Abstract
Epithelial-mesenchymal transition (EMT) and the mesenchymal-epithelial transition (MET) are processes required for embryo organogenesis. Liver develops from the epithelial foregut endoderm from which the liver progenitors, hepatoblasts, are specified. The migrating hepatoblasts acquire a mesenchymal phenotype to form the liver bud. In mid-gestation, hepatoblasts mature into epithelial structures: the hepatocyte cords and biliary ducts. While EMT has been associated with liver bud formation, nothing is known about its contribution to hepatic specification. We previously established an efficient protocol from human embryonic stem cells (hESC) to generate hepatic cells (Hep cells) resembling the hepatoblasts expressing alpha-fetoprotein (AFP) and albumin (ALB). Here we show that Hep cells express both epithelial (EpCAM and E-cadherin) and mesenchymal (vimentin and SNAI-1) markers. Similar epithelial and mesenchymal hepatoblasts were identified in human and mouse fetal livers, suggesting a conserved interspecies phenotype. Knock-down experiments demonstrated the importance of SNAI-1 in Hep cell hepatic specification. Moreover, ChIP assays revealed direct binding of SNAI-1 in the promoters of AFP and ALB genes consistent with its transcriptional activator function in hepatic specification. Altogether, our hESC-derived Hep cell cultures reveal the dual mesenchymal and epithelial phenotype of hepatoblast-like cells and support the unexpected transcriptional activator role of SNAI-1 in hepatic specification.
Keywords: Human embryonic stem cell, Liver development, Hepatic differentiation, Liver specification, SNAI-1
1. Introduction
The epithelial-mesenchymal transition (EMT) defines a series of orchestrated events during which epithelial cells lose the majority of epithelial characteristics and acquire properties typical of mesenchymal cells (Thiery and Sleeman, 2006). This process requires complex changes in the architecture and transcriptional program of the cell, which are tightly regulated by the transcription factor SNAI (Cano et al., 2000). The analogous but reverse mechanism gives mesenchymal cell characteristics of an epithelial cell, which is defined as a mesenchymal-epithelial transition (MET). EMT and MET are two major processes associated with embryogenesis from the blastula formation (Rossant and Tam, 2009; Stephenson et al., 2010; Takaoka and Hamada, 2012), gastrulation during which the embryonic epithelium gives rise to the three germ layers and subsequent organogenesis (Dressler, 2009; Nieto, 2001; Schluter and Margolis, 2009). More specifically, organogenesis of endoderm derived-tissues including the pancreas and liver requires EMT and MET (Lemaigre, 2009). For example, liver bud formation results from delamination of pseudostratified hepatic foregut endoderm cells through an EMT. Hepatic endoderm cells are known as hepatoblasts, and are the fetal progenitors for hepatocytes and cholangiocytes of the adult liver. As the fetal liver grows, mesenchymal hepatoblasts proliferate and differentiate into epithelial hepatocytes or biliary duct cells (Gordillo et al., 2015). Observations of developing fetal livers suggested that MET accompanies the differentiation of human and mouse liver progenitors to hepatocytes and cholangiocytes (Bort et al., 2006; Li et al., 2011; Vestentoft et al., 2011).
Here we asked whether the EMT process is recapitulated during early liver specification from human embryonic stem cells (hESCs), and whether EMT directly controls liver specification. We previously identified two types of cells generated during differentiation of hESCs toward the hepatic lineage (Goldman et al., 2013): the KDR (VEGFR2/FLK-1)-expressing hepatic progenitors and the hepatoblast-like hepatic cells (Hep cells) that are negative for KDR. We characterized the developing Hep cells as having a mixed phenotype since they express both epithelial (E-cadherin, EpCAM) and mesenchymal (Vimentin, SNAI-1) markers. Most interestingly, in contrary to its conventional role in promoting mesenchymal characteristics during EMT, our data support the notion that SNAI-1 acts autonomously as a transcriptional activator by instructing hepatic cell fate of the developing Hep cells.
2. Material and methods
2.1. Human hepatic differentiation and cell sorting
Human ESCs (HES2) were differentiated into hepatic cells following embryoid body formation in Serum Free Differentiation (SFD) medium with 1 mM Ascorbic Acid, 4 × 10−4 monothioglycerol, 2 mM glutamine and BMP4 (10 ng/ml) (Goldman et al., 2013). At day 1 of differentiation, medium was changed to SFD complemented with 1 mM Ascorbic Acid, 4 × 10−4 monothioglycerol, 2 mM glutamine, BMP4 (0.5 ng/ml), bFGF (2.5 ng/ml) and a high dose of Activin-A (100 ng/ml). At day 4 of differentiation the medium was changed to SFD complemented with 1 mM Ascorbic Acid, 4 × 10−4 monothioglycerol, 2 mM glutamine, VEGF (10 ng/ml), bFGF (2.5 ng/ml) and Activin-A (100 ng/ml) (Goldman et al., 2013). At day 5 of differentiation, CXCR4+ cKIT+ KDR-PDGFRα-endoderm cells were purified by FACS using an Aria cell sorter (BD Bioscience, CA) and cultured in hepatic media as previously defined (Han et al., 2011). At days 9, 12 and 17 of differentiation KDR-CD31-Hep cells were purified by FACS and either cultured in hepatic media as previously defined or analyzed by flow cytometry, real time qPCR or immunostaining.
2.2. Flow cytometry analyses
Cells were trypsinized and resuspended in PBS1X/BSA 0.3%. For cell surface protein cells were stained with specific antibodies listed in Supplemental Table 1 for 20 min at room temperature and analyzed using a LSRII flow cytometer (Becton Dickinson).
2.3. Quantitative real time PCR analyses
Total RNA was extracted using the RNeasy micro kit (Qiagen). Reverse transcription was performed with the Super Script-III First-strand Synthesis System kit (Invitrogen). qPCR was performed using the Roche SYBR Green master mix and analyzed with the Roche system LC480. Primers sequences used are listed in supplemental Table 2.
2.4. Immunostaining
Cells were fixed with 4% paraformaldehyde for 10 min at room temperature, permeabilized in 0.3% triton for 10 min and then blocked with the protein block serum-free kit from DAKO. Incubation with primary (overnight, 4C) and secondary antibodies (1 h, room temperature) (listed in Table 1) was performed in PBS1X/BSA 0.3%. DAPI was used to stain the cell nuclei. Images of immunostained cells were acquired using a Fluorescent inverted microscope (Leica).
2.5. Immunohistochemistry
Human fetal livers, 7–22 weeks old, or E9.5 mouse embryos or E13.5 mouse fetal livers were fixed overnight with 4% paraformaldehyde, dehydrated with 30% sucrose and embedded in OCT. 7-μm sections were permeabilized with 0.3% Triton X-100 for 10 min at room temperature and then blocked for 20 min with 2% donkey serum at room temperature. Sections were immunostained at 4 °C overnight with primary antibodies (listed in supplemental Table 1) and then incubated for 1 h at room temperature with secondary antibodies (listed in supplemental Table 1) and counterstained with DAPI. A fluorescent confocal microscope (Leica) was used to visualize sections.
2.6. siRNA assays
Pools of four siRNA against SNAI-1 (L-017386-00-0005), SNAI-2 (L017386-00-0005) or control non-target (D-001810-10-05) were purchased from Thermo Scientific. siRNA transfection was performed at day 10 and 12 of differentiation on purified Hep cells. Briefly, according to the manufacturer's protocol, 5 μM of each siRNA pool were incubated with OptiMEM for 5 min at room temperature. In parallel, 2 μl of Dharmafect transfection reagent 1 (Thermo T2005-01) was incubated with OptiMEM for 5 min at room temperature. The two reactions were then mixed together for 20 min at room temperature and then mixed with the hepatic culture media in the absence of antibiotics onto the cells. 24 h prior to transfection, cells were cultured in hepatic medium without antibiotics. The following day of each transfection, the medium was changed with regular hepatic medium without antibiotics.
2.7. Chromatin immunoprecipitation (ChIP)
ChIP was performed as previously described (Lien et al., 2011). Briefly, at day 9 of differentiation, hESC-derived Hep cells were fixed 10 min with 1% formaldehyde at room temperature. Nuclei where isolated and subjected to 30 cycles of sonication in the presence of Triton X-100 using Diagenode Bioruptor (30 s on, 30 s off at 4C). Sonicated chromatin from approximately nine 35 mm plates where incubated rotating overnight with 4 μl of two different antibodies against SNAI (1 and 2) (SNAI1/2 abcam Ab78105 or Snail1 Cell Signaling 3879S) or non-specific IgGs (IgG rabbit polyclonal ChIP Grade, ab171870) in DNA LoBind Eppendorf tubes at 4C. 10% of the sample was saved as input. Immunoprecipitation was performed by adding Protein G (Dynabeads 10003D, Life Technologies) for an additional 6 h at 4C. Beads where washed with Low Salt, High Salt, LiCl and TE buffers for 10 min each, after which the chromatin was eluted in 100 μl of 1% SDS buffer. Inputs and eluted chromatin were incubated overnight at 65C to revert the crosslinking. Samples were then treated with RNaseA and Proteinase K, and DNA was purified using Zymogen ChIP DNA Clean and Concentrator columns. QPCR was performed with Sybergreen in a Roche Lightcycler 480. Primers sequences used are listed in supplemental Table 3. The percentage of input recovery was calculated from the input signal of each primer in each experiment.
2.8. Statistical analysis
Results are expressed as mean ± SD. For each group, at least 3 independent experiments were analyzed and different groups were compared using Student's t-test analysis. p < 0.05 was considered statistically significant *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
3. Results
3.1. hESC-derived hepatic cells (Hep cells) are epithelial cells expressing the mesenchymal markers SNAI and vimentin
As described in our previous work, Hep cells were generated from hESCs by first inducing endoderm formation with a high dose of Activin-A (Goldman et al., 2013). At day 5 of differentiation, endoderm cells were purified by fluorescence-activated cell sorting (FACS) (with purity >95%) based on the expression of CXCR4 and cKIT and exclusion of the mesendodermal marker PDGFRα (platelet-derived growth factor) and the receptor KDR (VEGFR2 or FLK-1) (Goldman et al., 2013). The purified endoderm cell population was subsequently differentiated into Hep cells together with hepatic progenitors expressing KDR (Goldman et al., 2013). Both populations were negative for the endothelial marker CD31 (Goldman et al., 2013). As a first approach to investigate whether EMT occurs during hepatic differentiation, Hep cells, defined as cells negative for both KDR and CD31, were analyzed over time for expression of mesenchymal and epithelial markers (Fig. 1A). The hepatic phenotype of the purified KDR-CD31-Hep cells during hepatic differentiation was confirmed by alpha-fetoprotein (AFP) expression as early as day 9 of differentiation, which was maintained until day 17 (Fig. 1B). Detection of albumin (ALB) protein in most purified KDR-CD31-Hep cells by day 17 of differentiation was indicative of further hepatic maturation (Fig. 1B). The hepatic phenotype and functional characterization of Hep cells was reported in our previous work (Goldman et al., 2013). In line with a hepatic phenotype, all Hep cells expressed the epithelial marker EpCAM (epithelial cell adhesion molecule) (Trzpis et al., 2007) at days 9, 12 and 17 of differentiation (Fig. 1C). Interestingly, a subset of Hep cells also expressed the mesenchymal marker CD90 (Thy-1) (Delorme et al., 2006) with the percentage of positive cells varying from 3.2% at day 9 to 15% at later stages of differentiation (Fig. 1C). Protein expression of two additional mesenchymal markers SNAI (1 and 2) (Kalluri and Weinberg, 2009) and vimentin was detected in all Hep cells (99 and 95% respectively of total Hep cells) following purification at day 9 and further culture for one day (Fig. 1D). EpCAM protein in virtually all Hep cells (98% of total Hep cells) was also confirmed in this assay (Fig. 1D), indicating that Hep cells co-express both epithelial and mesenchymal markers at day 9 of differentiation as they initiate hepatic specification.
Fig. 1.
Developing hESC-derived Hep cells express both epithelial and mesenchymal markers. (A) Timeline of hepatic differentiation of hESC and analyses. (B) Immunostaining for hepatic markers AFP and ALB on Hep cells purified and cytospun at days 9, 12 and 17 of differentiation (×200). (C) Flow cytometry analysis of Hep cells (KDR-CD31−) at days 9, 12 and 17 of differentiation (one representative experiment out of 2, n = 2 independent experiments). (D) Immunostaining in the dish for the mesenchymal markers vimentin and SNAI (1 and 2) and the epithelial marker EpCAM in Hep cells purified at day 9 of differentiation and cultured for one more day (×200). Graphs indicate the means ± SD of the percentage of positive cells for each marker (vimentin, EpCAM and SNAI-1/2) among the total number of Hep cells. Three different fields for each staining were examined for n = 3 independent differentiations. (E) Relative transcript levels in Hep cells purified at days 9, 12 and 17 of differentiation. Gene expression from day 5 CXCR4+ cKIT+ PDGFRα-KDR-cells (End d5, black columns) was set to 1 and Huvecs (white columns) were used as negative control. Purple columns represent Hep cells at different time points. Data are represented as mean ± SD (n = 3 independent experiments). ND: not detectable (cycle number above 40).
Concomitant detection of both mesenchymal and epithelial markers in Hep cells was validated by quantitative real time PCR (qPCR) (Fig. 1E). The epithelial EpCAM and E-cadherin (CDH1) transcript levels in purified Hep cells were as high as those in endoderm cells regardless of the time of differentiation, and significantly much higher than measured in Huvec cells that served as negative control for both epithelial and mesenchymal phenotype. Transcript levels for the mesenchymal marker vimentin remained high following hepatic specification of Hep cells, while undetectable in Huvec cells. Interestingly, the mesenchymal markers SNAI-1 and SNAI-2 were expressed in Hep cells in an opposite pattern over time, with decreasing levels of SNAI-1 and increasing levels of SNAI-2 transcripts as Hep cells specify and mature (Fig. 1E), suggesting their distinct roles in liver differentiation. Specifically, detection of high levels of SNAI-1 in endoderm cells and day 9 Hep cells suggested a role for SNAI-1 in an early hepatic fate decision of endoderm. Altogether, protein and transcript level analyses of mesenchymal and epithelial markers defined developing hESC-derived Hep cells as cells with dual mesenchymal and epithelial characteristics.
3.2. Identification of mesenchymal and epithelial hepatoblasts in human and mouse fetal livers
To examine whether the dual mesenchymal and epithelial features of Hep cells are relevant to human fetal liver development and conserved in the mouse fetal liver in vivo, we performed immunostaining for HNF4α or ALB to identify hepatoblasts together with SNAI (1 and 2) in human and murine developing livers (Fig. 2). A small number of hepatoblasts co-expressing either HNF4α and SNAI (2% cells among HNF4α+ cells) or ALB and SNAI were identified in 7 weeks old human fetal livers (Fig. 2A, arrows). Few HNF4α+ SNAI+ hepatoblasts (2% cells among HNF4α+ cells) were also detected later in 22 weeks old human fetal liver samples (Fig. 2A, arrows) as well as in the liver bud of E9.5 mouse embryos and mouse fetal livers at E13.5 (Fig. 2B, arrows). No specific anatomical location of the HNF4α+SNAI+ hepatoblasts within the human or murine fetal liver was identified. Altogether, a small subset of hepatoblasts positive for SNAI was detected in human and mouse fetal livers, suggesting that similar mesenchymal hepatoblasts, as observed in hESC-derived Hep cells, are relevant to human and mouse fetal liver development.
Fig. 2.
Identification of mesenchymal and epithelial hepatoblasts in human and mouse fetal livers. (A) Co-immunostaining for HNF4α and SNAI (1 and 2) or ALB and SNAI (1 and 2) of 7 weeks old human fetal livers (upper panels) or 22 weeks old human fetal livers (lower panels) (×200). The graph indicates the means ± SD of the percentage of SNAI-1/2+ cells among the total number of HNF4α+ hepatoblast cells. (B) Co-immunostaining for HNF4α and SNAI (1 and 2) of mouse E9.5 liver bud and E13.5 fetal liver (×200).
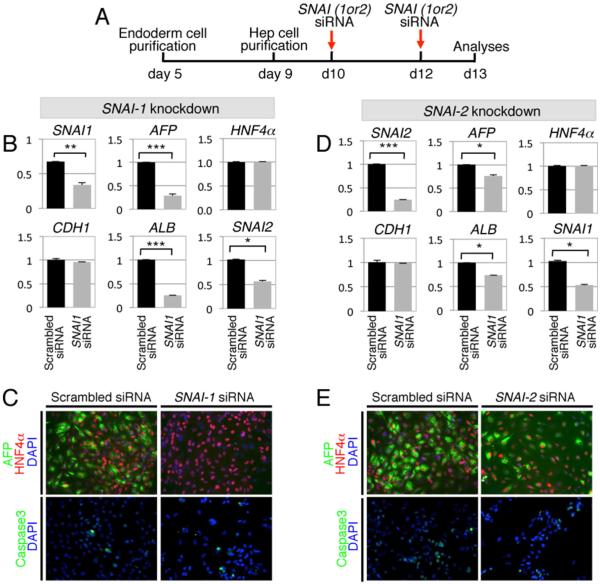
3.3. SNAI-1 controls hepatic specification in a cell autonomous fashion in developing hESC-derived Hep cells
Given the transient expression of SNAI-1 in endoderm cells and in day 9 Hep cells as they acquire AFP expression (Fig. 1), we investigated the function of SNAI-1 in instructing early hepatic specification of Hep cells. SNAI-1 expression was knocked-down using a pool of four siRNAs against SNAI-1 in Hep cells purified at day 9 and further cultured until day 13 (Fig. 3A). Hep cells were treated with SNAI-1 siRNAs at day 10 and day 12, and their hepatic fate was analyzed at day 13. SNAI-1 siRNAs reduced SNAI-1 transcript levels by 50% (Fig. 3B) and resulted in 75% reduction of levels of both hepatic markers AFP and ALB when compared to scrambled siRNA-treated cells (Fig. 3B, black columns for scrambled siRNA versus grey columns for SNAI-1 siRNAs). However, transcript levels of the epithelial marker E-cadherin (CDH1) and the transcription factor regulating endoderm hepatic specification, the hepatocyte nuclear factor 4 α (HNF4α), were not altered by SNAI-1 knock-down, most likely because their expression was induced prior to the knock-down assay (Fig. 1E). These results suggested that SNAI-1 is not required for the maintenance of CDH1 and HNF4α expression. The role of SNAI-1 on hepatic specification was confirmed at the AFP protein level, as virtually all Hep cells lost AFP expression while HNF4α expression was maintained following SNAI-1 knock-down (Fig. 3C). Altogether, SNAI-1 knock-down assays indicate that SNAI-1 induces hepatic marker expression in early committed Hep cells without modifying the epithelial phenotype of Hep cells that was also observed in culture (not shown). Curiously, SNAI-1 siRNAs also decreased transcript levels of SNAI-2 by 50% (Fig. 3B). To thus determine whether SNAI-2 knock-down alters AFP and ALB expression, Hep cells were treated with a pool of SNAI-2 siRNAs. Similarly, siRNAs against SNAI-2 reduced both SNAI-2 (by 75%) and SNAI-1 (by 50%) transcript levels (Fig. 3D), and levels of CDH1 and HNF4α were not altered. Although the presence of SNAI-2 siRNAs led to a 50% decrease of SNAI-1 levels, there was no synergistic decrease of AFP and ALB levels but rather a milder diminution of these hepatic genes (by 25% versus 75% in the presence of SNAI-1 siRNAs), supporting the notion that SNAI-1 and SNAI-2 have opposite effects on AFP and ALB expressions. Indeed, SNAI-1 knock-down in the presence of SNAI-1 siRNAs completely abrogated AFP protein levels (Fig. 3C), while SNAI-2 knock-down in the presence of SNAI-2 siRNAs partially diminished them (Fig. 3E). Altogether, the SNAI-1 and SNAI-2 knock-down assays indicated that, whether each other regulation is mediated by siRNA cross-specificity or by indirect gene regulation, SNAI-1 is the main positive regulator of hepatic specification.
Fig. 3.
SNAI-1 regulates positively hepatic specification of Hep cells. (A) Timeline of siRNA transfection and analyses of purified Hep cells. (B) Relative transcript levels at day 13 of differentiation following SNAI-1 knock-down assays (n = 3 independent experiments). (C) Co-immunostaining for HNF4α and AFP or Caspase 3 following SNAI-1 knock-down assays. (D) Relative transcript levels at day 13 of differentiation following SNAI-2 knock-down assays (n = 3 independent experiments). (E) Co-immunostaining for HNF4α and AFP or Caspase 3 DAPI following SNAI-2 knock-down assays.
As EMT has been reported to confer resistance to the apoptotic effects of transforming growth factor beta in fetal rat hepatocytes (Valdes et al., 2002), we examined Hep cell apoptosis upon SNAI knock-down assays (Fig. 3C and E) by caspase 3 immunostaining. Neither of the siRNAs against SNAI-1 or SNAI-2 led to an increase of Hep cell apoptosis, indicating that SNAI knock-downs did not alter cell apoptosis that could have indirectly affected hepatic fate decision.
Immuno-precipitation (ChIP) assays of day 9 Hep cells using 2 separate antibodies recognizing both SNAI-1 and 2 demonstrated further that SNAI (1 and/or 2) binds directly to two separate upstream promoter regions of AFP and ALB genes as well as of CDH1 gene previously reported to be transcriptionally repressed by SNAI-1. These data suggest a direct transcriptional activator function of SNAI in hepatic specification of developing Hep cells in a cell autonomous manner (Fig. 4A, B).
Fig. 4.
SNAI-1/2 binds to promoter areas of hepatic genes. (A,B) ChIP-qPCR assays using two different antibodies: (A) Cell Signaling and (B) Abcam, shows the binding of the transcription factor SNAI-1/2 to upstream sequences of ALB and AFP genes in purified day 9 Hep cells. Note that the percentage of recovery in (A) is higher than in (B). As negative control the open reading frame of the β-Actin (ACTB) gene and the promoter of the non-hepatic gene Atoh1 are shown. The binding of SNAI-1/2 to human E-Cadherin (CDH1) is shown as a positive control. Diagrams in the bottom panel show the amplified regions for each gene (red). Each chart is representative of 3 independent differentiations.
4. Discussions and conclusion
The zinc-finger transcription factors SNAI (1 and 2) are conventionally reported to induce EMT when expressed in epithelial cells during organogenesis (Cano et al., 2000) by maintaining the mesenchymal phenotype and directly repressing epithelial gene expression (Thiery et al., 2009). However, recent studies provided evidence that SNAI factors paradoxically enhance reprogramming of mesenchymal fibroblasts to induced pluripotent stem cells (iPSCs) characterized with an epithelial phenotype (Gingold et al., 2014; Unternaehrer et al., 2014). In this system, SNAI-1 together with the pluripotent factor NANOG cooperatively function as transcriptional activators of pluripotency-associated genes (Gingold et al., 2014). The transitioning pre-iPSCs express both the mesenchymal marker SNAI-1 and the epithelial marker E-cadherin, and are thus reminiscent of the developing hESC-derived Hep cells with dual mesenchymal and epithelial characteristics. The impact of SNAI-1 in reprogramming mouse fibroblasts to epithelial iPSCs introduced the notion that SNAI-1 can paradoxically instructs epithelial cell fate. Here, we discovered a similar function of SNAI-1 as a transcriptional activator instructing hepatic specification of hESC-derived Hep cells.
Interestingly, in contrast to the common roles of SNAI-1 and 2 in repressing E-cadherin mediated EMT, SNAI-1 is the only SNAI factor that is expressed in endoderm and Hep cells, and that instructs positively hepatic cell fate. The unique function of SNAI-1 compared to SNAI-2 was also noted during the cell reprogramming process, in which solely SNAI-1 is a direct target of NANOG during the last step of fibroblast reprogramming (Gingold et al., 2014). The differential function of SNAI factors is clearly shown by the analysis of knock-out mice. Deletion of SNAI-1 causes embryonic lethality due to gastrulation defects (Carver et al., 2001), while mice with SNAI-2 deletion are viable and show only postnatal defects (Parent et al., 2010; Perez-Losada et al., 2002).
The hESC-derived Hep cell differentiation cultures described in this manuscript constitute a dynamic in vitro platform recapitulating the transient development of hepatoblasts with dual mesenchymal and epithelial phenotype as previously observed in E11.5 mouse fetal livers with the vast majority of hepatoblasts expressing vimentin and AFP (Li et al., 2011). Specific expression of SNAI in a much smaller subset of hHNF4α+ hepatoblasts in human and murine fetal livers may represent for instance a small progenitor pool that persists in adult either to play a role in homeostatic hepatocyte renewal as defined as axin+ cells within the adult central vein areas (Wang et al., 2015) or to be activated to become mesenchymal-epithelial oval cells within the portal vein areas when the liver is injured (Yovchev et al., 2008).
In conclusion, this study reveals a unique function of the EMT factor SNAI-1 in promoting hepatic cell fate in differentiating human Hep cells derived from hESC cultures. As Hep cells differentiate from endoderm cells, they express both epithelial (E-cadherin, EpCAM) and mesenchymal (Vimentin, SNAI-1) markers. In contrast to its conventional role in promoting mesenchymal phenotype during EMT, our work introduces a novel role for SNAI-1 as a transcriptional activator to instruct cell autonomously hepatic cell fate in differentiating Hep cells.
Supplementary Material
Acknowledgements
We are grateful to Drs. Georges Uzan, Adam Jacobs and Tamara Talir for providing us with human fetal livers. We thank Drs. Carolina Perdigoto, Karen Handschuh and Todd Evans for editing the manuscript, and Dr. Henia Darr for her technical advice for siRNA assays. This work was supported by the Black Family Stem Cell Institute, the National Institute of Diabetes and Digestive and Kidney Diseases for the mouse study (R01DK087867-01 to VGE), and March of Dimes (FY113-969 to VGE), and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01AR063724 to EE), V.J.V. is a Pew Latin American Fellow in the Biomedical Sciences, supported by The Pew Charitable Trusts (00026199).
Abbreviations
- hESC
human embryonic stem cell
- Hep cells
hESC-derived hepatic cells
- FACS
fluorescence-activated cell sorting
- PDGFRα
platelet-derived growth factor receptor alpha
- KDR (VEGFR2 or FLK-1)
kinase insert domain receptor
- SNAI-1
Snail-1
- SNAI-2
Snail-2
- EMT
epithelial-mesenchymal transition
- MET
mesenchymal-epithelial transition
- AFP
alpha fetoprotein
- ALB
albumin
- EpCAM
epithelial cell adhesion molecule
- CDH1
E-cadherin
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.scr.2016.05.007.
References
- Bort R, Signore M, Tremblay K, Martinez Barbera JP, Zaret KS. Hex homeobox gene controls the transition of the endoderm to a pseudostratified, cell emergent epithelium for liver bud development. Dev. Biol. 2006;290:44–56. doi: 10.1016/j.ydbio.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol. Cell. Biol. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme B, Chateauvieux S, Charbord P. The concept of mesenchymal stem cells. Regen. Med. 2006;1:497–509. doi: 10.2217/17460751.1.4.497. [DOI] [PubMed] [Google Scholar]
- Dressler GR. Advances in early kidney specification, development and patterning. Development. 2009;136:3863–3874. doi: 10.1242/dev.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold JA, Fidalgo M, Guallar D, Lau Z, Sun Z, Zhou H, Faiola F, Huang X, Lee DF, Waghray A, et al. A genome-wide RNAi screen identifies opposing functions of Snai1 and Snai2 on the Nanog dependency in reprogramming. Mol. Cell. 2014;56:140–152. doi: 10.1016/j.molcel.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman O, Han S, Sourrisseau M, Dziedzic N, Hamou W, Corneo B, D'Souza S, Sato T, Kotton DN, Bissig KD, et al. KDR identifies a conserved human and murine hepatic progenitor and instructs early liver development. Cell Stem Cell. 2013;12:748–760. doi: 10.1016/j.stem.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordillo M, Evans T, Gouon-Evans V. Orchestrating liver development. Development. 2015;142:2094–2108. doi: 10.1242/dev.114215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Dziedzic N, Gadue P, Keller GM, Gouon-Evans V. An endothelial cell niche induces hepatic specification through dual repression of wnt and notch signaling. Stem Cells. 2011;29:217–228. doi: 10.1002/stem.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaigre FP. Mechanisms of liver development: concepts for understanding liver disorders and design of novel therapies. Gastroenterology. 2009;137:62–79. doi: 10.1053/j.gastro.2009.03.035. [DOI] [PubMed] [Google Scholar]
- Li B, Zheng YW, Sano Y, Taniguchi H. Evidence for mesenchymal-epithelial transition associated with mouse hepatic stem cell differentiation. PLoS One. 2011;6:e17092. doi: 10.1371/journal.pone.0017092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien WH, Guo X, Polak L, Lawton LN, Young RA, Zheng D, Fuchs E. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell. 2011;9:219–232. doi: 10.1016/j.stem.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA. The early steps of neural crest development. Mech. Dev. 2001;105:27–35. doi: 10.1016/s0925-4773(01)00394-x. [DOI] [PubMed] [Google Scholar]
- Parent AE, Newkirk KM, Kusewitt DF. Slug (Snai2) expression during skin and hair follicle development. J. Investig. Dermatol. 2010;130:1737–1739. doi: 10.1038/jid.2010.22. [DOI] [PubMed] [Google Scholar]
- Perez-Losada J, Sanchez-Martin M, Rodriguez-Garcia A, Sanchez ML, Orfao A, Flores T, Sanchez-Garcia I. Zinc-finger transcription factor Slug contributes to the function of the stem cell factor c-kit signaling pathway. Blood. 2002;100:1274–1286. [PubMed] [Google Scholar]
- Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- Schluter MA, Margolis B. Apical lumen formation in renal epithelia. J. Am. Soc. Nephrol. 2009;20:1444–1452. doi: 10.1681/ASN.2008090949. [DOI] [PubMed] [Google Scholar]
- Stephenson RO, Yamanaka Y, Rossant J. Disorganized epithelial polarity and excess trophectoderm cell fate in preimplantation embryos lacking E-cadherin. Development. 2010;137:3383–3391. doi: 10.1242/dev.050195. [DOI] [PubMed] [Google Scholar]
- Takaoka K, Hamada H. Cell fate decisions and axis determination in the early mouse embryo. Development. 2012;139:3–14. doi: 10.1242/dev.060095. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Trzpis M, McLaughlin PM, de Leij LM, Harmsen MC. Epithelial cell adhesion molecule: more than a carcinoma marker and adhesion molecule. Am. J. Pathol. 2007;171:386–395. doi: 10.2353/ajpath.2007.070152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unternaehrer JJ, Zhao R, Kim K, Cesana M, Powers JT, Ratanasirintrawoot S, Onder T, Shibue T, Weinberg RA, Daley GQ. The epithelial-mesenchymal transition factor SNAIL paradoxically enhances reprogramming. Stem Cell Rep. 2014;3:691–698. doi: 10.1016/j.stemcr.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes F, Alvarez AM, Locascio A, Vega S, Herrera B, Fernandez M, Benito M, Nieto MA, Fabregat I. The epithelial mesenchymal transition confers resistance to the apoptotic effects of transforming growth factor Beta in fetal rat hepatocytes. Mol. Cancer Res. 2002;1:68–78. [PubMed] [Google Scholar]
- Vestentoft PS, Jelnes P, Hopkinson BM, Vainer B, Mollgard K, Quistorff B, Bisgaard HC. Three-dimensional reconstructions of intrahepatic bile duct tubulogenesis in human liver. BMC Dev. Biol. 2011;11:56. doi: 10.1186/1471-213X-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhao L, Fish M, Logan CY, Nusse R. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature. 2015;524:180–185. doi: 10.1038/nature14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yovchev MI, Grozdanov PN, Zhou H, Racherla H, Guha C, Dabeva MD. Identification of adult hepatic progenitor cells capable of repopulating injured rat liver. Hepatology. 2008;47:636–647. doi: 10.1002/hep.22047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.