caps and presence of necrotic cores. Currently, in-vivo imaging techniques such as OCT and IVUS are unable to distinguish between lipid and necrotic cores and the ability to identify necrosis will be important for identifying vulnerable plaques and the risk of a potential rupture.
In summary the results presented in this work highlight the ability of Raman spectroscopy to complement features observed in FLIm maps as discussed above. Since Raman signals are generated from a larger depth in tissue, it is less sensitive in detecting surface features pertaining to DIT or PIT and includes signal originating from the adventitial fat beyond the medial layer. However, these surface features can be easily identified by FLIm. Because fibrous tissue is present in all plaques identified as FT, ThCFA, ThCFAM and FC, these features may be misclassified on the FLIm map. Additional morphological information employing IVUS or OCT would allow an even better classification as demonstrated in our previous work [17]. It should be noted that there are several false-positive regions for TCFA on the FLIm map. These may be attributed to extracellular lipid or lipid-rich macrophages (foam cells) present in the fibrous tissue. Nonetheless, these results are in good agreement with the sensitivity and specificity of the FLIm classifier reported previously.[17] A new SVM classifier can also be trained using the same data by defining simpler features (such as collagen rich tissue, elastin rich tissue and lipid rich tissue) to enable generating classified maps with lower false positive rate. This is because any given pathological feature is complex and cannot be assessed entirely from FLIm data alone.
In histology section 2 (Fig. 3) the ThCFAs appears larger in the FLIm classification map than in the Raman map, where the lipid in the ThCFAs appears as a scattered lipid signal. In this example, FLIm is showing that there are large ThCFAs present (as is validated by histology) and Raman is able to provide information about the specific type of lipid present.
In summary, Raman can be used to distinguish lipid from necrotic cores since the penetration depth of this modality is rather high while FLIm shows information from the fibrous cap. This will be clinically relevant as we learn more about identifying and treating vulnerable plaques. These techniques would be of great use for characterizing plaque composition to learn more about why some vulnerable plaques rupture compared to other non-vulnerable plaques that are stable and to study the effects of new pharmaceutical agents designed to promote plaque stabilization. While morphological imaging techniques such as IVUS or OCT were not used in this study, further addition of such modalities could enable the distinction between Raman signal coming from the adventitia of thin arteries or lipid pools in thicker arteries.
Acknowledgement
The presented study was supported in part by NIH grant R01 HL067377. Financial support of the Bundesministerium für Bildung und Forschung for the project “Fiber Health Probe” is highly acknowledged (FKZ: 13N1225, 3N12526), as well as the financial sponsorship of the Carl-Zeiss-Stiftung.
References
- 1.Manfrini O, Mont E, Leone O, Arbustini E, Eusebi V, Virmani R, Bugiardini R. The American journal of cardiology. 2006;98:156–159. doi: 10.1016/j.amjcard.2006.01.097. [DOI] [PubMed] [Google Scholar]
- 2.Huisman J, Hartmann M, Birgelen C. v. Int J Cardiovasc Imaging. 2011;27:209–214. doi: 10.1007/s10554-011-9797-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcu L, Jo JA, Fang Q, Papaioannou T, Reil T, Qiao JH, Baker JD, Freischlag JA, Fishbein MC. Atherosclerosis. 2009;204:156–164. doi: 10.1016/j.atherosclerosis.2008.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komachi Y, Sato H, Aizawa K, Tashiro H. Appl. Optics. 2005;44:4722–4732. doi: 10.1364/ao.44.004722. [DOI] [PubMed] [Google Scholar]
- 5.Motz JT, Fitzmaurice M, Miller A, Gandhi SJ, Haka AS, Galindo LH, Dasari RR, Kramer JR, Feld MS. J Biomed Opt. 2006;11:021003. doi: 10.1117/1.2190967. [DOI] [PubMed] [Google Scholar]
- 6.Matthäus C, Dochow S, Bergner G, Lattermann A, Romeike BF, Marple ET, Krafft C, Dietzek B, Brehm BR, Popp J. Anal. Chem. 2012;84:7845–7851. doi: 10.1021/ac301522d. [DOI] [PubMed] [Google Scholar]
- 7.Bec J, Ma DM, Yankelevich DR, Liu J, Ferrier WT, Southard J, Marcu L. J Biophotonics. 2014;7:281–285. doi: 10.1002/jbio.201200220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phipps JE, Sun Y, Fishbein MC, Marcu L. Lasers in surgery and medicine. 2012;44:564–571. doi: 10.1002/lsm.22059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buschman HP, Motz JT, Deinum G, Romer TJ, Fitzmaurice M, Kramer JR, van der Laarse A, Bruschke AV, Feld MS. Cardiovasc Pathol. 2001;10:59–68. doi: 10.1016/s1054-8807(01)00063-1. [DOI] [PubMed] [Google Scholar]
- 10.Lattermann A, Matthäus C, Bergner N, Beleites C, Romeike BF, Krafft C, Brehm BR, Popp J. Journal of BIOPHOTONICS. 2013;6:110–121. doi: 10.1002/jbio.201200146. [DOI] [PubMed] [Google Scholar]
- 11.van de Poll SW, Kastelijn K, Bakker Schut TC, Strijder C, Pasterkamp G, Puppels GJ, van der Laarse A. Heart. 2003;89:1078–1082. doi: 10.1136/heart.89.9.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marzec KM, Wróbel TP, Fedorowicz A, Mateuszuk Ł, Maślak E, Jasztal A, Chlopicki S. Vibrational Microspectroscopy for Analysis of Atherosclerotic Arteries. In: Baranska M, editor. Optical Spectroscopy and Computational Methods in Biology and Medicine. Vol. 14. Springer; Netherlands: 2014. pp. 505–535. [Google Scholar]
- 13.Yang YC, Chang WT, Huang SK, Liau I. Anal Chem. 2014;86:3863–3868. doi: 10.1021/ac404051f. [DOI] [PubMed] [Google Scholar]
- 14.Cicchi R, Baria E, Matthaus C, Lange M, Lattermann A, Brehm BR, Popp J, Pavone FS. J Biophotonics. 2015;8:347–356. doi: 10.1002/jbio.201400142. [DOI] [PubMed] [Google Scholar]
- 15.Dochow S, Ma D, Latka I, Bocklitz T, Hartl B, Bec J, Fatakdawala H, Marple E, Urmey K, Wachsmann-Hogiu S, Schmitt M, Marcu L, Popp J. Analytical and Bioanalytical Chemistry. 2015:1–11. doi: 10.1007/s00216-015-8800-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yankelevich DR, Ma D, Liu J, Sun Y, Sun Y, Bec J, Elson DS, Marcu L. The Review of Scientific Instruments. 2014;85:034303. doi: 10.1063/1.4869037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fatakdawala H, Gorpas D, Bishop J, Bec J, Ma D, Southard J, Margulies K, Marcu L. Journal of Cardiovascular Translational Research. 2015;8:253–263. doi: 10.1007/s12265-015-9627-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Virmani R, Kolodgie FD, Burke AP, Farb A, Schwartz SM. Arteriosclerosis Thrombosis and Vascular Biology. 2000;20:1262–1275. doi: 10.1161/01.atv.20.5.1262. [DOI] [PubMed] [Google Scholar]
- 19.Jing L, Yang S, Jinyi Q, Laura M. Physics in Medicine and Biology. 2012;57:843. doi: 10.1088/0031-9155/57/4/843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bocklitz T, Walter A, Hartmann K, Rosch P, Popp J. Anal Chim Acta. 2011;704:47–56. doi: 10.1016/j.aca.2011.06.043. [DOI] [PubMed] [Google Scholar]
- 21.Team RDC. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: [Google Scholar]
- 22.Ryan CG, Clayton E, Griffin WL, Sie SH, Cousens DR. SNIP, a statistics-sensitive background treatment for the quantitative analysis of pixe spectra in geosience aplications. Nuclear Instruments and Methods in Physics Research B34. 1988;34:396–402. [Google Scholar]
- 23.Bec J, Ma DM, Yankelevich DR, Liu J, Ferrier WT, Southard J, Marcu L. Journal of Biophotonics. 2014;7:281–285. doi: 10.1002/jbio.201200220. [DOI] [PMC free article] [PubMed] [Google Scholar]