Abstract
Epigenetic changes, such as alteration of DNA methylation patterns, have been proposed as a molecular mechanism underlying the effect of alcohol on the maintenance of adult stem cells. We have performed genome-wide gene expression microarray and DNA methylome analysis to identify molecular alterations via DNA methylation changes associated with exposure of human dental pulp stem cells (DPSCs) to ethanol (EtOH). By combined analysis of the gene expression and DNA methylation, we have found a significant number of genes that are potentially regulated by EtOH-induced DNA methylation. As a focused approach, we have also performed a pathway-focused RT-PCR array analysis to examine potential molecular effects of EtOH on genes involved in epigenetic chromatin modification enzymes, fibroblastic markers, and stress and toxicity pathways in DPSCs. We have identified and verified that lysine specific demethylase 6B (KDM6B) was significantly dysregulated in DPSCs upon EtOH exposure. EtOH treatment during odontogenic/osteogenic differentiation of DPSCs suppressed the induction of KDM6B with alterations in the expression of differentiation markers. Knockdown of KDM6B resulted in a marked decrease in mineralization from implanted DPSCs in vivo. Furthermore, an ectopic expression of KDM6B in EtOH-treated DPSCs restored the expression of differentiation-related genes. Our study has demonstrated that EtOH-induced inhibition of KDM6B plays a role in the dysregulation of odontogenic/osteogenic differentiation in the DPSC model. This suggests a potential molecular mechanism for cellular insults of heavy alcohol consumption that can lead to decreased mineral deposition potentially associated with abnormalities in dental development and also osteopenia/osteoporosis, hallmark features of fetal alcohol spectrum disorders.
Keywords: alcohol, dental pulp stem cells (DPSCs), epigenetics, DNA methylation, osteogenic differentiation, odontogenic differentiation, mineralization
INTRODUCTION
Heavy alcohol consumption could result in a range of health, social and behavioral problems. Studies have demonstrated the potential toxic effects of alcohol in many organs and have shown deleterious molecular effects on cellular physiology, including the potency and maintenance of stem cells. Based on our previous study demonstrating the epigenetic effect of alcohol on embryonic stem cell pluripotency [1], we tested alcohol’s epigenetic effect on the potency and differentiation capability of adult stem cells.
Alcohol consumption has been shown to have detrimental effects on the brain, liver, muscles, skeleton and fetal development [2]. While some studies have indicated that low doses of alcohol consumption may increase bone mass and density [3, 4], chronic heavy alcohol use can dramatically affect bone health and increase the risk of osteoporosis later in life. Different mechanisms have been hypothesized to cause changes related to alcohol abuse, including a direct effect on osteoblasts and osteoclasts, changes in signaling due to oxidative stress, increased fat accumulation in the bone marrow, modulation of regulatory hormones, and/or indirectly through decreased caloric intake [5]. The effects of alcohol consumption on bone health are multifactorial and are related to the duration and dosage of consumption [6]. It has been demonstrated that Fetal Alcohol Syndrome (FAS) patients show high incidences of dentofacial and temporomandibular joint disorders, including midfacial underdevelopment with shortage of bone, delayed dental development, enamel anomalies, and cleft palate or cleft lip [7]. In an animal model, maternal alcohol ingestion before and during gestation caused retardation in cell differentiation within the tooth germ and in calcification of the dentin matrix [8]. The dental anomalies observed may stem from cellular alterations in the basal layer of the epithelium of the tooth germ during odontogenesis [8–11].
Dental pulp stem cells (DPSCs), also known as dental pulp-derived mesenchymal stem cells, are a multipotent adult stem cell population that has a high proliferative potential. DPSCs in this study isolated from adult teeth are easily accessible and cryopreservable for long periods [12, 13]. DPSCs were used as a model of a mineralizing system, in vitro and in vivo, to study the effects of EtOH. It has been shown in numerous studies that under various conditions, dental pulp stem cells can be induced towards odontogenic and osteogenic lineages. One study demonstrated that vascular endothelial growth factor (VEGF) gene could promote odontogenic differentiation in DPSCs in vitro [14] while another identified that DPSCs undergo osteogenic differentiation through the NF-kB signaling pathway [15]. DPSCs had the ability to differentiate toward both odontogenic and osteogenic lineages in presence of a carboxymethyl cellulose-hydroxyapatite hybrid hydrogel [16]. Furthermore, medium modification with bone morphogenetic protein 2 was shown to stimulate odontogenic differentiation and formation of an osteo-dentin matrix [17]. Although DPSCs have been long studied for their regenerative capabilities in both dentistry and orthopedics, the molecular mechanisms controlling their stem cell potency have yet to be discovered.
It has been shown that KDM6B, a lysine-specific demethylase, plays a key role in osteogenic differentiation by removing H3K27me3 from the promoters of osteogenic genes in human bone marrow stromal cells (BMSCs) [18]. It has also identified KDM6B in controlling HOX expression through the removal of H3K27me3 in human BMSCs [18]. A recent study has shown KDM6B to play a critical role in the epigenetic regulation of odontogenic differentiation in human DPSCs [19]. In DPSCs, KDM6B knockdown studies resulted in decreased alkaline phosphatase activity and alizarin red staining, and reduced expression levels of marker genes, including osterix (OSX), osteocalcin (OCN), and osteopontin (OPN) [19]. Decreased levels of these markers and mineralization parallel effects observed in postmenopausal osteoporotic patients, who exhibit significant reductions in the number of osteocytes and osteoblasts, and demineralization of compact and cancellous bone [20]. While DPSCs primarily differentiate to dentin and BMSCs differentiate to bone, both dentin and bone formation share similar mineralization matrix components. Using DPSC model of mineralization will give us insight on the molecular effects of EtOH on mineralization matrix that is similar to bone mineralization model of BMSCs. In this study, we explore the epigenetic effects of alcohol on DPSCs and a possible link between alcohol and mineralization through the dysregulation of KDM6B.
MATERIALS AND METHODS
Culture of human DPSCs and EtOH treatment
Early passage DPSCs (P1-P2) were isolated from adult teeth (molars) were cultured in α-MEM supplemented with 10% fetal bovine serum (v/v), 2 mM L-glutamine, 100 µM L-ascorbate-2-phosphate, 50 unit/ml penicillin and 50 µg/ml streptomycin. Exponentially growing DPSCs were treated with different concentrations of EtOH diluted from absolute EtOH (FW=21.7 M). For acute exposure, cells were fed with media containing given concentrations of EtOH (20 or 50 mM) for 24 or 48 hrs. For chronic exposure, cells were intermittently exposed to EtOH by oneday exposure and one-day withdrawal for up to 2 weeks.
Transcriptomic analysis
Total RNA was isolated with RNeasy kit (Qiagen, Carlsbad, CA). Samples were prepared in biological duplicates. Equal amount from each sample was subjected to biotinylation using BioArray High Yield RNA Transcript Labeling System (Enzo Life Sciences, Farmingdale, NY, USA). Equal amount cRNA from each sample was labeled, purified and fragmented by using GeneChip Sample Cleanup Module (Affymetrix, Santa Clara, CA, USA). Following the manufacturer’s protocols, the Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA) was applied for gene expression analysis (UCLA Clinical Microarray Core Facility).
Methyl-DNA Immunoprecipitation and DNA Methylation Array Analysis
Methyl-DNA immunoprecipitation was performed according to a previous standard protocol from our laboratory by using antibody against 5-methylcytidine (Eurogentec, San Diego, CA) [21]. Methylation array analysis was performed according to a previous standard protocol from our laboratory [21]. Genomic profiling was performed by NimbleGen Systems (100718 HG18 CpG Refseq Promoter MeDIP). 3 µg of sonicated total DNA as input and 4 µg of DNA sample immunoprecipitated with anti-5-methylcytidine were sent to the UCLA Clinical Microarray core for differential random labeling by priming with Cy3 or Cy5 and hybridization to arrays. From the scaled log2 ratio data, a fixed-length window (750 bp) was placed around each consecutive probe, and the one-sided Kolmogorov-Smirnov (KS) test was applied to determine whether the probes were drawn from a significantly more positive distribution of intensity log ratios than those in the rest of the array. Resulting score for each probe was −log10 (p value) from the windowed KS test around the probe and was assigned as “p-value.” NimbleScan software (NimbleGen Systems) detects peaks by searching for at least two probes above a p value minimum cutoff (−log10) of 2. Using a custom Unix code, we aligned “ratio peak p-values” to human genome18 (RefSeq.hg18) and created a matrix file.
Bioinformatics analysis
The fastq files generated have been processed using GALAXY and aligned to the human genome, build hg19 (https://main.g2.bx.psu.edu/). Further analysis was done using R and bioconductor. After alignment to the genome perl scripts was used to delineate regions of interest on proximal and distal promoters using the reference sequence obtained from the UCSC genome browser (http://genome.ucsc.edu/). Subsequently, these regions have been clustered by K-means clustering using Gene Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/). Clusters identified were visualized using JavaTreeView (http://jtreeview.sourceforge.net/). Furthermore, data has been collapsed and correlated to gene expression data within R.
Pathway focused RT-PCR array analysis
Total RNA was purified and the quality of RNA was determined by using Agilent Bioanalyzer. The samples were processed for RT2 Profiler PCR array analysis (SABiosciences, Valencia, CA). Three different pathway specific arrays were used- Epigenetic chromatin modification enzyme array, Stress and toxicity array, and Fibroblastic marker array. These arrays also contain genomic DNA contamination control, positive PCR controls, and RT controls. To ensure inter-well and intra-plate consistency, these controls were considered for determining Ct value for each well. Results (Ct values) were normalized against five internal controls (ex: Actin B, GAPDH, 18S rRNA, etc.). Results have been analyzed by using the company’s RT2 Profiler PCR Array Data Analysis version 3.5 (SABiosciences, Valencia, CA).
Odontogenic/osteogenic differentiation of human DPSCs and EtOH treatment
DPSCs were grown in mineralization-inducing media containing 100 μM ascorbic acid, 2 mM β-glycerophosphate and 50 nM dexamethasone. Exponentially growing DPSCs were treated with different concentrations of EtOH diluted from absolute EtOH (FW=21.7 M). For acute exposure, cells were fed with media containing given concentrations of EtOH (20 or 50 mM) for 24 or 48 hrs. For chronic exposure, cells were intermittently exposed to EtOH (one-day exposure and one-day withdrawal) for up to 2 weeks.
Transduction of human DPSCs and EtOH treatment
GP2-293 cells were grown in DMEM containing 10% FBS and transiently transfected with Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA) to produce retroviral MSCV-KDM6B. DPSCs were cultured to 60% confluency and stably transduced with either control retrovirus or retroviral MSCV-KDM6B to generate KDM6B expressing DPSCs. Exponentially growing DPSCs were grown in mineralization-inducing media as described above and treated with different concentrations of EtOH diluted from absolute EtOH (FW=21.7M). Cells were fed with differentiation media containing given concentrations of EtOH (50 or 100 mM) for indicated period of time.
Quantitative RT-PCR analysis of differentiation markers
For validation of gene expression by quantitative RT-PCR analysis, total RNA was first subjected to DNase digestion with Turbo DNA-free kit (Life Technologies, Grand Island, NY). 2 µg of total RNA treated with Turbo DNA-free kit (Thermo Fisher Scientific, Waltham, MA) was used to synthesize cDNA by using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) in 40 µl reaction mixture. The resulting cDNA was diluted (mixed with H2O by 1:4) and 1.5 µl of diluted cDNA was used per well (in 10 µl reaction volume) in a 384 well plate using LightCycler 480 SYBR Green I master mix (Roche Diagnostics, Indianapolis, IN). PCR was done with specific set of primers (Supplemental Table 1) at annealing temperature of 60°C.
Alkaline phosphatase and Alizarin Red staining
For alkaline phosphatase (ALP) staining, after induction, cells were fixed with 4% paraformaldehyde and incubated with a solution of 0.25% naphthol AS-BI phosphate and 0.75% Fast Blue BB (Sigma-Aldrich, St Louis, MO, USA) dissolved in 0.1 M Tris buffer (pH 9.3). ALP activity assay was performed using an ALP kit according to the manufacturer's protocol (Sigma-Aldrich, St Louis, MO, USA) and normalized based on protein concentrations. To detect mineralization potential, cells were induced for 2–3 weeks, fixed with 4% paraformaldehyde and stained with 2% Alizarin Red (Sigma-Aldrich, St Louis, MO, USA). To quantify the calcium mineral deposition, Alizarin Red was destained with 10% cetylpyridinium chloride in 10 mM sodium phosphate for 30 min at room temperature.
Implantation of DPSCs into immunocompromised mice
6.0×106 DPSCs were mixed with 40 mg of hydroxyapatite/tricalcium phosphate (HA/TCP) ceramic powder (Zimmer Inc.) and then implanted into the dorsal surface of 10-week-old immunocompromised mice (Beige nude/nude Xid (III) mice) as previously described (Miura et al., 2004). The implants were harvested at 8 weeks post-implantation, fixed in 4% paraformaldehyde, and then decalcified with 10% EDTA (pH 8.0) for paraffin embedding. Paraffin sections were deparaffinized, rehydrated, and stained with hematoxylin and eosin (H&E). New mineralized matrix formation was observed under microscope. For quantification, the NIH software Image J was used as previously described [22].
RESULTS
EtOH treatment resulted in significant gene dysregulation and DNA methylomic alterations in human DPSCs
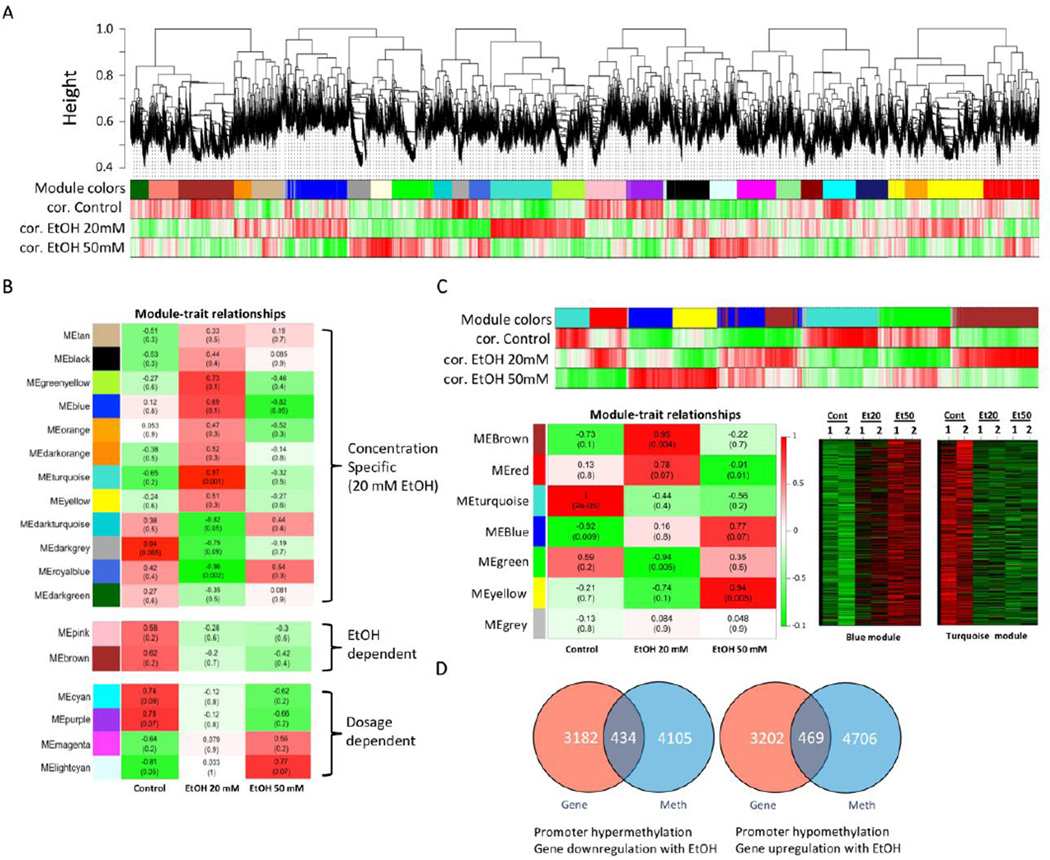
To test our hypothesis that alcohol may have deleterious molecular effects on the maintenance and regulation of adult stem cells, we have used multipotent dental pulp stem cells (DPSCs) and acutely exposed them (24 or 48 hrs) to defined concentration of EtOH (20 or 50 mM). Cell growth analysis showed that 20 or 50 mM EtOH treatment did not induce significant reduction in cell growth (Supplemental Figure 1). To profile molecular signatures that are affected by EtOH treatment in DPSCs, we have performed gene expression microarray analysis on DPSCs after 24 hr EtOH treatment. A genome-wide microarray (Affymetrix Human Genome U133 Plus 2.0 Array platform) was used on biological duplicates to generate gene expression data for DPSCs in order to compare the concentration-dependent effects of EtOH (0, 20 or 50 mM) (GEO Accession: GSE57255). Weighted Gene Correlation Analysis (WGCNA) identified several gene modules that are significantly correlated to EtOH treatment and revealed a certain level of complexity in gene expression level changes in response to EtOH treatment (Fig. 1A). We identified modules with dose-dependent changes in gene expression upon EtOH treatment (Fig. 1B). Also there were modules that showed changes only specific to certain EtOH concentration. We observed differential molecular effects by different concentrations of EtOH, such as 20 mM EtOH showed different pattern of expression changes compared to 50 mM EtOH. This implies that EtOH at different concentration levels may have different physiological impacts through differential molecular responses. Recently, epigenetic effects, such as DNA methylation, of alcohol on gene regulation have been documented [1, 23]. To test our hypothesis that alcohol induces epigenetic alterations and leads to deregulation of gene signatures in DPSCs, we have profiled DNA methylomic changes in DPSCs that are potentially affected by EtOH exposure. We have performed CpG Promoter microarray analysis coupled with methylated DNA immunoprecipitation (MeDIP) on the same set of samples used for transcriptomic profiling. The blue module consisting of genes whose promoters are hypermethylated with EtOH treatment and the turquoise module represents genes whose promoters are hypomethylated upon EtOH exposure (Fig. 1C). Heatmaps show changes in DNA methylation in biological duplicates of DPSCs upon exposure to different concentrations of EtOH (0, 20 and 50 mM) for 24 hrs. We have examined potential effects of EtOH-mediated DNA methylomic changes on actual transcriptomic alterations. We have performed combinatory analysis on genes that were (1) hypermethylated with genes that were downregulated and (2) hypomethylated with genes that were upregulated (Fig. 1D). We have identified genes that show concordant changes in promoter methylation and gene expression. With 20 mM EtOH treatment, we found that 434 genes were hypermethylated on the promoter whose expressions were downregulated. We also found 469 genes that were hypomethylated and upregulated. The results suggest that the molecular repertoire of gene expression in DPSCs is widely affected by EtOH-mediated DNA methylomic alterations.
Figure 1. Transcriptome analysis by gene expression microarray.
A. Weighted Gene Co-Expression Network Analysis (WGCNA) was performed to identify transcriptomic changes in DPSCs induced by EtOH. Expression microarray data was processed by WGCNA and modules of genes that are correlated together were identified. Modules are clusters of highly interconnected genes. In an unsigned co-expression network, modules correspond to clusters of genes with high absolute correlations. In a signed network, modules correspond to positively correlated genes. A group of genes (module) correlated to Control (cor. Control without EtOH treatment), EtOH 20 mM (cor. EtOH 20 mM) or EtOH 50 mM (cor. EtOH 50 mM) was depicted by different colors (module colors). B. Intramolecular connectivity measures how connected, or co-expresses, a given gene is with respect to the genes of a particular module. The intramolecular connectivity is interpreted as a measure of module membership. Module-trait relationship map of all modules identified from analysis demonstrating specific EtOH concentration-dependent, EtOH treatment-dependent, or EtOH dosage-dependent gene association modules. C. WGCNA analysis on DNA methylomic changes to identify modules associated with EtOH treatment and epigenetically regulated by EtOH-induced DNA methylation in DPSCs. Most significant modules were selected from the module-trait detection and corresponding heat-maps (blue and turquois modules) are shown. D. Venn diagram analysis of transcriptome vs. DNA methylation to display the number of epigenetically regulated genes. Dosage-dependent hypermethylation and correlated decrease in gene expression due to EtOH. Dosage-dependent hypomethylation and correlated increase in gene expression due to EtOH.
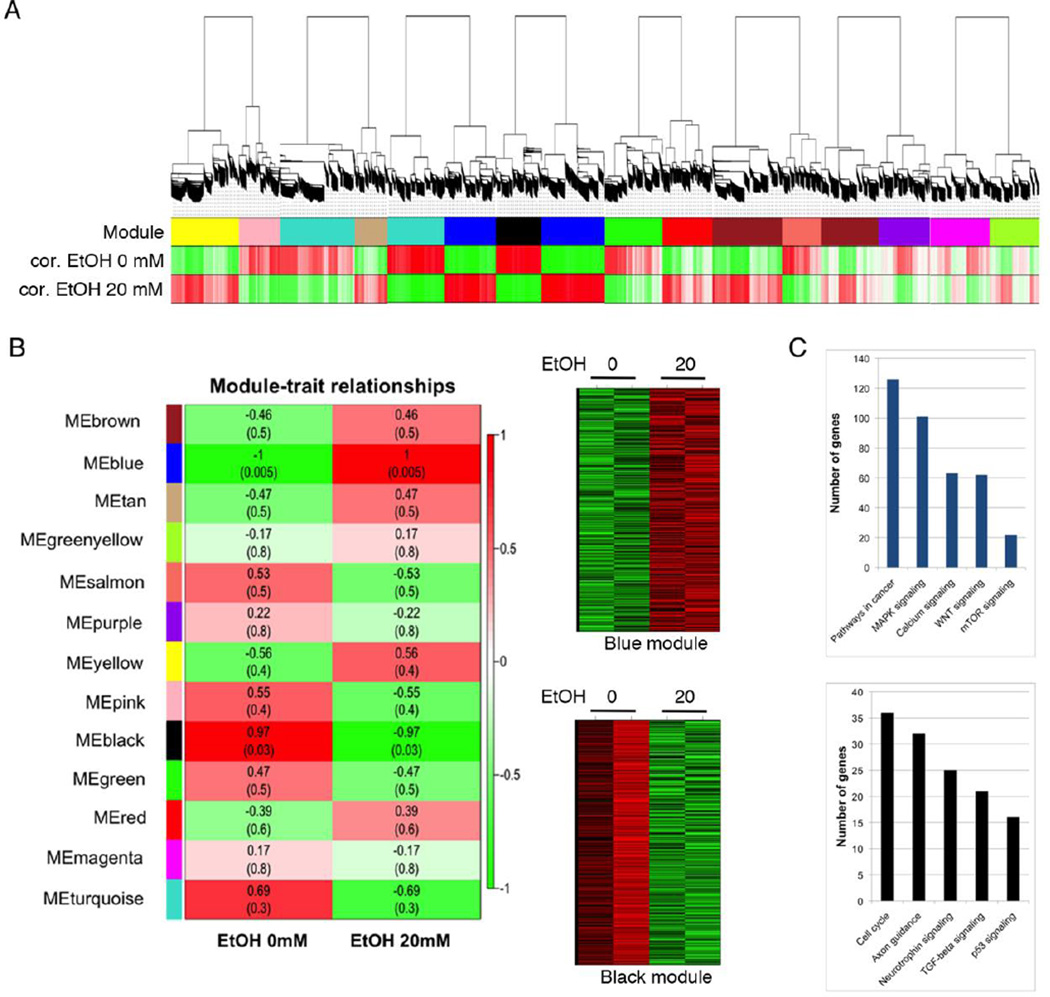
Since we were interested in more physiologically relevant effects of EtOH, we focused on 20 mM EtOH’s effect that is known to be the DUI level of alcohol in blood (Fig. 2A). We identified two gene modules that were most significantly associated with EtOH treatment- the blue module for genes upregulated with EtOH exposure and the black module for genes downregulated with EtOH treatment (Fig. 2B). To examine potential effects of EtOH on biological process, we have performed DAVID (The Database for Annotation, Visualization and Integrated Discovery) on genes from the blue and black modules. The blue module of upregulated genes upon EtOH treatment was associated with several important signaling pathways, such as MAPK, calcium, WNT and mTOR signaling (Fig. 2C). On the other hand, the black module of downregulated genes showed association with cell cycle, axon guidance, neurotrophin signaling, TGF-beta signaling and p53 signaling (Fig. 2C). To examine if there is any concordance in molecular networks affected by EtOH treatment, we have performed basic expression analysis in Cytoscape. We established the core gene interaction network by combining the genes in the blue or the black module with the network data (Supplemental Figure 2). The blue module contains CTBP1, FAM82A1 (RMD4), CCDC149, MYO10, FCGR2C, SRGAP2, DLGAP1, PPP2R5C and CGLF2. The black module consists of STAG2, CCT2, PKG1, MAGIX, PDZD3, HNRNPD, INSIG1, FCRL5, HLA-E and PDLIM5. We have ranked genes based on fold changes (>2-fold) and p values (<0.05) for each EtOH concentration treatment (20 mM or 50 mM) for further selection and validation (Table 1).
Figure 2. WGCNA on DPSCs treated with 20 mM EtOH.
A. WGCNA for transcriptomic changes induced by 20 mM EtOH treatment that is comparable to a 0.08% blood alcohol concentration (BAC) of the DUI level, leads to EtOH-induced gene expression changes in DPSCs. B. Module-trait relationship map and heatmap analysis of the black and blue modules, or gene expression profiles, where red indicates up-regulation and green indicates down-regulation. C. The Database for Annotation, Visualization and Integrated Discovery (DAVID) gene functional analysis on the blue and the black module.
Table 1.
List of genes from DPSCs treated with 20 or 50 mM EtOH for 24 hrs (p < 0.0 5).
| EtOH 20 mM | |||
|---|---|---|---|
| Gene Symbol |
Fold change |
Gene Symbol |
Fold change |
| ELK4 | 4.4 | DDR2 | −3.2 |
| MGC24103 | 2.9 | ITPRIPL2 | −2.8 |
| ZCCHC7 | 2.9 | MALAT1 | −2.7 |
| SPATS2L | 2.8 | TMPO | −2.6 |
| SPEN | 2.7 | ZYG11B | −2.6 |
| SATB2 | 2.7 | KDM6B | −2.5 |
| PPP1R9B | 2.5 | CALD1 | −2.5 |
| NCRNA00182 | 2.4 | ZNF655 | −2.5 |
| CLCN5 | 2.3 | C13orf37 | −2.4 |
| LRP1 | 2.3 | RPL37A | −2.3 |
| MCTP2 | 2.3 | APOOL | −2.2 |
| RNF167 | 2.2 | LOC100288675 | −2.2 |
| RAB3B | 2.2 | PTPN11 | −2.1 |
| KIAA0754 | 2.2 | EIF5B | −2.1 |
| QSER1 | 2.2 | ATG3 | −2.1 |
| MAP4K4 | 2.2 | MALAT1 | −2.1 |
| NFAT5 | 2.2 | KPNA5 | −2.1 |
| LOC286272 | 2.1 | DHFR | −2.0 |
| ANKRD28 | 2.1 | WSB1 | −2.0 |
| LOC100289230 | 2.1 | C6orf62 | −2.0 |
| RALGPS2 | 2.1 | N4BP2L2 | −2.0 |
| RANBP2 | 2.1 | ESCO1 | −2.0 |
| ITGA8 | 2.1 | EHBP1L1 | −2.0 |
| PIK3C2A | 2.0 | PTPRO | −2.0 |
| MYCBP2 | 2.0 | ERCC8 | −2.0 |
| LOC572558 | 2.0 | GNG12 | −2.0 |
| LRCH3 | 2.0 | ||
| PARVA | 2.0 | ||
| CD44 | 2.0 |
| EtOH 50 mM | |||
|---|---|---|---|
| Gene Symbol |
Fold change |
Gene Symbol |
Fold change |
| ELK4 | 2.9 | RPL10L | −4.3 |
| CALD1 | 2.9 | MALAT1 | −2.9 |
| SUZ12P | 2.3 | ATG3 | −2.7 |
| MCTP2 | 2.2 | RBM5 | −2.3 |
| RASEF | 2.1 | SNX11 | −2.3 |
| GUSBP3 | 2.0 | SSFA2 | −2.2 |
| LRP1 | 2.0 | LAMA3 | −2.1 |
| COL12A1 | 2.0 | METTL7A | −2.1 |
| LRRC27 | −2.1 | ||
| SLAIN2 | −2.1 | ||
| MALAT1 | −2.1 | ||
| SEC62 | −2.1 | ||
| C11orf31 | −2.0 | ||
| C13orf31 | −2.0 | ||
| USP28 | −2.0 | ||
| DDR2 | −2.0 | ||
| LARP4 | −2.0 |
EtOH treatment triggered changes in epigenetic modifiers in human DPSCs
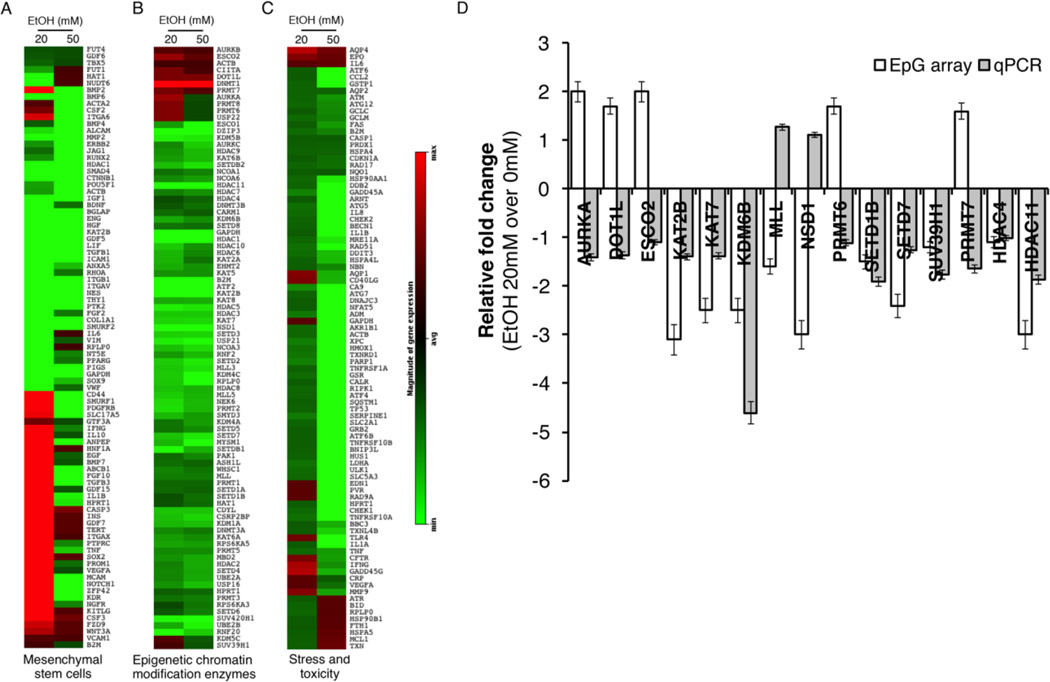
To examine EtOH-induced changes in known epigenetic modifiers in DPSCs we have performed a pathway focused RT-PCR analysis and assessed the effect of EtOH on the levels of known epigenetic chromatin modification enzymes, fibroblastic markers, and stress/toxicity genes (Qiagen Inc., Valencia, CA). We have used two different EtOH concentrations (20 and 50 mM) and treated cells acutely for 24 hrs. Heatmaps show the level of genes upon exposure to EtOH relative to non-treatment control (Fig. 3). We observed both EtOH concentration-dependent and concentration-independent changes in the level of genes. We then further attempted to validate the acute changes in gene expression, especially of epigenetic chromatin modification enzymes, by quantitative PCR analysis (Fig. 3D). Among 15 genes we tested (AURKA, DOT1L, ESCO2, KAT2B, KAT7, KDM6B, MLL, NSD1, PRMT6, SETD1B, SETD7, SUV39H1, FCHO, PRMT7, DNMT1, HDAC4 and HDAC11), we found that 8 genes showed the levels of expression concordant between the array result and qRT-PCR result. We found lysine demethylase 6B (KDM6B) showed the most noticeable change upon EtOH treatment (2.5-fold downregulated in the RT array analysis and 4.6-fold downregulated in the qRT-PCR analysis) (Fig. 3D). Previously KDM6B has been demonstrated to play a role in the control of DPSC and BMSC [18, 19], which suggests that alcohol-mediated dysregulation of KDM6B may have a functional link to the effect of alcohol exposure on osteogenic differentiation of DPSCs.
Figure 3. Pathway focused RT-PCR array analysis for genes affected in DPSCs by EtOH treatment.
For acute exposure DPSCs were treated for 24hrs with 20 or 50mM EtOH. For chronic exposure cells were treated every other day for 10 days with 20 or 50mM EtOH. Total RNA was prepared and subjected to RT-PCR array analysis. A. Fibroblastic marker array, B. Epigenetic chromatin modification enzymes array. C. Stress and toxicity pathway finder array. Data was analyzed and fold changes against no treatment are presented. D. Quantitative RT-PCR analysis was done to validate the result from Epigenetic modifier RT array. Error bar shows the standard error margin (SEM).
EtOH treatment induced dysregulation of KDM6B and odontogenic/osteogenic differentiation
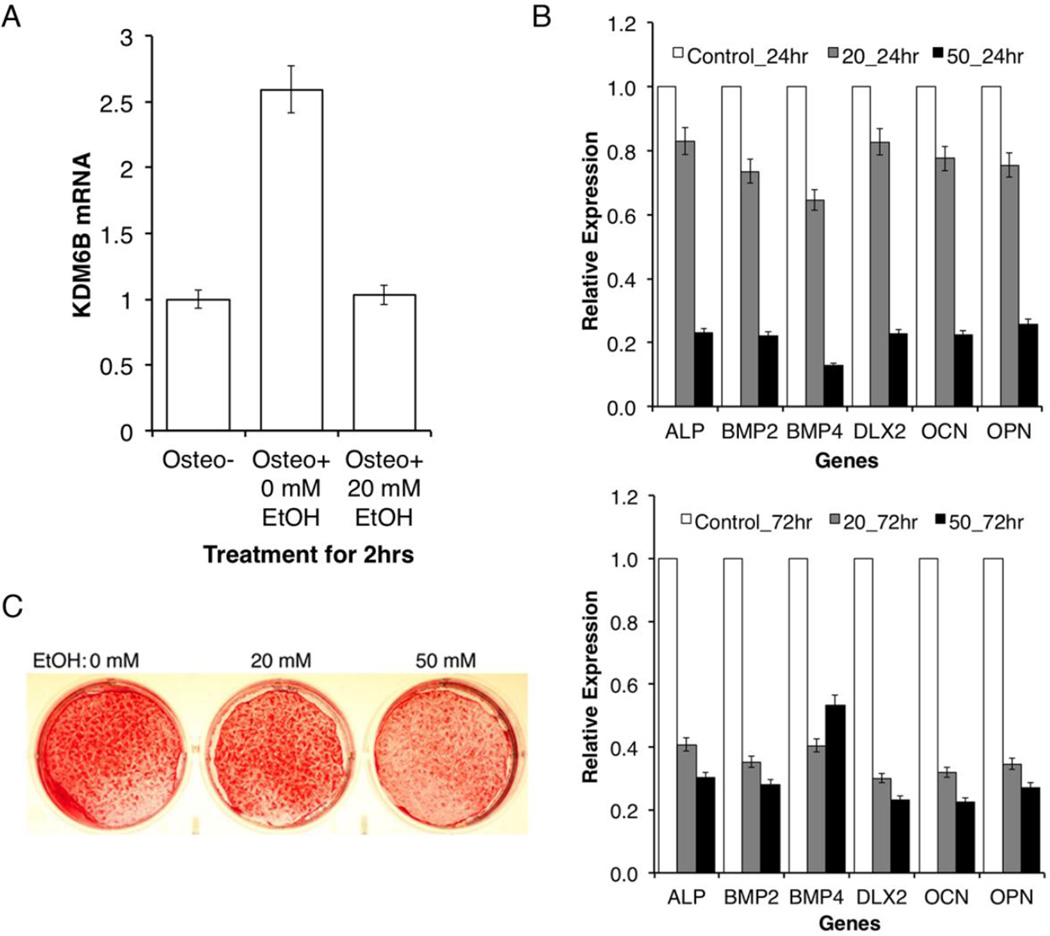
To examine if EtOH treatment has any functional effect on the mineralization of DPSCs, we have cultured DPSCs under odontogenic/osteogenic differentiation conditions and examined the molecular effect of EtOH on the expression of differentiation-related lineage markers (Fig. 4). It has been previously demonstrated that the induction of KDM6B during odontogenic differentiation is immediate. Since KDM6B was previously demonstrated as an early responder [19], the fold changes of KDM6B expression with and without induction of differentiation treatment at 20 mM EtOH were analyzed at the 2hr time point (Fig. 4A). Upon analysis, EtOH treatment resulted in a significant reduction of KDM6B expression level during odontogenic/osteogenic differentiation (2.5-fold downregulated in the qRT-PCR analysis). Furthermore, relative expression levels of the mineralization-associated markers we tested (ALP, BMP2, BMP4, DLX2, OCN, and OPN) at the 24 and 72 hr time periods all exhibited concentration dependent reductions to EtOH, with more significant decreases at the 72 hr time point (Fig. 4B). As KDM6B has been demonstrated to play an early role in the control of DPSC fate, we suggest that these differentiation-related markers may be functioning downstream of the epigenetic modifier.
Figure 4. Effect of EtOH on molecular regulation of odontogenic/osteogenic differentiation.
A. Comparison of the fold changes of KDM6B expression with and without odontogenic/osteogenic differentiation treatment in DPSCs. In response to EtOH, KDM6B expression is significantly reduced during differentiation. Error bar shows the standard error margin (SEM). Statistical significance was determined by Student t-test (p<0.05). B. Comparison of the relative expression levels of osteomarkers in DPSCs cells under differentiation with or without treatment (20 mM or 50 mM EtOH) for 24 hr and 72 hr. At both 24 hr and 72 hr, reduced levels of various osteomarkers were observed. Statistical significance was determined by the one-way ANOVA (p<0.05). Error bar shows the standard error margin (SEM). C. Disruptive effects of EtOH on mineralization in vitro. Reduced levels of mineralization assessed by Alizarin staining were observed in response to EtOH.
Following the analysis of the acute effects of EtOH in DPSCs, we investigated the chronic effects of EtOH on mineralization. We investigated these long-term effects on differentiation in vitro by culturing DPSCs in odontogenic/osteogenic differentiation media for 2 weeks with 20 mM or 50 mM level of EtOH treatment. The effects of EtOH on mineralization were qualitatively visualized through Alizarin Red staining. At the 2-week time point, Alizarin red staining, indicative of calcium accumulation by cells of an odontogenic/osteogenic lineage, was highest in the control group differentiated without EtOH treatment (Fig. 4C). As the concentration of EtOH treatment increased from 20 mM to 50 mM, a progressive level of the decreased staining was observed. Similarly, alkaline phosphatase staining was highest in the control group differentiated without EtOH treatment (data not shown). Much like the results observed with the Alizarin Red staining, the density of the staining progressively decreased with increasing concentrations of EtOH at 20 mM and 50 mM. These results suggest that EtOH has both acute and chronic effects on the mineralization potential of DPSCs.
Suppression of KDM6B altered odontogenic/osteogenic potency in DPSCs
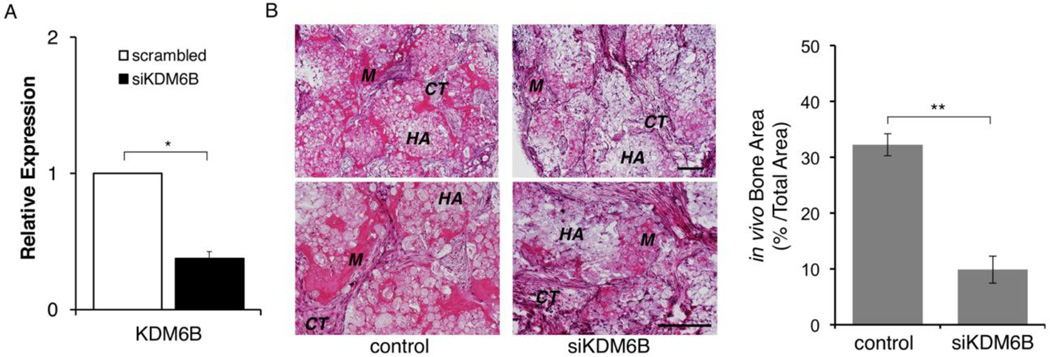
Our findings support that EOH treatment resulted in suppression of KDM6B along with inhibition of mineralization in DPSCs. We next investigated whether the expression level of KDM6B was directly correlated with odontogenic/osteogenic differentiation potential through associated osteomarkers. It has recently been demonstrated that a knockdown of KDM6B in BMSCs and DPSCs resulted in dysregulation of osteogenic and odontogenic differentiation in vitro and in vivo [18, 19]. As KDM6B was significantly related to the odontogenic/osteogenic potential of DPSCs in vitro, we performed an in vivo study in mice to further investigate KDM6B ability to induce mineralization. DPSCs in the control group were transfected with non-targeting siRNA while the experimental group was transfected with siKDM6B (Fig. 5A). Subsequently, these cells were implanted into immunocompromised mice and allowed to differentiate. Following an 8-week time period, histological analysis of the H&E staining in the control group indicated the formation of mineralized tissue and minimal connective tissue around tricalcium phosphate/hydroxyapatite (TCP-HA) bone scaffold (Fig. 5B). In contrast, DPSCs transfected with siKDM6B showed significant reductions in the formation of mineralized tissue and more prominent formation of connective tissue. When a quantitative analysis of the total in vivo mineralized matrix area was performed, the DPSCs transfected with siKDM6B showed a 66% reduction in mineral deposition as compared to the control (Fig. 5C). Our results suggest that KDM6B is necessary for odontogenic/osteogenic differentiation potential and formation of mineralized tissue.
Figure 5. Knockdown of KDM6B resulted in a reduced mineralization potential in vivo.
A. DPSCs were transiently transfected with a scrambled siRNA or siKDM6B construct (Thermo Scientific, Waltham, MA). Quantitative RT-PCR analysis showed >2-fold reduction in KDM6B in siKDM6B cells. Error bar shows the standard error margin (SEM). Statistical significance was determined by Student t-test (*: p<0.05). B. DPSCs transfected with control (scrambled siRNA) or siKDM6B cells were implanted into mice as described in Materials and Methods. Formation of mineralized tissue (M) and connective tissue (CT) around HA/TCP (HA) are indicated in H&E staining section. Quantitative measurement showed that siKDM6B resulted in about 66% reduction in bone area compared to the control. Statistical significance was determined by Student t-test (**: p<0.05). Error bar shows the standard error margin (SEM).
Ectopic expression of KDM6B restored odontogenic/osteogenic differentiation in EtOH-treated DPSCs
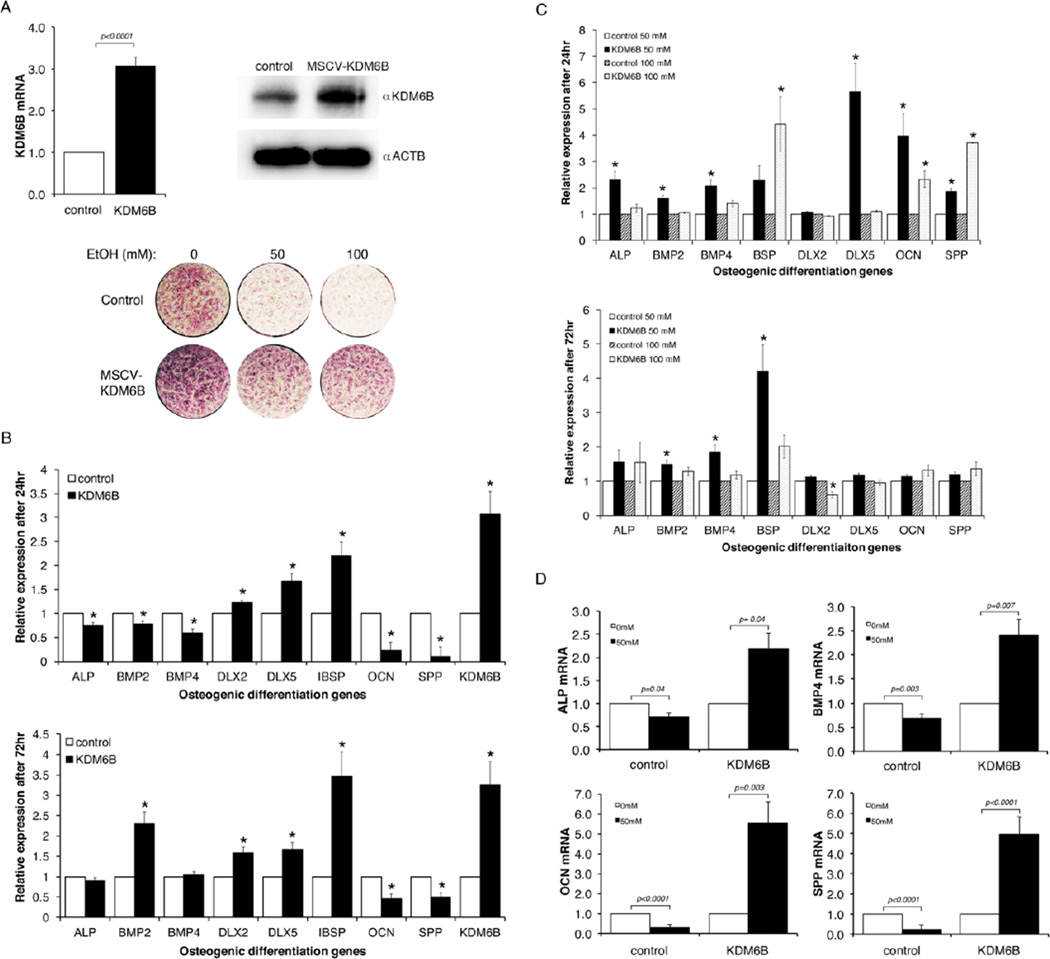
To further assess the direct functional significance of EtOH-induced suppression of KDM6B in the effect of alcohol exposure on odontogenic/osteogenic differentiation, we examined the restorative effect of the ectopic expression of KDM6B on EtOH-induced dysregulation of mineralization. DPSCs in culture were stably transduced with retroviral KDM6B expression construct and control virus. The expression of transduced KDM6B was determined by qRT-PCR and Western analysis (Fig. 6A). In addition, the transduced cells were subjected to odontogenic/osteogenic differentiation for 2 weeks. We analyzed the odontogenic/osteogenic differentiation potential of transduced DPSCs through the expression of alkaline phosphatase (ALP), which is frequently used to characterize activity of mineralizing cells [24–28]. Following ALP staining, we found that DPSCs transduced with the control vector showed a significant, progressive reduction in ALP staining at 50 and 100 mM EtOH treatment. In contrast, DPSCs transduced with the MSCV-KDM6B construct and treated with 50 and 100 mM EtOH showed enhanced levels of staining as compared to the control cells. Our results suggest that KDM6B was able to adequately restore mineralization potential of DPSCs in the presence of varying levels of EtOH.
Figure 6. Expression of KDM6B restored differentiation potency in EtOH-treated DPSCs.
A. DPSCs were transduced with retroviral KDM6B expression construct (MSCV-KDM6B) or control virus. Three days after transduction cells were lysed and total RNA or protein was purified. The expression of transduced KDM6B was monitored by qRT-PCR or Western analysis using anti-KDM6B (1:1,1000, OriGene Technologies, Inc., Rockville, MD). For protein loading control anti-actin antibody (1:3,000, Sigma, St. Louis, MO) was used. Transduced DPSC cells were plated into 24 well plates and treated with mineralization media containing 0, 50 or 100 mM EtOH for 2 weeks with daily medium change. Differentiation of DPSCs was monitored by alkaline phosphatase staining. B. The effect of overexpressing KDM6B in DPSCs on the expression of differentiation-related genes was examined. DPSCs transduced with either control or MSCV-KDM6B were differentiated in osteogenic media for 24 or 72 hrs and the level of differentiation-related genes was assessed by qRT-PCR in triplicates. Ectopic expression of KDM6B resulted in a differential regulation of lineage marker genes. For both 24 and 72 hr differentiation, DLX2, DLX5 and IBSP were consistently induced by KDM6B, but OCN and SPP were consistently suppressed. Statistical significance was determined by the one-way ANOVA and p value <0.05 is considered as significant (with asterisk). Error bar shows the standard error margin (SEM). C. Transduced cells were plated into 6 well plates and >90% confluent cells were treated with odontogenic/osteogenic differentiation media containing 50 or 100 mM EtOH for 24 and 72 hrs. Total RNA was isolated and qRT-PCR analysis was performed for various osteogenic marker genes. Comparison was made within samples for control cells and KDM6B expressing cells by determining the relative fold change for 50 or 100mM EtOH treatment. Statistical analysis was done on triplicate samples using the one-way ANOVA and p value <0.05 is considered as significant (with asterisk). Error bar shows the standard error margin (SEM). D. The effect of EtOH treatment on mineralization-associated gene expression was compared between DPSCs transduced with the control virus and overexpressing KDM6B. DPSCs transduced with either the control virus or MSCV-KDM6B were differentiated for 24 hrs in the presence of 0 or 50 mM EtOH. The levels of mineralization-associated genes were determined by qRT-PCR analysis. Statistical significance was determined by the one-way ANOVA. Error bar shows the standard error margin (SEM).
The resulting transduced cells were subjected to odontogenic/osteogenic differentiation for 24 or 72 hrs and the level of osteogenic marker expression was quantitatively assessed. It was found that the forced expression of KDM6B in differentiating DPSCs in the absence of EtOH treatment caused alterations in mineralization-related genes (Fig. 6B). Some of the markers (DLX2, DLX5, and IBSP) were further induced by the expression of exogenous KDM6B compared to the control vector. However, the expression of OCN and SPP were suppressed by the ectopic expression of KDM6B. There were slight differences at the level of changes between the 24 and 72 hr differentiation, but the trend of the effect of KDM6B expression on marker gene expression was similar between the two time points.
The effect of KDM6B expression on the EtOH-induced suppression of odontogenic/osteogenic differentiation genes was determined. As shown in Fig. 6C, the forced expression of KDM6B in DPSCs suppressed EtOH-induced inhibition of mineralization in vitro. Compared to DPSCs transduced with the control virus, KDM6B expressing DPSCs showed significant induction of differentiation-related genes even in the presence of either 50 or 100 mM EtOH (Fig. 6C). It was noted that the suppressive effect of 50 mM EtOH treatment on gene expression was more readily reversed by the expression of KDM6B than cells with 100 mM EtOH treatment (ALP, BMP2, BMP4, IBSP, DLX5 and OCN). The DLX2 gene was least affected by KDM6B. A higher dose of EtOH (100 mM compared to 50 mM) and a longer exposure (72 hr compared to 24 hr) to EtOH resulted in a reduction of the effect of exogenous KDM6B expression on the recovery of odontogenic/osteogenic gene expression.
We found that the forced expression of KDM6B significantly altered the response of DPSCs to EtOH treatment. As shown in Fig. 6D, we found that EtOH treatment induced the expression of mineralization-related genes in KDM6B expressing cells. The expression of markers was significantly inhibited by 50 mM EtOH treatment when compared within control group (0 mM EtOH) for genes ALP (p=0.04), BMP4 (p=0.003), OCN (p<0.0001), and SPP (p<0.0001) (Fig. 6D). The ectopic expression of KDM6B resulted in significant increases in expression levels of these genes when treated with EtOH (50 mM) compared to non-treated cells (0 mM), particularly at 5.53 fold (p=0.003) and 4.94 fold (p<0.0001) increases for OCN and SPP, respectively. It seems that molecular dynamics and timing in signaling between mineralization signals and the regulation of key regulators such as KDM6B is important to ensure proper mineralization of DPSCs. A prolonged expression of KDM6B at a non-physiological level as shown in KDM6B expression cells could cause dysregulation in cellular response and downstream molecular signaling pathways. Overall, our results suggest that the expression of KDM6B in DPSCs can counteract the effects of EtOH-induced inhibition of odontogenic/osteogenic differentiation and promote mineralization in the presence of EtOH.
DISCUSSION
Alcohol abuse appears to lead to periodontal disease, tooth decay and mouth sores that are potentially precancerous [29–32]. Persons who abuse alcohol are at high risk of having seriously deteriorated teeth, gums and compromised oral health in general [29–32]. It is generally believed that alcohol may have toxic effects on various cellular functions, but we are in lack of detailed information about molecular and cellular effects of alcohol on stem cell maintenance and the differentiation process. Our recent publication demonstrated that EtOH exposure induced significant transcriptomic alterations in human embryonic stem cells (hESCs) through DNA methylomic deregulation [1]. Several studies have demonstrated effects of EtOH on DNA methylation, resulting in genetic and phenotypic changes [33–42]. It has been shown that EtOH induced alterations in DNA methylation patterns and inhibited neural stem cell (NSC) differentiation [43]. EtOH induced the hypermethylation of multiple cell cycle genes and increased the expression of DNA methyltransferases in NSCs. These alterations affected growth factor signaling, in conjunction with the down regulation of associated mRNAs and cell cycle proteins [44]. In another study, EtOH exposure prevented the methylation of specific genes related to neural development, including insulin-like growth factor 1 (IGF1), epidermal growth factor-containing fibulin-like extracellular matrix protein (EFEMP1), and SRY-box-containing gene 7 (SOX7) [43]. The hyper- or hypomethylation of specific genes have been shown to significantly affect NSCs differentiation and proliferation. However, it is important to note that many studies related to EtOH exposure and adult stem cell potency have shown that the intrinsic genetic and epigenetic mechanisms that control cellular fate are potentially of equal significance.
The negative, long term effects of alcoholism on bone mass have been well-established [3]. Studies have shown that heavy chronic alcohol consumption compromises bone quality and increases the risk for osteoporosis. Other studies have shown that alcoholism is associated with a variety of risk factors that may contribute to the pathogenesis of bone disease, including poor nutrition, liver disease, malabsorption, vitamin D deficiency, hypogonadism, parathyroid dysfunction and tobacco use [45]. Recent studies have reported that alcohol may affect bone formation through osteocyte apoptosis, oxidative stress, and Wnt signaling pathway modulation [6]. One study involving alcohol-binged rats suggested that following administration of 20% alcohol/saline solution for 1, 2 or 3 weeks, a stimulation of bone resorption and decrease in bone mineral density was observed. However, it was interesting to note that concurrent administration of risedronate, a bisphosphonate, mitigated the response and maintained trabecular architectural indices [46]. The duration of alcohol treatment resulted in the modulation of expression profiles of RANKL and OPG, genes that regulate osteoclastogenesis. This study sheds light on the deleterious effects of bone metabolism in response to binge drinking. However, the exact mechanisms of how alcohol is related to bone loss remains unknown, but may include both direct and indirect actions, and be related to diet and other lifestyle factors.
While chronic alcohol abuse has been linked to an increase in the risk for osteoporosis, light to moderate consumption has been correlated with a reduction in osteoporotic risk. Some studies have identified that light to moderate concentrations of EtOH exposure has resulted in higher bone mineral density and a reduced risk of osteoporosis [3, 4]. Another study found that light alcohol consumption resulted in increased lumbar spine bone mineral density (BMD) and whole body BMD in postmenopausal women [47]. In animal studies, low alcohol consumption of ethanol (5%, 2h per day) in 4-week old rats showed higher BMD and greater trabecular thickness than compared to the control groups [48]. However, there are few animal models related to the effects of light alcohol consumption in the literature and further investigative studies in animal models should be performed in the future to shed light on these results. In contrast, at longer durations of moderate to high alcoholic exposure, there is a dose dependent effect on bone to increase risk for stress fractures. In a rat study investigating the long-term effects of heavy alcohol consumption on cancellous and cortical bone microarchitecture, ethanol consumption resulted in lower bone mineral density and content, reduced cortical thickness, and a lower femur length as compared to controls [49]. These results suggest that chronic, heavy alcohol consumption results in a decrease in bone size, mass, and density, and negatively alters cancellous bone microarchitecture resulting in decreased skeletal integrity.
In this study, we have used DPSCs as a model to examine the effect of EtOH on mineralization process. Our findings show that EtOH treatment resulted in altered mineralization in DPSCs. The dysregulation of odontogenic/osteogenic differentiation in DPSCs treated with EtOH is reminiscent of previous findings with reductions in bone mineral density and volume. We discovered that EtOH treatment results in the reduction of several known mineralization-related gene expression profiles, including ALP, BMP2, BMP4, DLX2, OCN, and OPN. These findings appear to mirror potential biological mechanisms that are significantly dysregulated in the etiology of alcohol-induced osteoporotic events. In vitro studies are often difficult to adapt to in vivo studies due to their inability to fully simulate in vivo conditions. There are no in vitro models to study the effects of EtOH on bone formation and resorption [50]. Furthermore, one study identified a significant discrepancy in body weight gain and bone measurements due to the delivery method of EtOH. In this study, intraperitoneal injection resulted in reduced body weight, suppression of periosteal and cancellous bone formation, and decreased mRNA levels for bone matrix proteins as compared to intragastric administration [51]. This study suggests that treatment of DPSCs in vitro using EtOH-containing media may provide a dosage that is not replicable physiologically. In addition, the effects of alcohol on epigenetic modifiers and bone loss may not be fully adaptable to human studies, as it fails to take into account co-morbidity factors including poor nutrition, vitamin deficiencies, mechanical loading, weight and smoking [50]. The effects of alcohol related bone loss is controversial, with human studies reporting bone loss as well as no bone loss.
Based on our findings on genome-wide, epigenetic effects of EtOH on DPSCs, we analyzed known epigenetic modifiers to elucidate a potential epigenetic link between alcohol and osteoporosis. Consistent with our conclusion, KDM6B, a known epigenetic modifier involved in osteogenic and odontogenic differentiation, was found to be significantly dysregulated in response to EtOH. Knockdown of KDM6B resulted in similar phenotype as EtOH treatment in vitro while concurrently showing similar reductions in the gene expression profiles of known osteomarkers. Thus, it is suggested that dysregulation of KDM6B by EtOH reduces odontogenic/osteogenic potency of DPSCs, however the molecular pathways behind the interactions of KDM6B and known mineralization-associated markers remains to be discovered.
Recent studies show that KDM6B is involved in the control of calcium-induced differentiation [52], regulates osteoblast differentiation [53], contributes to neuronal survival and differentiation [54], odontogenic differentiation of DPSCs [19], and osteogenic differentiation of human BMSCs [18]. It has been reported that calcium-induced differentiation leads to increased binding of KDM6B and erasure of repressive marks such as H3K27me3 [52]. It has been demonstrated that BMP4/7-mediated activation of SMAD1/4 may induce the expression of KDM6B and trigger osteogenic pathway [18]. Mechanistically, KDM6B is recruited to bone morphogenic protein 2 (BMP2) and HOX (homeotic genes) promoters and activate the odontogenic differentiation-related gene expression [18]. KDM6B removes epigenetic marks, H3K27me3, from the promoter of osteogenic genes to promote osteogenic commitment [18]. In addition, a recent study has demonstrated the facilitating role of KDM6B on odontogenic differentiation [19]. It needs to be experimentally demonstrated if EtOH triggers dysregulation of KDM6B via altered regulation of differentiation signal-induced activation of SMAD1/4 pathway and the control of H3K27me3 mark as demonstrated in BMSCs [18].
In conclusion, our findings confirmed that EtOH has a significant impact on the dysregulation of KDM6B and provides a potential epigenetic effect of heavy alcohol exposure in DPSCs. These findings will be helpful in identifying molecular mechanisms associated with alcohol induced osteoporosis in a proper model, as it suggests a potential mechanism of EtOH-induced suppression of mineralization. Further study is needed to evaluate the significance of our findings in fetal development where maternal alcohol consumption results in craniofacial and dental abnormalities, which are hallmark features of fetal alcohol spectrum disorders (FASD).
Supplementary Material
CytoEdge analysis of the blue (genes upregulated with 20 mM EtOH treatment) and the black module (genes downregulated with 20 mM EtOH treatment) from Figure 2 visually present the interaction network of genes in the given module closely associated with EtOH treatment.
Effect of EtOH treatment on the growth of DPSCs was examined. Cells (5,000 cells per well) were seeded on 24 well plates in triplicates and treated with EtOH (0, 20, 50 and 100 mM) intermittently by one-day exposure and one-day withdrawal up to six days. Live cell number was determined by MTT assay with measurement at 570 nm. The error bar shows standard error margin (SEM) from triplicates (p value <0.05). It was noticed that 100 mM EtOH treatment induced a reduction in cell number, but other concentrations didn’t have significant effect on the cell growth.
Highlights.
Molecular effects of alcohol on transcriptome and DNA methylome in human DPSCs
Identification of KDM6B as a target in alcohol-induced disruption of DPSC potency
Validation of the role of KDM6B in alcohol-induced disruption in vitro and in vivo
Acknowledgments
This work was supported by a research grant award from NIH/NIAAA (R01AA21301) and UCLA School of Dentistry Faculty Seed Grant Award to Y.K. CIRM CSUN-UCLA Bridges to Stem Cell Research Award to B.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Khalid O, Kim JJ, Kim HS, et al. Gene expression signatures affected by alcohol-induced DNA methylomic deregulation in human embryonic stem cells. Stem Cell Res. 2014;12:791–806. doi: 10.1016/j.scr.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuckit MA. Alcohol-use disorders. Lancet. 2009;373:492–501. doi: 10.1016/S0140-6736(09)60009-X. [DOI] [PubMed] [Google Scholar]
- 3.Feskanich D, Korrick SA, Greenspan SL, et al. Moderate alcohol consumption and bone density among postmenopausal women. J Womens Health. 1999;8:65–73. doi: 10.1089/jwh.1999.8.65. [DOI] [PubMed] [Google Scholar]
- 4.Jugdaohsingh R, O'Connell MA, Sripanyakorn S, et al. Moderate alcohol consumption and increased bone mineral density: potential ethanol and non-ethanol mechanisms. Proc Nutr Soc. 2006;65:291–310.e. doi: 10.1079/pns2006508. [DOI] [PubMed] [Google Scholar]
- 5.Sampson HW. Alcohol, osteoporosis, and bone regulating hormones. Alcohol Clin Exp Res. 1997;21:400–403. doi: 10.1111/j.1530-0277.1997.tb03782.x. [DOI] [PubMed] [Google Scholar]
- 6.Maurel DB, Boisseau N, Benhamou CL, et al. Alcohol and bone: review of dose effects and mechanisms. Osteoporos Int. 2012;23:1–16. doi: 10.1007/s00198-011-1787-7. [DOI] [PubMed] [Google Scholar]
- 7.Church MW, Eldis F, Blakley BW, et al. Hearing, language, speech, vestibular, and dentofacial disorders in fetal alcohol syndrome. Alcohol Clin Exp Res. 1997;21:227–237. [PubMed] [Google Scholar]
- 8.Sant'Anna LB, Tosello DO, Pasetto S. Effects of maternal ethanol intake on immunoexpression of epidermal growth factor in developing rat mandibular molar. Arch Oral Biol. 2005;50:625–634. doi: 10.1016/j.archoralbio.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 9.Kieser JA. Fluctuating odontometric asymmetry and maternal alcohol consumption. Ann Hum Biol. 1992;19:513–520. doi: 10.1080/03014469200002342. [DOI] [PubMed] [Google Scholar]
- 10.Jimenez-Farfan D, Guevara J, Zenteno E, et al. Alteration of the sialylation pattern of the murine tooth germ after ethanol exposure. Birth Defects Res A Clin Mol Teratol. 2005;73:980–988. doi: 10.1002/bdra.20198. [DOI] [PubMed] [Google Scholar]
- 11.Jimenez-Farfan D, Guevara J, Zenteno E, et al. EGF-R and erbB-2 in murine tooth development after ethanol exposure. Birth Defects Res A Clin Mol Teratol. 2005;73:65–71. doi: 10.1002/bdra.20113. [DOI] [PubMed] [Google Scholar]
- 12.Papaccio G, Graziano A, d'Aquino R, et al. Long-term cryopreservation of dental pulp stem cells (SBP-DPSCs) and their differentiated osteoblasts: a cell source for tissue repair. J Cell Physiol. 2006;208:319–325. doi: 10.1002/jcp.20667. [DOI] [PubMed] [Google Scholar]
- 13.Laino G, Carinci F, Graziano A, et al. In vitro bone production using stem cells derived from human dental pulp. J Craniofac Surg. 2006;17:511–515. doi: 10.1097/00001665-200605000-00021. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Liu W, Ling J, et al. Odontogenic differentiation of vascular endothelial growth factor-transfected human dental pulp stem cells in vitro. Mol Med Rep. 2014;10:1899–1906. doi: 10.3892/mmr.2014.2481. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Yan M, Wang Z, et al. Dental pulp stem cells from traumatically exposed pulps exhibited an enhanced osteogenic potential and weakened odontogenic capacity. Arch Oral Biol. 2013;58:1709–1717. doi: 10.1016/j.archoralbio.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Teti G, Salvatore V, Focaroli S, et al. In vitro osteogenic and odontogenic differentiation of human dental pulp stem cells seeded on carboxymethyl cellulose-hydroxyapatite hybrid hydrogel. Front Physiol. 2015;6:297. doi: 10.3389/fphys.2015.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atalayin C, Tezel H, Dagci T, et al. Medium modification with bone morphogenetic protein 2 addition for odontogenic differentiation. Braz Oral Res. 2016;30 doi: 10.1590/1807-3107BOR-2016.vol30.0020. [DOI] [PubMed] [Google Scholar]
- 18.Ye L, Fan Z, Yu B, et al. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell. 2012;11:50–61. doi: 10.1016/j.stem.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu J, Yu B, Hong C, et al. KDM6B epigenetically regulates odontogenic differentiation of dental mesenchymal stem cells. Int J Oral Sci. 2013;5:200–205. doi: 10.1038/ijos.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pavel OR, Popescu M, Novac L, et al. Postmenopausal osteoporosis - clinical, biological and histopathological aspects. Rom J Morphol Embryol. 2016;57:121–130. [PubMed] [Google Scholar]
- 21.Kim JJ, Khalid O, Vo S, et al. A novel regulatory factor recruits the nucleosome remodeling complex to wingless integrated (Wnt) signaling gene promoters in mouse embryonic stem cells. J Biol Chem. 2012;287:41103–41117. doi: 10.1074/jbc.M112.416545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miura M, Chen XD, Allen MR, et al. A crucial role of caspase-3 in osteogenic differentiation of bone marrow stromal stem cells. J Clin Invest. 2004;114:1704–1713. doi: 10.1172/JCI20427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khalid O, Kim JJ, Duan L, et al. Genome-wide transcriptomic alterations induced by ethanol treatment in human dental pulp stem cells (DPSCs) Genom Data. 2014;2:127–131. doi: 10.1016/j.gdata.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benoit DS, Durney AR, Anseth KS. Manipulations in hydrogel degradation behavior enhance osteoblast function and mineralized tissue formation. Tissue Eng. 2006;12:1663–1673. doi: 10.1089/ten.2006.12.1663. [DOI] [PubMed] [Google Scholar]
- 25.Chen B, Lin H, Zhao Y, et al. Activation of demineralized bone matrix by genetically engineered human bone morphogenetic protein-2 with a collagen binding domain derived from von Willebrand factor propolypeptide. J Biomed Mater Res A. 2007;80:428–434. doi: 10.1002/jbm.a.30900. [DOI] [PubMed] [Google Scholar]
- 26.George J, Kuboki Y, Miyata T. Differentiation of mesenchymal stem cells into osteoblasts on honeycomb collagen scaffolds. Biotechnol Bioeng. 2006;95:404–411. doi: 10.1002/bit.20939. [DOI] [PubMed] [Google Scholar]
- 27.Roostaeian J, Carlsen B, Simhaee D, et al. Characterization of growth and osteogenic differentiation of rabbit bone marrow stromal cells. J Surg Res. 2006;133:76–83. doi: 10.1016/j.jss.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Yang M, Lin L, et al. Runx2 overexpression enhances osteoblastic differentiation and mineralization in adipose--derived stem cells in vitro and in vivo. Calcif Tissue Int. 2006;79:169–178. doi: 10.1007/s00223-006-0083-6. [DOI] [PubMed] [Google Scholar]
- 29.Tezal M, Grossi SG, Ho AW, et al. The effect of alcohol consumption on periodontal disease. J Periodontol. 2001;72:183–189. doi: 10.1902/jop.2001.72.2.183. [DOI] [PubMed] [Google Scholar]
- 30.Amaral Cda S, Luiz RR, Leao AT. The relationship between alcohol dependence and periodontal disease. J Periodontol. 2008;79:993–998. doi: 10.1902/jop.2008.070525. [DOI] [PubMed] [Google Scholar]
- 31.Yusko DA, Buckman JF, White HR, et al. Risk for excessive alcohol use and drinkingrelated problems in college student athletes. Addict Behav. 2008;33:1546–1556. doi: 10.1016/j.addbeh.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park JB, Han K, Park YG, et al. Association between alcohol consumption and periodontal disease: the 2008 to 2010 Korea National Health and Nutrition Examination Survey. J Periodontol. 2014;85:1521–1528. doi: 10.1902/jop.2014.130782. [DOI] [PubMed] [Google Scholar]
- 33.Pandey SC, Ugale R, Zhang H, et al. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008;28:3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oberlander TF, Weinberg J, Papsdorf M, et al. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- 35.Shukla SD, Velazquez J, French SW, et al. Emerging role of epigenetics in the actions of alcohol. Alcohol Clin Exp Res. 2008;32:1525–1534. doi: 10.1111/j.1530-0277.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- 36.Ouko LA, Shantikumar K, Knezovich J, et al. Effect of alcohol consumption on CpG methylation in the differentially methylated regions of H19 and IG-DMR in male gametes: implications for fetal alcohol spectrum disorders. Alcohol Clin Exp Res. 2009;33:1615–1627. doi: 10.1111/j.1530-0277.2009.00993.x. [DOI] [PubMed] [Google Scholar]
- 37.Haycock PC. Fetal alcohol spectrum disorders: the epigenetic perspective. Biol Reprod. 2009;81:607–617. doi: 10.1095/biolreprod.108.074690. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Balaraman Y, Wang G, et al. Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics. 2009;4:500–511. doi: 10.4161/epi.4.7.9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moonat S, Starkman BG, Sakharkar A, et al. Neuroscience of alcoholism: molecular and cellular mechanisms. Cell Mol Life Sci. 2010;67:73–88. doi: 10.1007/s00018-009-0135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiang M, Denny A, Chen J, et al. The site specific demethylation in the 5'-regulatory area of NMDA receptor 2B subunit gene associated with CIE-induced up-regulation of transcription. PLoS One. 2010;5:e8798. doi: 10.1371/journal.pone.0008798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miranda RC, Pietrzykowski AZ, Tang Y, et al. MicroRNAs: master regulators of ethanol abuse and toxicity? Alcohol Clin Exp Res. 2010;34:575–587. doi: 10.1111/j.1530-0277.2009.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaminen-Ahola N, Ahola A, Maga M, et al. Maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model. PLoS Genet. 2010;6:e1000811. doi: 10.1371/journal.pgen.1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou FC, Balaraman Y, Teng M, et al. Alcohol alters DNA methylation patterns and inhibits neural stem cell differentiation. Alcohol Clin Exp Res. 2011;35:735–746. doi: 10.1111/j.1530-0277.2010.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hicks SD, Middleton FA, Miller MW. Ethanol-induced methylation of cell cycle genes in neural stem cells. J Neurochem. 2010;114:1767–1780. doi: 10.1111/j.1471-4159.2010.06886.x. [DOI] [PubMed] [Google Scholar]
- 45.Kim MJ, Shim MS, Kim MK, et al. Effect of chronic alcohol ingestion on bone mineral density in males without liver cirrhosis. Korean J Intern Med. 2003;18:174–180. doi: 10.3904/kjim.2003.18.3.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Callaci JJ, Himes R, Lauing K, et al. Binge alcohol-induced bone damage is accompanied by differential expression of bone remodeling-related genes in rat vertebral bone. Calcif Tissue Int. 2009;84:474–484. doi: 10.1007/s00223-009-9240-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ilich JZ, Brownbill RA, Tamborini L, et al. To drink or not to drink: how are alcohol, caffeine and past smoking related to bone mineral density in elderly women? J Am Coll Nutr. 2002;21:536–544. doi: 10.1080/07315724.2002.10719252. [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto A, Sekino A, Tajima M, et al. Effect of long-term alcohol administration on bone metabolism in rats. J Nutr Sci Vitaminol (Tokyo) 1997;43:369–375. doi: 10.3177/jnsv.43.369. [DOI] [PubMed] [Google Scholar]
- 49.Johnson TL, Gaddini G, Branscum AJ, et al. Effects of chronic heavy alcohol consumption and endurance exercise on cancellous and cortical bone microarchitecture in adult male rats. Alcohol Clin Exp Res. 2014;38:1365–1372. doi: 10.1111/acer.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turner RT. Skeletal response to alcohol. Alcohol Clin Exp Res. 2000;24:1693–1701. [PubMed] [Google Scholar]
- 51.Iwaniec UT, Turner RT. Intraperitoneal injection of ethanol results in drastic changes in bone metabolism not observed when ethanol is administered by oral gavage. Alcohol Clin Exp Res. 2013;37:1271–1277. doi: 10.1111/acer.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sen GL, Webster DE, Barragan DI, et al. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang D, Okamura H, Nakashima Y, et al. Histone demethylase Jmjd3 regulates osteoblast differentiation via transcription factors Runx2 and osterix. J Biol Chem. 2013;288:33530–33541. doi: 10.1074/jbc.M113.497040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wijayatunge R, Chen LF, Cha YM, et al. The histone lysine demethylase Kdm6b is required for activity-dependent preconditioning of hippocampal neuronal survival. Mol Cell Neurosci. 2014;61:187–200. doi: 10.1016/j.mcn.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CytoEdge analysis of the blue (genes upregulated with 20 mM EtOH treatment) and the black module (genes downregulated with 20 mM EtOH treatment) from Figure 2 visually present the interaction network of genes in the given module closely associated with EtOH treatment.
Effect of EtOH treatment on the growth of DPSCs was examined. Cells (5,000 cells per well) were seeded on 24 well plates in triplicates and treated with EtOH (0, 20, 50 and 100 mM) intermittently by one-day exposure and one-day withdrawal up to six days. Live cell number was determined by MTT assay with measurement at 570 nm. The error bar shows standard error margin (SEM) from triplicates (p value <0.05). It was noticed that 100 mM EtOH treatment induced a reduction in cell number, but other concentrations didn’t have significant effect on the cell growth.