Summary
The growth hormone/insulin‐like growth factor (IGF) axis can be manipulated in animal models to promote longevity, and IGF‐related proteins including IGF‐I and IGF‐binding protein‐3 (IGFBP‐3) have also been implicated in risk of human diseases including cardiovascular diseases, diabetes, and cancer. Through genomewide association study of up to 30 884 adults of European ancestry from 21 studies, we confirmed and extended the list of previously identified loci associated with circulating IGF‐I and IGFBP‐3 concentrations (IGF1, IGFBP3,GCKR,TNS3, GHSR, FOXO3, ASXL2, NUBP2/IGFALS, SORCS2, and CELSR2). Significant sex interactions, which were characterized by different genotype–phenotype associations between men and women, were found only for associations of IGFBP‐3 concentrations with SNPs at the loci IGFBP3 and SORCS2. Analyses of SNPs, gene expression, and protein levels suggested that interplay between IGFBP3 and genes within the NUBP2 locus (IGFALS and HAGH) may affect circulating IGF‐I and IGFBP‐3 concentrations. The IGF‐I‐decreasing allele of SNP rs934073, which is an eQTL of ASXL2, was associated with lower adiposity and higher likelihood of survival beyond 90 years. The known longevity‐associated variant rs2153960 (FOXO3) was observed to be a genomewide significant SNP for IGF‐I concentrations. Bioinformatics analysis suggested enrichment of putative regulatory elements among these IGF‐I‐ and IGFBP‐3‐associated loci, particularly of rs646776 at CELSR2. In conclusion, this study identified several loci associated with circulating IGF‐I and IGFBP‐3 concentrations and provides clues to the potential role of the IGF axis in mediating effects of known (FOXO3) and novel (ASXL2) longevity‐associated loci.
Keywords: aging, genomewide association study, growth hormone axis, IGF‐I, IGFBP‐3, longevity
Introduction
The insulin‐like growth factor (IGF) axis is an evolutionarily conserved system that plays important biologic roles in embryonic development, growth, and adulthood (Le Roith, 1997). IGF‐I mediates most of the activity of growth hormone (GH). The GH/IGF system consists of two ligands (IGF‐I and IGF‐II), six IGF‐binding proteins (IGFBP‐1‐6), and three IGF receptor subtypes (IGF‐I receptor, IGF‐II receptor, and insulin receptor) (Jones & Clemmons, 1995). IGF‐I promotes mitosis and cell cycle progression and is involved in human postnatal growth and development. Circulating IGF‐I is mainly bound to IGF‐binding proteins (IGFBPs) (Fowlkes, 1997), which affect activity (Lee et al., 1997) and half‐life of IGF‐I (Guler et al., 1989). From the clinical point of view, the measurement of IGF‐I and IGFBP‐3 blood concentrations is an important tool in establishing the diagnosis as well as monitoring treatment of GH‐related diseases (Ho & Participants, 2007; Cohen et al., 2008; Melmed et al., 2009).
Circulating concentrations of IGF‐I and IGFBP‐3 have been associated with risk of type 2 diabetes, cardiovascular diseases, cancer, and mortality in epidemiological studies (Juul et al., 2002; Vasan et al., 2003; Renehan et al., 2004; Kaplan et al., 2007; Friedrich et al., 2009; Burgers et al., 2011; Rajpathak et al., 2012). In animal models, diminished IGF‐I/insulin signaling has been associated with extended lifespans (Ziv & Hu, 2011), although the role of the IGF axis in human longevity remains inconclusive. Human genetic studies have suggested an association between polymorphisms in IGF‐I signaling pathway genes and longevity (Willcox et al., 2008; Ziv & Hu, 2011; Di Bona et al., 2014). Heritability studies have provided evidence for a substantial genetic contribution to circulating concentrations of IGF‐I (with heritability estimates ~40–60%) and IGFBP‐3 (80%) (Harrela et al., 1996; Hong et al., 1996; Souren et al., 2007). Identifying genetic determinants of circulating IGF‐I and IGFBP‐3 could lead to a better understanding of the biological basis of these factors in relation to human health and might identify pathways or susceptibility biomarkers that may assist with the development of interventions that target IGF‐I, its receptors, and binding proteins.
Our prior genomewide association study (GWAS) (N = 10 280) identified four loci with genomewide significant (P < 5 × 10−8) associations with circulating IGF‐I and IGFBP‐3 concentrations, including SNPs in or near IGFBP3, TNS3, SORCS2, and NUBP2/IGFALS, as well as three additional loci with suggestive associations (P < 1 × 10−6) in or near RPA3, SPOCK2, and FOXO3 (Kaplan et al., 2011). Some of these genes are involved in well‐described IGF regulatory or signaling pathways (such as IGFBP3 and IGFALS) (Deal et al., 2001; Gu et al., 2010; Schumacher et al., 2010) and are believed to influence traits that are also associated with concentrations or bioactivity of IGFs (e.g., FOXO3 locus associated with longevity (Willcox et al., 2008) and IGFBP3 locus associated with hip osteoarthritis (Evans et al., 2015)). To identify additional genetic variants with smaller effect sizes and enable sex specific analyses, we expanded our GWAS meta‐analysis to include up to a total of 30 884 individuals of European ancestry from 21 studies with measured circulating concentrations of IGF‐I and IGFBP‐3. In addition, using published GWAS data, we also performed lookups of associations of identified IGF‐I and IGFBP‐3 loci with survival beyond 90 years and other age‐related clinical traits.
Results
Characteristics of study samples
An overview of the study samples and data collection methods can be found in Tables S1 and S2 (Supporting information). Analyses of IGF‐I included up to 30 884 individuals (14 424 men and 16 460 women) from 21 studies and analyses of IGFBP‐3 included up to 18 995 individuals (8053 men and 10 942 women) from 13 studies.
Loci associated with circulating IGF‐I and IGFBP‐3 concentrations
An overview of the GWAS meta‐analysis results is given by the Manhattan plots in Fig. 1 and in Fig. S1 (Supporting information). There was no indication of inflated test statistics (i.e., due to unaccounted population stratification) as seen by the quantile–quantile (QQ) plots, and genomic control lambda ranged from 1.02 to 1.08 for the meta‐analysis results (Fig. S2, Supporting information) and from 0.98 to 1.08 (median 1.01) for the individual GWAS results. All lead SNPs (independent SNPs with the smallest P‐value within a locus, see Methods) had a good imputation quality across the studies (median imputation quality >0.9).
Figure 1.
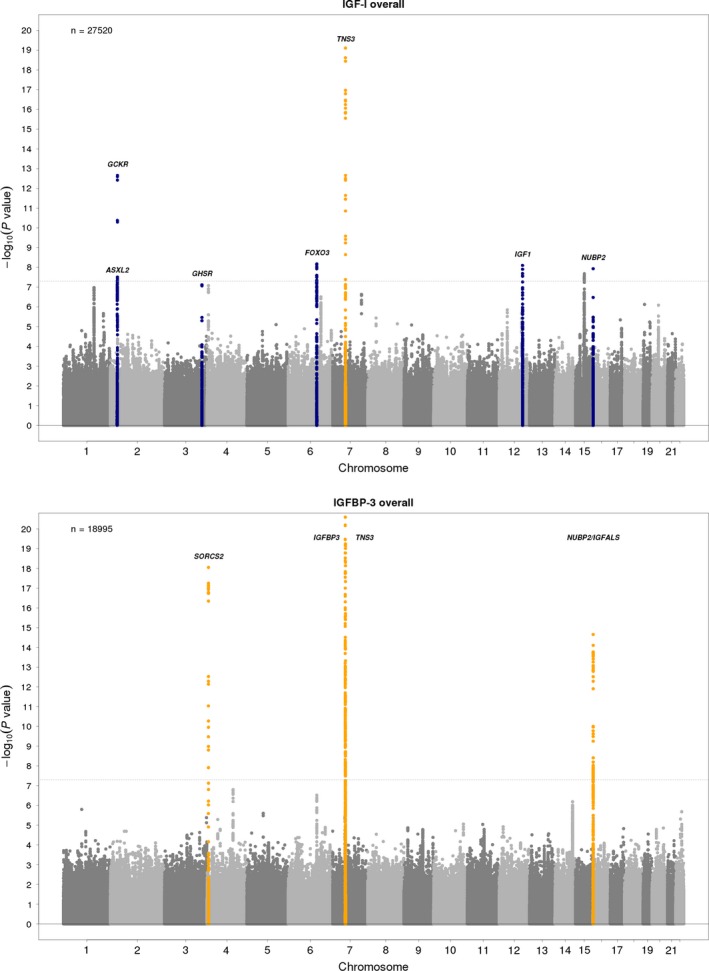
Manhattan plots of the combined stage 1 and 2 meta‐analysis results of IGF‐I and IGFBP‐3 traits in the men and women combined sample. SNPs are plotted on the x‐axis according to their position on each chromosome with the ‐log10 association P‐value on the y‐axis. The upper solid horizontal line indicates the threshold for genomewide significance. Known hits are colored in orange and new findings in blue. Plots are truncated on the y‐axis to 20.
After the final stage, which combines results of stages 1 and 2 plus de novo genotyping in stage 3, we found seven genomewide significant loci (P < 5.0 × 10−8) associated with circulating IGF‐I concentration (Table 1). In addition to the known locus near TNS3, we identified new loci in or near GCKR, IGF1, FOXO3, ASXL2, NUBP2, and GHSR associated with IGF‐I concentrations.
Table 1.
Loci associated with IGF‐I and IGFBP‐3 concentrations in men and women combined samples at genomewide significance (P < 5 × 10−8) after final stage
| Trait | SNP | A1 | A2 | F1 | P | I² | Chr | Position | Nearest gene | Gene distance | Direction effect | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IGF‐I | IGFBP‐3 | |||||||||||
| IGF‐Ia | rs700753 | C | G | 0.35 | 1.60E‐23 | 4.2 | 7 | 46,720,209 | TNS3 | 561067 | – | – |
| IGF‐I | rs780093 | T | C | 0.41 | 2.19E‐13 | 24.5 | 2 | 27,596,107 | GCKR | 0 | – | + |
| IGF‐I | rs978458 | T | C | 0.26 | 1.56E‐10 | 0.0 | 12 | 101,326,369 | IGF1 | 0 | + | – |
| IGF‐I | rs2153960 | A | G | 0.69 | 5.16E‐09 | 22.5 | 6 | 109,082,339 | FOXO3 | 0 | + | + |
| IGF‐I | rs934073 | C | G | 0.71 | 6.48E‐09 | 21.8 | 2 | 25,790,669 | ASXL2 | 25087 | – | – |
| IGF‐I | rs1065656 | C | G | 0.31 | 1.17E‐08 | 47.9 | 16 | 1,778,837 | NUBP2 | 0 | – | – |
| IGF‐I | rs509035 | A | G | 0.31 | 2.09E‐08 | 0.0 | 3 | 173,646,143 | GHSR | 0 | + | + |
| IGFBP‐3a | rs11977526 | A | G | 0.41 | 4.16E‐161 | 51.5 | 7 | 45,974,635 | IGFBP3 | 47239 | ||
| IGFBP‐3a | rs700753 | C | G | 0.35 | 1.11E‐46 | 26.7 | 7 | 46,720,209 | TNS3 | 561067 | – | + |
| IGFBP‐3a | rs1065656 | C | G | 0.31 | 8.55E‐23 | 24.1 | 16 | 1,778,837 | NUBP2 | 0 | – | – |
| IGFBP‐3a | rs4234798 | T | G | 0.39 | 8.86E‐19 | 0.0 | 4 | 7,270,834 | SORCS2 | 0 | – | – |
| Bivariate analysis | rs646776 | T | C | 0.78 | 6.87E‐9 | 26.1/43.1 | 1 | 109,620,053 | CELSR2 | 152 | – | – |
‘−’ = coding allele associated with lower IGF‐1 and IGFBP‐3 levels (indicated by bold italicized text were genomewide significant); ‘+’ = coding allele associated with higher IGF‐1 and IGFBP‐3 levels (indicated by bold italicized text were genomewide significant); Chr = chromosome; A1 = coding allele; A2 = other allele; F1 = frequency of coding allele.
Known association.
We found genomewide significant associations with IGFBP‐3 concentration for SNPs in or near IGFBP3, TNS3, NUBP2, and SORCS2, thus confirming all four previously known loci (Table 1). The SNPs at TNS3 and NUBP2 were genomewide significantly associated with both IGF‐I and IGFBP‐3 concentrations and had the same direction of effect for each circulating protein.
For six of ten genomewide significant SNPs, effects were in the same direction of association for IGF‐I concentrations and IGFBP‐3 concentrations (Table 1).
Detailed results of the significant associations after the final stage can be found in Table 1. Results of the individual analysis stages appear in Table S3 (Supporting information). Regional association plots are shown in Figs S3 and S4 (Supporting information).
Bivariate analysis of IGF‐I and IGFBP‐3
We performed a bivariate analysis of IGF‐I and IGFBP‐3. By leveraging shared variance between the two outcomes, this analysis can have improved power to identify SNPs associated with both IGF‐I and IGFBP‐3 concentrations, especially in the case of SNPs that have opposite effects on positively correlated traits (Aschard et al., 2014). The bivariate analysis identified a new locus at CELSR2 (Table 1), which had nominal association with IGFBP‐3 (P = 2.08 × 10−05) and IGF‐1 (P = 0.0096) in the univariate analysis. SNP rs646776 at CELSR2 had opposite effects on the two traits, being negatively associated with IGF‐1 and positively associated with IGFBP‐3 (Table 1, Table S4 and Fig. S5, Supporting information). In addition, SNPs at IGFBP3, TNS3, NUBP2, SORCS2, GCKR, IGF1, and FOXO3, identified in the univariate analysis, also showed genomewide significant associations in the bivariate analysis.
Interaction by sex
The sex‐stratified analyses revealed no additional discoveries that were not detected in the overall population. Although the direction of effect was similar for IGFBP3 and SORCS2 SNPs within sex subgroups, these two SNPs were found to have significantly different association effect sizes between men and women for IGFBP‐3, consistent with stronger associations in women. These findings of sex interaction maintained statistical significance after Bonferroni correction for the 12 tested genomewide significant lead SNPs (P < 0.004) (Table S5, Supporting information).
Gene‐based analysis (vegas)
Gene‐based analyses showed several significant IGF‐I‐associated genes within or close to the GCKR GWAS locus: EIF2B4, FNDC4, GCKR, IFT172, PPM1G, SNX17, ZNF513, GTF3C2, KRTCAP3, MPV17, and NRBP1 (associated with IGF‐I). New gene‐based associations that were not covered by a single SNP GWAS association were found for C6orf173 (chromosome 6) on IGF‐I concentration. The following genes of the NUBP2 GWAS locus were associated with circulating IGFBP‐3 concentration: EME2, IGFALS, MAPK8IP3, MRPS34, NME3, NUBP2, HS3ST6, RPL3L, SEPX1, and SPSB. Two genes, IGFBP1 and IGFBP3, at the IGFBP3 locus were associated IGFBP‐3 concentration.
Lookup for expression quantitative trait loci associations
A lookup of the lead SNPs for cis expression quantitative trait loci (eQTL) was performed in the publicly available database of whole blood eQTL associations (Westra et al., 2013). For the following SNPs, one or more cis eQTL associations were found: rs1065656 (NUBP2), rs11977526 (IGFBP3), rs2153960 (FOXO3), rs509035 (GHSR), rs780093 (GCKR), rs934073 (ASXL2), and rs978458 (IGF1) (Table 2).
Table 2.
Results of significant whole blood eQTL associations of the genomewide significant lead SNPs
| SNP | GWAS locus | eQTL p‐value | Chr | Probe center position | Probe name | SNP alleles | Effect allele | Effect direction | EQTL gene |
|---|---|---|---|---|---|---|---|---|---|
| rs1065656 | NUBP2 | 3.66E‐04 | 16 | 1,829,861 | 1710332 | C/G | C | + | FAHD1 |
| rs1065656 | NUBP2 | 4.28E‐12 | 16 | 1,799,259 | 4900333 | C/G | C | − | HAGH |
| rs1065656 | NUBP2 | 3.78E‐04 | 16 | 1,809,162 | 1780356 | C/G | C | − | HAGH |
| rs1065656 | NUBP2 | 9.35E‐04 | 16 | 1,760,172 | 5270575 | C/G | C | + | MAPK8IP3 |
| rs1065656 | NUBP2 | 3.34E‐05 | 16 | 1,762,997 | 6110307 | C/G | C | + | MRPS34 |
| rs1065656 | NUBP2 | 7.75E‐34 | 16 | 1,760,510 | 6450424 | C/G | C | + | NME3 |
| rs1065656 | NUBP2 | 1.09E‐14 | 16 | 1,778,738 | 6960730 | C/G | C | − | NUBP2 |
| rs1065656 | NUBP2 | 9.87E‐05 | 16 | 1,766,816 | 2850433 | C/G | C | − | SPSB3 |
| rs2153960 | FOXO3 | 2.95E‐06 | 6 | 109,129,272 | 7200189 | G/A | G | − | HS.133419 |
| rs509035 | GHSR | 5.97E‐05 | 3 | 173,706,575 | 870202 | G/A | A | + | TNFSF10 |
| rs780093 | GCKR | 2.46E‐04 | 2 | 27,440,911 | 5960546 | T/C | T | − | EIF2B4 |
| rs780093 | GCKR | 5.70E‐04 | 2 | 27,440,904 | 6370494 | T/C | T | − | EIF2B4 |
| rs780093 | GCKR | 2.69E‐05 | 2 | 27,518,384 | 430239 | T/C | T | + | NRBP1 |
| rs780093 | GCKR | 1.00E‐10 | 2 | 27,453,289 | 3360468 | T/C | T | + | SNX17 |
| rs934073 | ASXL2 | 5.96E‐04 | 2 | 25,816,038 | 650075 | G/C | G | + | ASXL2 |
| rs978458 | IGF1 | 1.71E‐03 | 12 | 101,115,257 | 990136 | T/C | T | + | C12ORF48 |
| rs11977526 | IGFBP3 | 1.84E‐05 | 7 | 45,918,692 | 6840372 | G/A | A | − | IGFBP3 |
Chr, chromosome; eQTL, expression quantitative trait loci; GWSD, genomewide association study.
mRNA of probe and gene names marked in bold showed also significant association with circulating IGFBP‐3 levels (P < 3.5 × 10−4).
Additionally, using a similar strategy we performed lookup in the MuTHER consortium (Grundberg et al., 2012) for cis eQTL associations found in fat cell, skin cell, and lymphoblastic cell lines (LCL). After Bonferroni correction for 297 lookups of three different traits (P < 5.6 × 10−5), rs1065656 (NUBP2) showed significant associations, specifically with FAHD1 (all tissues) and HAGH (fat cells and LCL) (Table S6, Supporting information). Furthermore, both genes were also significant in whole blood cis eQTL for the SNP rs1065656 (NUBP2) (Table 2).
Associations of gene expression with circulating IGF‐I and IGFBP‐3 concentrations
We next sought to link associations between SNPs and circulating IGF‐I and IGFBP‐3 concentrations, with cis eQTL associations of the same SNPs. In the 986 samples of the SHIP‐TREND cohort, we examined associations between whole blood mRNA expression levels of the genes located in a 500‐kb vicinity of our significant lead SNPs and circulating IGF‐I and IGFBP‐3 concentrations. Significance of the 323 array probe trait associations was defined by a false discovery rate (FDR) <0.05. Only mRNA levels of genes in vicinity of the NUBP2 GWAS locus were significantly associated with IGF‐I concentration (gene SEPX1) or IGFBP‐3 concentration (genes HAGH and RPS2). Of note, HAGH was the gene on which the corresponding lead SNP (rs1065656) had also a significant cis eQTL. The complete gene expression association results are listed in Table S7 (Supporting information).
Association with plasma protein levels
In 197 samples of the SHIP‐TREND cohort, peptides of the following proteins that were encoded by genes in a 500‐kb vicinity of the lead SNPs were examined for protein quantitative trait analyses (pQTL): insulin‐like growth factor‐binding protein complex acid labile subunit (ALS encoded by IGFALS at the NUBP2 locus), 28S ribosomal protein S34, mitochondrial (RT34 encoded by MRPS34 at the NUBP2 locus), insulin‐like growth factor‐binding protein 3 (IBP3 encoded by IGFBP3 at the IGFBP3 locus), and coiled‐coil domain‐containing protein 121 (CC121 encoded by CCDC121 at the GCKR locus). Of the 32 tested SNP peptide pairs, peptides of the ALS protein had significant pQTL at FDR <0.05 (Table S8, Supporting information). Furthermore, strong associations were found in the same samples for the pQTL‐associated peptides of ALS and IBP3 with circulating levels of IGF‐I and IGFBP‐3 (P < 1.0 × 10−6).
Allelic heterogeneity of the NUBP2 locus
To further examine the NUBP2 locus, we performed a conditional analysis of this locus based on the meta‐analysis results adjusting for the lead SNP rs1065656. The analyses revealed an independent genomewide significant association of a second SNP rs11644716 with IGFBP‐3 (P = 6.3 × 10−14) and an opposite effect direction of the minor allele C (MAF = 0.05) compared with the lead SNP rs1065656 (MAF = 0.31) (r² = 0.03 between these two SNPs based on the HapMap R22 reference data). Although not genomewide significant, rs11644716 was also associated with circulating IGF‐I concentration (P = 1.8 × 10−6) and has an eQTL for HAGH (probe 4900333: P = 3.2 × 10−5; probe 1780356: P = 0.003) and a pQTL with ALS (eight peptides with P‐value <0.01). In all cases, the effect directions based on the minor allele of rs11644716 were opposite of that for the minor allele of rs1065656.
Figure 2 summarizes the relationship between the lead SNPs at the NUBP2 locus and gene expression levels, protein levels, and circulating IGF‐I and IGFBP‐3 concentrations.
Figure 2.
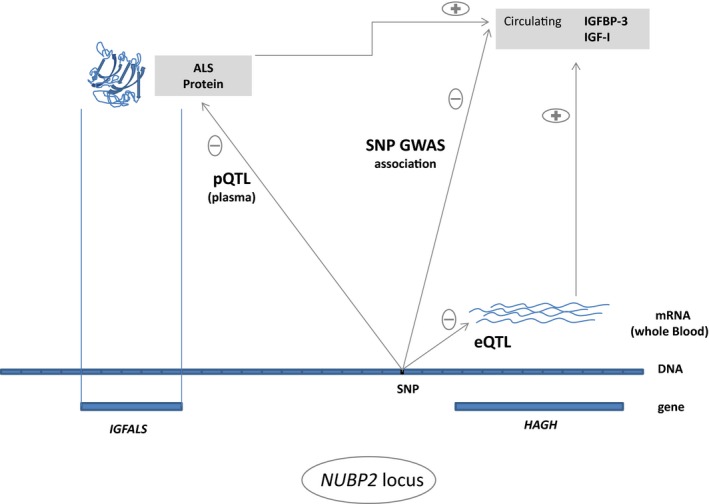
Overview of the NUBP2 locus that shows the relations of gene expression levels, protein levels and circulating IGF‐I and IGFBP‐3 with respect to the SNP. Effect directions (±) are given for the minor C‐allele of SNP rs1065656 (MAF 31%). Note: the second signal rs11644716 (MAF 5%) has opposite expression quantitative trait loci (eQTL) and protein quantitative trait analyses (pQTL) effect directions to rs1065656, but they are consistent.
Associations with serum metabolites
Lead SNPs associated with IGF‐I and IGFBP‐3 concentrations were examined in a published metabolite‐SNP association database (Suhre et al., 2011; Shin et al., 2014). The IGF‐I‐associated SNP rs780093 at GCKR locus was associated with glucose/mannose ratio (P = 9.4 × 10−143), and the IGFBP‐3‐associated SNP rs4234798 at SORCS2 locus was associated with caprylate (8:0)/phenylalanine ratio (P = 7.3 × 10−7).
Associations of top loci with age‐related traits
We also examined the associations of the IGF‐I‐ and IGFBP‐3‐associated SNPs with anthropometric traits (height, BMI, waist‐to‐hip ratio, and fat percentage) (Heid et al., 2010; Lango Allen et al., 2010; Speliotes et al., 2010), bone mineral density (Estrada et al., 2012), risk of type 2 diabetes (Voight et al., 2010; Morris et al., 2012) and related traits (fasting glucose, 2‐h glucose, HbA1c, fasting insulin, proinsulin, HOMA‐IR, and HOMA‐B) (Dupuis et al., 2010; Saxena et al., 2010; Soranzo et al., 2010), and coronary artery disease (Coronary Artery Disease Genetics C, 2011; Schunkert et al., 2011; Consortium CAD and Deloukas, 2013) (Table S9, Supporting information). Many nominal associations were expected because of the known influence of the IGF system on these traits. Of note is the finding that for rs780093 in the GCKR locus, the allele associated with higher IGF‐I concentration was already known to be associated with elevated risk of type 2 diabetes (P = 3.7 × 10−6), as well as higher levels of fasting glucose, fasting insulin, and HOMA‐IR (all P < 2.0 × 10−4), lower 2‐h glucose levels (P = 1.7 × 10−6), increased height (P = 2.0 × 10−4), lower waist‐to‐hip ratio (P = 0.0003), and higher lumbar spine bone mineral density (P = 0.002).
Three additional loci (GHSR, CELSR2, and FOXO3) showed strong associations with height (all P < 1.0 × 10−4). The IGFBP‐3‐increasing allele of SNP rs646776 (CELSR2 locus) was associated with increased risk of coronary artery disease (P = 9.4 × 10−15). The IGF‐I‐decreasing allele of SNP rs934073 at ASXL2 showed a nominal association with survival beyond 90 years (P = 0.018) as well as higher levels of BMI (P = 0.008) and fat percentage (P = 9.4 × 10−5) and lower lumbar spine bone mineral density (P = 0.004).
We further performed lookups of top IGF‐I‐ and IGFBP‐3‐associated SNPs (P < 10−6 in the meta‐analysis of stage 1 and stage 2) for associations with survival beyond 90 years using published GWAS data (Broer et al., 2015) (Tables S10 and S11, Supporting information). Among 15 independent circulating IGF‐I‐associated SNPs defined based on linkage disequilibrium (LD) (settings r 2 > 0.01, 1 Mb distance), the SNP rs10457180 (r 2 = 0.96 with the lead SNP rs2153960) at FOXO3 (P = 8.6 × 10 −5 ) and SNP rs11892454 (r 2 = 0.71 with the lead SNP rs934073) at ASXL2 (P = 0.003) reached statistical significance after Bonferroni correction for 15 independent tests. Among 13 independent circulating IGFBP‐3‐associated SNPs, the SNP rs9398172 (r 2 = 1 with the lead SNP rs2153960) at FOXO3 (P = 2.5 × 10−4) remained significantly associated with survival beyond 90 years after Bonferroni correction.
Enrichment of putative regulatory elements among loci associated with circulating IGF‐1 and IGFBP‐3 concentrations
We examined whether the identified SNPs fall within regulatory elements in the epigenetic encode and roadmap data for associated SNPs using Haploreg (http://www.broadinstitute.org/mammals/haploreg/haploreg.php) (Ward & Kellis, 2012) and RegulomeDB (http://regulomedb.org/) (Boyle et al., 2012) (Table S11, Supporting information). Lower scores indicate stronger evidence for the presence of a regulatory element. To determine whether these IGF‐I‐ and IGFBP‐3‐associated loci are enriched for regions likely to affect gene expression, we further examined the distribution of scores among these SNPs compared with all RegulomeDB SNPs. We found that these identified SNPs are highly enriched for low Regulome scores (P < 2.2 × 10−16 by multinomial method, Fig. 3A). The genomic and representative epigenetic context surrounding rs646776 (CELSR2 locus), a SNP with a Regulome score of 1f, is shown as an example (Fig. 3). SNP rs646776 localizes to a genomic region of high LD (Fig. 3B) and lies within peaks of histone marks associated with regulatory elements, and a DNase hypersensitivity region (Fig. 3C)(Kent et al., 2002). In addition, SNP rs646776 falls in ChIP identified binding regions for CTCF, POLR2A, REST, and TAF7 (data not shown).
Figure 3.
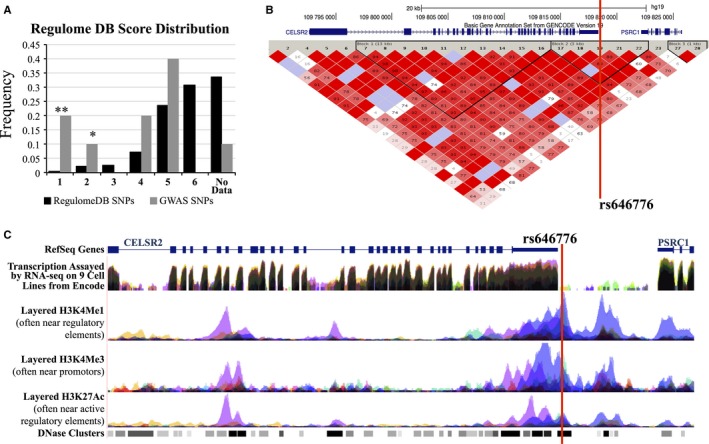
Analysis of regulatory element marks in loci associated with serum IGF‐1 and IGFBP‐3 concentrations. (A) IGF‐1‐ and IGFBP‐3‐associated SNPs are enriched for putative regulatory elements compared with all SNPs in RegulomeDB. **P < 0.005, *P < 0.05 (Monte Carlo). Overall distribution of genomewide association study (GWAS) SNPs vs. RegulomeDB SNPs P < 2.2E‐16, multinomial method. (B) Genomic context surrounding rs646776 showing R 2 values. (C) Representative regulatory motif tracks from USCS Genome Browser and encode showing histone mark peaks and DNase hypersensitivity at the location of rs646776.
Discussion
Our second GWAS report from the CHARGE IGF Working Group, here expanded to include more than >30 000 individuals, revealed several SNPs influencing circulating levels of IGF‐I and IGFBP‐3 which also have been associated with other metabolic and age‐related traits. These included several loci already implicated in the biology of GH/IGF‐I (IGF1, IGFBP3, IGFALS, GHSR, FOXO3) as well other novel findings including rs934073 SNP on chr 2, which is an eQTL for polycomb group gene ASXL2 associated with reduced circulating IGF‐I. Cross‐reference of IGF‐I‐ and IGFBP‐3‐associated SNPs against published GWAS of age‐related traits identified the ASXL2 SNP as a (to our knowledge) novel locus for longevity as defined as survival beyond 90 years. All genomewide significant associations with IGFBP‐3 and IGF‐I levels that were reported in our preceding study (Kaplan et al., 2011) could be confirmed here using a larger sample. Additionally, our preliminary finding of an association of circulating IGF‐I level with rs2153960 in the FOXO3 gene reached genomewide significance using this larger sample size. Moreover, bioinformatics analysis also suggests enrichment of putative regulatory elements among these IGF‐I‐ and IGFBP‐3‐associated loci, particularly of rs646776 at CELSR2.
Our study reveals clues about mechanisms of IGF system regulation through the interplay of IGFBP‐3 and genes within the NUBP2 locus, including IGFALS and HAGH. The IGFALS encodes the insulin‐like growth factor‐binding protein complex, acid labile subunit protein (ALS) which forms a ternary complex with IGF‐I and IGFBP‐3 (Firth et al., 1998; Twigg & Baxter, 1998). Like in most GWAS, our analyses cannot establish which is the causative SNP or gene of a locus. However, IGFALS seems to be a strong candidate supported by the associations of SNPs in the vicinity of IGFALS with both circulating IGFBP‐3 and plasma levels of the protein coded by IGFALS. Although the sample size for the plasma proteome analyses was restricted to 197 probands of SHIP‐TREND, the observed associations of the second signal rs11644716 achieved a moderately high level of statistical significance (P < 0.0015). IGFALS was not abundantly expressed in whole blood cells, and its transcript levels were neither associated with IGF‐I nor IGFBP‐3 concentration. Certainly, the protein ALS encoded by IGFALS is abundant in serum, but the gene is translated in liver. In contrast to IGFALS, HAGH (hydroxyacylglutathione hydrolase) was sufficiently expressed in whole blood cells and the amount of its mRNA was strongly correlated with circulating IGFBP‐3 serum concentration. Taking into account the eQTL association of the lead SNP rs1065656 with HAGH and the correlation with the mRNA and IGFBP‐3, this SNP might influence the IGFBP‐3 levels by modulating the amount of HAGH mRNA, whereas both the levels of mRNA and IGFBP‐3 are reduced per copy of the minor allele. Although this chain of associations was revealed in whole blood, it might be present in other tissues as well because significant eQTL for rs1065656 with HAGH were observed in other tissues studied in the MuTHER dataset (Grundberg et al., 2012). Given the more pronounced association with ALS and the less significant eQTL with HAGH of the second signal compared with the lead SNP, rs11644716 might reduce the level of circulating IGFBP‐3 indirectly by reducing the amount of ALS per minor allele. Of note, there was no nonsynonymous SNP in LD in the 1000 Genomes v3 dataset (R² > 0.8, SHIP cohort) for both SNPs which could have helped narrow down the functional mechanism.
Taken together with genotype–phenotype association data assembled by others, our study revealed that IGF‐I‐ and IGFBP‐3‐associated SNPs had expected associations with anthropometric and age‐related chronic disease traits (e.g., bone mineral density, disordered carbohydrate metabolism). We also found that SNPs associated with reduced IGF‐I levels tended to be associated with longer survival defined as death after 90 years (Broer et al., 2015). This is consistent with an observation from prior analysis of candidate genes associated with the insulin and IGF signaling axis (van Heemst et al., 2005). In addition, rs934073, an eQTL for additional sex comb‐like 2 (ASXL2), was a genomewide significant SNP associated with lower IGF‐I level which also has enriched frequency in adults older than 90 years of age (Broer et al., 2015). Other traits associated with the IGF‐I‐decreasing allele of rs934073 included greater adiposity and reduced lumbar spine (but not femoral neck) bone mineral density. ASXL2 is a polycomb group protein with known functions in development that has also been associated with pediatric cancer, but to our knowledge has not before been suggested as a longevity gene (Huether et al., 2014). The SNP in the known longevity gene FOXO3 has not only been previously associated with reduced fasting insulin and HOMA‐IR (Willcox et al., 2008), but here it was found to produce lower circulating IGF‐I levels. Multiple genetic determinants for circulating IGF‐I in normal and IGF1R resistance states might partially explain the U‐shaped association of circulating IGF‐I concentration with mortality (Suh et al., 2008; Burgers et al., 2011).
While our meta‐analysis encompasses a large number of samples from multiple cohorts, this may lead to limitations. Given the different origin of the cohorts (Table S1, Supporting information), heterogeneity in the association results might occur due to the different genetic background and the patterns of intake of nutrients across the individual studies.
In summary, this project extends our prior work (Kaplan et al., 2011) through the identification of several new loci related to circulating IGF‐I and IGFBP‐3 levels that also may affect aging. While the effects of insulin/IGF‐I signaling on survival often displays sex dimorphism in humans and other organisms, we found similar genetic determinants of IGF protein levels in men and women, even with relatively large sample size finding interaction by sex only for two loci (IGFBP3 and SORCS2) in association with circulating IGFBP‐3. Finally, a novel identified gene candidate for long‐term survival, ASXL2, requires further study. Taking into account the design of our study, most of the findings should be considered as important and well‐grounded hypotheses to work on.
Experimental procedures
Participating studies
In total, the CHARGE IGF Working Group included 21 and 13 studies that participated in the association analysis for IGF‐I (N = 27 520, 53% women) and IGFBP‐3 (N = 18 995, 58% women), respectively. Four of the cohorts (N = 10 280) were previously included in a GWAS meta‐analysis of IGF‐I and IGFBP‐3 levels (Kaplan et al., 2011). Imputed SNPs for chromosome X were available for 16 670 and 11 959 individuals with IGF‐I and IGFBP‐3 measurements, respectively. Additionally, up to 3364 individuals (55% women) with IGF‐I from one study were available for de novo genotyping of selected SNPs. Detailed information on participant characteristics, IGF‐I and IGFBP‐3 measurements, and genotyping of all studies participated in the different analyses and stages is given in Table S2 (Supporting information). All participants provided informed consent, and human subjects’ research review was obtained from each participating cohort.
Statistical analyses
GWAS in individual studies
Each study of the GWAS stages performed genotyping on genomewide arrays and imputed SNPs using the HapMap2 reference panel. Detailed information on genotyping and imputation is provided in Table S2 (Supporting information). Association analyses in individual studies were performed on IGF‐I and IGFBP‐3 levels measured in ng mL−1 using a multiple linear regression with an additive genetic model based on allele dosages adjusted for age and stratified by sex. All cohorts accounted for relatedness, population substructure using genetic principal components, study center, and laboratory batch of IGF measurement where applicable. Individuals of non‐European ancestry, with missing phenotypic data, diagnosed growth hormone deficiency, or known use of human growth hormones were excluded prior to the analyses.
Meta‐analysis
From each cohort's result file, monomorphic SNPs as well as SNPs with an imputation quality below 0.3 were excluded prior to the meta‐analysis. All study‐specific GWAS results were corrected by the genomic inflation factor λGC if >1. Due to the IGF‐I and IGFBP‐3 assay‐based differences in both effect sizes and variances of measurements across cohorts, a sample size‐weighted z‐score‐based meta‐analysis implemented in METAL (Willer et al., 2010) was conducted, and the meta‐analysis P‐values were corrected for genomic inflation. After meta‐analysis, SNPs with a MAF ≤1% were removed from subsequent analyses.
Our multistage design had two GWAS stages (stages 1 and 2) and an additional stage (stage 3) with de novo genotyping data (N = 3364 individuals) to confirm novel loci. After stage 1 GWAS, all 19 lead SNPs from all traits with a P < 10−6 were taken forward to stage 2. All IGF‐I lead SNPs of novel loci that had a combined stage 1 and stage 2 P < 10−8 (except GCKR) were selected for de novo replication in an additional cohort. An overview of the design and the significantly associated loci at each stage is provided in Fig. S6 (Supporting information). Details on SNP selection and quality control are given in the Appendix S1 (Supporting information). Regional association plots were generated using LocusZoom (Pruim et al., 2010).
Assessment of independent signals
To define a lead SNP of each locus, the association results of a GWAS stage with P‐values <1 × 10−5 were grouped based on the LD structure of the HapMap release 28 CEU dataset using PLINK (settings r 2 >0.01, 1 Mb distance) (Purcell et al., 2007). Due to the strong association of the IGFBP3 locus with IGFBP‐3, only one lead SNP was selected regardless of several grouped results.
The analysis of secondary signals in the NUBP2 locus was performed using the software gcta (Yang et al., 2011) and the genotypes of the SHIP cohort as a reference, and was verified by an analysis using the genotypes of the NHS/HPFS cohorts as a reference.
Sex interaction analysis
Sex interactions on IGF‐I and IGFBP‐3 levels were obtained by comparing, for each SNP, the stage 2 meta‐analysis z‐scores from men (z_men) (IGF‐I: N = 12 917, IGFBP‐3: N = 8052) and women (z_women) (IGF‐I: N = 14 602, IGFBP‐3: N = 10 942) using the formula z_interaction=(z_men‐z_women)/√2, assuming independent effect sizes between men and women, and matched to a common effect allele.
Bivariate meta‐analysis of IGF‐I and IGFBP‐3
The stage 2 meta‐analysis z‐scores of the combined samples IGF‐I and IGFBP‐3 were used to calculate a bivariate meta‐analysis implemented in the function multipheno.T2 of the r‐package gtx (version 0.0.8. http://CRAN.R-project.org/package=gtx). The function corresponds closely with Hotelling's T 2 test and calculates a multiphenotype association test for each marker based on the meta‐analysis result z‐statistics that is equivalent to using the subject‐specific data to perform a multivariate analysis of variance.
Gene‐based analysis
Genomewide gene‐based tests which account for both gene length and LD between SNPs were performed by vegas 0.8.27 (Versatile Gene‐Based Association Study) (Liu et al., 2010) using SNP P‐value results from the overall meta‐analyses. SNPs were allocated to one or more autosomal genes using gene boundaries ±50 kb. We performed 1 × 107 permutations and defined a gene‐based P‐value <1 × 10−6 as gene‐based genomewide significant.
Gene expression and eQTL analysis
For each of the lead SNPs of the significant loci after final stage, significant cis eQTL associations in whole blood, lymphocytes, subcutaneous fat, muscle, and skin were looked up in the publically available association result databases (Grundberg et al., 2012; Westra et al., 2013). Association analysis of whole blood gene expression data with serum IGF‐I and IGFBP‐3 levels was conducted in 986 samples of the SHIP‐TREND cohort (Schurmann et al., 2012).
Association with plasma protein levels
Plasma proteome data were obtained as described in Appendix S1 (Supporting information) using liquid chromatography–mass spectrometry (LC‐MS). mascot (in‐house mascot server v2.3.2; Matrix Science, London, GB) search algorithm was used to match the generated peak lists with a human fasta‐formatted database containing 20 268 unique sequence entries (reviewed human database, release of October 2011). Prior to data analyses, all peptide intensity values were log10‐transformed and median–median‐normalized. Association analyses between peptides and serum IGF‐I and IGFBP‐3 levels were performed by linear regression, adjusted for age, sex, and the MS processing batch. Associations of a SNP with the peptides were conducted by linear regression, adjusted for age, sex, and the first four principal components of a peptide‐level‐based principal component analysis. Protein intensities used for analyses were obtained by averaging the corresponding peptide intensities that passed the QC filter, and were put instead of the peptide intensities into the association model. All measured peptides that passed QC and that belonged to proteins which were encoded by genes located in a 500‐kb vicinity of our lead SNPs were selected for association analyses. The assignment of protein names (uniprot identifiers) to the corresponding genes was performed using the DAVID gene conversion tool (http://david.abcc.ncifcrf.gov/). Finally, after QC the following proteins measured in 197 SHIP‐TREND samples were available: ALS, CC121, IBP3, and RT34.
Lookups of top loci in association with IGF correlated traits
Top SNPs associated with levels of IGF‐I and IGFBP‐3 were examined in relationship to other phenotypes using published data on serum metabolites (Suhre et al., 2011; Shin et al., 2014), anthropometric traits (Heid et al., 2010; Lango Allen et al., 2010; Speliotes et al., 2010), bone mineral density (Estrada et al., 2012), diabetes (Voight et al., 2010; Morris et al., 2012) and glycemic traits (Dupuis et al., 2010; Saxena et al., 2010; Soranzo et al., 2010), coronary artery disease (Coronary Artery Disease Genetics C, 2011; Schunkert et al., 2011; Consortium CAD and Deloukas, 2013), and survival beyond 90 years (Broer et al., 2015). Detailed information of the published datasets used including its references is given in Table S9 (Supporting information).
Assessment of regulatory elements associated with identified loci
encode and roadmap data were assessed using haploreg (http://www.broadinstitute.org/mammals/haploreg/haploreg.php) and regulomedb (http://regulomedb.org/). Statistical analysis of individual Regulome scores was performed using Monte Carlo sampling of 10 SNPs (the size of our ‘observed data’ pool). RegulomeDB assigns scores to SNP loci based on the presence of histone marks, predicted and experimentally validated transcription factor binding, DNase hypersensitivity, and other evidence for regulatory function. Scores range from 1 to 7, with lower scores indicating stronger evidence for the presence of a regulatory element. For the purpose of this analysis, score subcategories (1a, 1b, etc.) were merged. A multinomial test was performed for a statistical comparison between the observed distribution and the background distribution. The LD plot in Fig. 3B was generated using haploview 4.2 with genetic data downloaded from version 3, Release 2, using the genomic region Chr1:109590000‐109630000, and analysis panel CEU + TSI. Histone mark and DNase tracks in Fig. 3C were downloaded from UCSC Genome Browser.
Conflict of interest
The authors do not have conflict of interests.
Supporting information
Appendix S1. Materials and methods.
Fig. S1 Manhattan plots of men and women strata.
Fig. S2 QQ plots of meta‐analysis results.
Fig. S3 Regional association plots for IGF‐I traits.
Fig. S4 Regional association plots for IGFBP‐3 traits.
Fig. S5 Results of bivariate analysis of IGF‐I and IGFBP‐3.
Fig. S6 Flow chart of the study's design.
Table S1 Characteristics of study cohorts.
Table S2 Assay and genotyping information of study cohorts.
Table S3 GWAS results with P‐value <10−6 in stage 1.
Table S4 Genome‐wide significant loci of bivariate analysis.
Table S5 Sex interaction results of the 12 genome‐wide significant SNPs.
Table S6 Results of eQTL lookup in the Muther dataset.
Table S7 Results of gene expression analysis of the genome‐wide significant loci with IGF‐I and IGFBP‐3 in the SHIP‐TREND cohort.
Table S8 Results of pQTL analysis in the SHIP‐TREND cohort.
Table S9 Lookup of genome‐wide significant lead SNPs in IGF‐I/IGFBP‐3 correlated traits.
Table S10 Lookup of Top IGF‐I associated SNPs in associations with survival beyond age 90 years old.
Table S11 Lookup of Top IGFBP‐3 associated SNPs in associations with survival beyond age 90 years old.
Table S12 Enrichment of putative regulatory elements among IGF‐1 and IGFBP‐3 associated loci.
Data S1 Design and funding of participating cohort studies.
Funding
Funding sources for the individual studies included in this work are listed in Supporting information.
References
- Aschard H, Vilhjalmsson BJ, Greliche N, Morange PE, Tregouet DA, Kraft P (2014) Maximizing the power of principal‐component analysis of correlated phenotypes in genome‐wide association studies. Am. J. Hum. Genet. 94, 662–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broer L, Buchman AS, Deelen J, Evans DS, Faul JD, Lunetta KL, Sebastiani P, Smith JA, Smith AV, Tanaka T, Yu L, Arnold AM, Aspelund T, Benjamin EJ, De Jager PL, Eirkisdottir G, Evans DA, Garcia ME, Hofman A, Kaplan RC, Kardia SL, Kiel DP, Oostra BA, Orwoll ES, Parimi N, Psaty BM, Rivadeneira F, Rotter JI, Seshadri S, Singleton A, Tiemeier H, Uitterlinden AG, Zhao W, Bandinelli S, Bennett DA, Ferrucci L, Gudnason V, Harris TB, Karasik D, Launer LJ, Perls TT, Slagboom PE, Tranah GJ, Weir DR, Newman AB, van Duijn CM, Murabito JM (2015) GWAS of Longevity in CHARGE Consortium Confirms APOE and FOXO3 Candidacy. J. Gerontol. A Biol. Sci. Med. Sci. 70, 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers AM, Biermasz NR, Schoones JW, Pereira AM, Renehan AG, Zwahlen M, Egger M, Dekkers OM (2011) Meta‐analysis and dose‐response metaregression: circulating insulin‐like growth factor I (IGF‐I) and mortality. J. Clin. Endocrinol. Metab. 96, 2912–2920. [DOI] [PubMed] [Google Scholar]
- Cohen P, Rogol AD, Deal CL, Saenger P, Reiter EO, Ross JL, Chernausek SD, Savage MO, Wit JM, Participants ISSCW (2008) Consensus statement on the diagnosis and treatment of children with idiopathic short stature: a summary of the Growth Hormone Research Society, the Lawson Wilkins Pediatric Endocrine Society, and the European Society for Paediatric Endocrinology Workshop. J. Clin. Endocrinol. Metab. 93, 4210–4217. [DOI] [PubMed] [Google Scholar]
- Consortium CAD , Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA, Stirrups K, Konig IR, Cazier JB, Johansson A, Hall AS, Lee JY, Willer CJ, Chambers JC, Esko T, Folkersen L, Goel A, Grundberg E, Havulinna AS, Ho WK, Hopewell JC, Eriksson N, Kleber ME, Kristiansson K, Lundmark P, Lyytikainen LP, Rafelt S, Shungin D, Strawbridge RJ, Thorleifsson G, Tikkanen E, Van Zuydam N, Voight BF, Waite LL, Zhang W, Ziegler A, Absher D, Altshuler D, Balmforth AJ, Barroso I, Braund PS, Burgdorf C, Claudi‐Boehm S, Cox D, Dimitriou M, Do R, Consortium D , Consortium C , Doney AS, El Mokhtari N, Eriksson P, Fischer K, Fontanillas P, Franco‐Cereceda A, Gigante B, Groop L, Gustafsson S, Hager J, Hallmans G, Han BG, Hunt SE, Kang HM, Illig T, Kessler T, Knowles JW, Kolovou G, Kuusisto J, Langenberg C, Langford C, Leander K, Lokki ML, Lundmark A, McCarthy MI, Meisinger C, Melander O, Mihailov E, Maouche S, Morris AD, Muller‐Nurasyid M, Mu TC, Nikus K, Peden JF, Rayner NW, Rasheed A, Rosinger S, Rubin D, Rumpf MP, Schafer A, Sivananthan M, Song C, Stewart AF, Tan ST, Thorgeirsson G, van der Schoot CE, Wagner PJ, Wellcome Trust Case Control C , Wells GA, Wild PS, Yang TP, Amouyel P, Arveiler D, Basart H, Boehnke M, Boerwinkle E, Brambilla P, Cambien F, Cupples AL, de Faire U, Dehghan A, Diemert P, Epstein SE, Evans A, Ferrario MM, Ferrieres J, Gauguier D, Go AS, Goodall AH, Gudnason V, Hazen SL, Holm H, Iribarren C, Jang Y, Kahonen M, Kee F, Kim HS, Klopp N, Koenig W, Kratzer W, Kuulasmaa K, Laakso M, Laaksonen R, Lee JY, Lind L, Ouwehand WH, Parish S, Park JE, Pedersen NL, Peters A, Quertermous T, Rader DJ, Salomaa V, Schadt E, Shah SH, Sinisalo J, Stark K, Stefansson K, Tregouet DA, Virtamo J, Wallentin L, Wareham N, Zimmermann ME, Nieminen MS, Hengstenberg C, Sandhu MS, Pastinen T, Syvanen AC, Hovingh GK, Dedoussis G, Franks PW, Lehtimaki T, Metspalu A, Zalloua PA, Siegbahn A, Schreiber S, Ripatti S, Blankenberg SS, Perola M, Clarke R, Boehm BO, O'Donnell C, Reilly MP, Marz W, Collins R, Kathiresan S, Hamsten A, Kooner JS, Thorsteinsdottir U, Danesh J, Palmer CN, Roberts R, Watkins H, Schunkert H, Samani NJ (2013) Large‐scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 45, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronary Artery Disease Genetics C (2011) A genome‐wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet. 43, 339–344. [DOI] [PubMed] [Google Scholar]
- Deal C, Ma J, Wilkin F, Paquette J, Rozen F, Ge B, Hudson T, Stampfer M, Pollak M (2001) Novel promoter polymorphism in insulin‐like growth factor‐binding protein‐3: correlation with serum levels and interaction with known regulators. J. Clin. Endocrinol. Metab. 86, 1274–1280. [DOI] [PubMed] [Google Scholar]
- Di Bona D, Accardi G, Virruso C, Candore G, Caruso C (2014) Association between genetic variations in the insulin/insulin‐like growth factor (Igf‐1) signaling pathway and longevity: a systematic review and meta‐analysis. Curr. Vasc. Pharmacol. 12, 674–681. [DOI] [PubMed] [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia‐Naji N, Gloyn AL, Lindgren CM, Magi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JR, Egan JM, Lajunen T, Grarup N, Sparso T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti‐Proenca C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O'Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, Ben‐Shlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch‐Johnsen K, Bottcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day IN, de Geus EJ, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer‐Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PR, Jorgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martinez‐Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orru M, Pakyz R, Palmer CN, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvanen AC, Tanaka T, Thorand B, Tichet J, Tonjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van Hoek M, Varma D, Visvikis‐Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC, Consortium D, Consortium G, Global BC, Borecki IB, Loos RJ, Meneton P, Magnusson PK, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano‐Rios M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WH, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP, Wichmann HE, Illig T, Rudan I, Wright AF, Stumvoll M, Campbell H, Wilson JF, Anders Hamsten on behalf of Procardis C , Investigators M , Bergman RN, Buchanan TA, Collins FS, Mohlke KL, Tuomilehto J, Valle TT, Altshuler D, Rotter JI, Siscovick DS, Penninx BW, Boomsma DI, Deloukas P, Spector TD, Frayling TM, Ferrucci L, Kong A, Thorsteinsdottir U, Stefansson K, van Duijn CM, Aulchenko YS, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Waterworth DM, Vollenweider P, Peltonen L, Mooser V, Abecasis GR, Wareham NJ, Sladek R, Froguel P, Watanabe RM, Meigs JB, Groop L, Boehnke M, McCarthy MI, Florez JC, Barroso I (2010) New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 42, 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, Oei L, Albagha OM, Amin N, Kemp JP, Koller DL, Li G, Liu CT, Minster RL, Moayyeri A, Vandenput L, Willner D, Xiao SM, Yerges‐Armstrong LM, Zheng HF, Alonso N, Eriksson J, Kammerer CM, Kaptoge SK, Leo PJ, Thorleifsson G, Wilson SG, Wilson JF, Aalto V, Alen M, Aragaki AK, Aspelund T, Center JR, Dailiana Z, Duggan DJ, Garcia M, Garcia‐Giralt N, Giroux S, Hallmans G, Hocking LJ, Husted LB, Jameson KA, Khusainova R, Kim GS, Kooperberg C, Koromila T, Kruk M, Laaksonen M, Lacroix AZ, Lee SH, Leung PC, Lewis JR, Masi L, Mencej‐Bedrac S, Nguyen TV, Nogues X, Patel MS, Prezelj J, Rose LM, Scollen S, Siggeirsdottir K, Smith AV, Svensson O, Trompet S, Trummer O, van Schoor NM, Woo J, Zhu K, Balcells S, Brandi ML, Buckley BM, Cheng S, Christiansen C, Cooper C, Dedoussis G, Ford I, Frost M, Goltzman D, Gonzalez‐Macias J, Kahonen M, Karlsson M, Khusnutdinova E, Koh JM, Kollia P, Langdahl BL, Leslie WD, Lips P, Ljunggren O, Lorenc RS, Marc J, Mellstrom D, Obermayer‐Pietsch B, Olmos JM, Pettersson‐Kymmer U, Reid DM, Riancho JA, Ridker PM, Rousseau F, Slagboom PE, Tang NL, Urreizti R, Van Hul W, Viikari J, Zarrabeitia MT, Aulchenko YS, Castano‐Betancourt M, Grundberg E, Herrera L, Ingvarsson T, Johannsdottir H, Kwan T, Li R, Luben R, Medina‐Gomez C, Palsson ST, Reppe S, Rotter JI, Sigurdsson G, van Meurs JB, Verlaan D, Williams FM, Wood AR, Zhou Y, Gautvik KM, Pastinen T, Raychaudhuri S, Cauley JA, Chasman DI, Clark GR, Cummings SR, Danoy P, Dennison EM, Eastell R, Eisman JA, Gudnason V, Hofman A, Jackson RD, Jones G, Jukema JW, Khaw KT, Lehtimaki T, Liu Y, Lorentzon M, McCloskey E, Mitchell BD, Nandakumar K, Nicholson GC, Oostra BA, Peacock M, Pols HA, Prince RL, Raitakari O, Reid IR, Robbins J, Sambrook PN, Sham PC, Shuldiner AR, Tylavsky FA, van Duijn CM, Wareham NJ, Cupples LA, Econs MJ, Evans DM, Harris TB, Kung AW, Psaty BM, Reeve J, Spector TD, Streeten EA, Zillikens MC, Thorsteinsdottir U, Ohlsson C, Karasik D, Richards JB, Brown MA, Stefansson K, Uitterlinden AG, Ralston SH, Ioannidis JP, Kiel DP, Rivadeneira F (2012) Genome‐wide meta‐analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 44, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DS, Cailotto F, Parimi N, Valdes AM, Castano‐Betancourt MC, Liu Y, Kaplan RC, Bidlingmaier M, Vasan RS, Teumer A, Tranah GJ, Nevitt MC, Cummings SR, Orwoll ES, Barrett‐Connor E, Renner JB, Jordan JM, Doherty M, Doherty SA, Uitterlinden AG, van Meurs JB, Spector TD, Lories RJ, Lane NE (2015) Genome‐wide association and functional studies identify a role for IGFBP3 in hip osteoarthritis. Ann. Rheum. Dis. 74, 1861–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth SM, Ganeshprasad U, Baxter RC (1998) Structural determinants of ligand and cell surface binding of insulin‐like growth factor‐binding protein‐3. J. Biol. Chem. 273, 2631–2638. [DOI] [PubMed] [Google Scholar]
- Fowlkes JL (1997) Insulinlike growth factor‐binding protein proteolysis an emerging paradigm in insulinlike growth factor physiology. Trends Endocrinol. Metab. 8, 299–306. [DOI] [PubMed] [Google Scholar]
- Friedrich N, Haring R, Nauck M, Ludemann J, Rosskopf D, Spilcke‐Liss E, Felix SB, Dorr M, Brabant G, Volzke H, Wallaschofski H (2009) Mortality and serum insulin‐like growth factor (IGF)‐I and IGF binding protein 3 concentrations. J. Clin. Endocrinol. Metab. 94, 1732–1739. [DOI] [PubMed] [Google Scholar]
- Grundberg E, Small KS, Hedman AK, Nica AC, Buil A, Keildson S, Bell JT, Yang TP, Meduri E, Barrett A, Nisbett J, Sekowska M, Wilk A, Shin SY, Glass D, Travers M, Min JL, Ring S, Ho K, Thorleifsson G, Kong A, Thorsteindottir U, Ainali C, Dimas AS, Hassanali N, Ingle C, Knowles D, Krestyaninova M, Lowe CE, Di Meglio P, Montgomery SB, Parts L, Potter S, Surdulescu G, Tsaprouni L, Tsoka S, Bataille V, Durbin R, Nestle FO, O'Rahilly S, Soranzo N, Lindgren CM, Zondervan KT, Ahmadi KR, Schadt EE, Stefansson K, Smith GD, McCarthy MI, Deloukas P, Dermitzakis ET, Spector TD, Multiple Tissue Human Expression Resource C (2012) Mapping cis‐ and trans‐regulatory effects across multiple tissues in twins. Nat. Genet. 44, 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F, Schumacher FR, Canzian F, Allen NE, Albanes D, Berg CD, Berndt SI, Boeing H, Bueno‐de‐Mesquita HB, Buring JE, Chabbert‐Buffet N, Chanock SJ, Clavel‐Chapelon F, Dumeaux V, Gaziano JM, Giovannucci EL, Haiman CA, Hankinson SE, Hayes RB, Henderson BE, Hunter DJ, Hoover RN, Johansson M, Key TJ, Khaw KT, Kolonel LN, Lagiou P, Lee IM, LeMarchand L, Lund E, Ma J, Onland‐Moret NC, Overvad K, Rodriguez L, Sacerdote C, Sanchez MJ, Stampfer MJ, Stattin P, Stram DO, Thomas G, Thun MJ, Tjonneland A, Trichopoulos D, Tumino R, Virtamo J, Weinstein SJ, Willett WC, Yeager M, Zhang SM, Kaaks R, Riboli E, Ziegler RG, Kraft P (2010) Eighteen insulin‐like growth factor pathway genes, circulating levels of IGF‐I and its binding protein, and risk of prostate and breast cancer. Cancer Epidemiol. Biomarkers Prev. 19, 2877–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guler HP, Zapf J, Schmid C, Froesch ER (1989) Insulin‐like growth factors I and II in healthy man. Estimations of half‐lives and production rates. Acta Endocrinol. (Copenh). 121, 753–758. [DOI] [PubMed] [Google Scholar]
- Harrela M, Koistinen H, Kaprio J, Lehtovirta M, Tuomilehto J, Eriksson J, Toivanen L, Koskenvuo M, Leinonen P, Koistinen R, Seppala M (1996) Genetic and environmental components of interindividual variation in circulating levels of IGF‐I, IGF‐II, IGFBP‐1, and IGFBP‐3. J Clin Invest. 98, 2612–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heemst D, Beekman M, Mooijaart SP, Heijmans BT, Brandt BW, Zwaan BJ, Slagboom PE, Westendorp RG (2005) Reduced insulin/IGF‐1 signalling and human longevity. Aging Cell 4, 79–85. [DOI] [PubMed] [Google Scholar]
- Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, Thorleifsson G, Zillikens MC, Speliotes EK, Magi R, Workalemahu T, White CC, Bouatia‐Naji N, Harris TB, Berndt SI, Ingelsson E, Willer CJ, Weedon MN, Luan J, Vedantam S, Esko T, Kilpelainen TO, Kutalik Z, Li S, Monda KL, Dixon AL, Holmes CC, Kaplan LM, Liang L, Min JL, Moffatt MF, Molony C, Nicholson G, Schadt EE, Zondervan KT, Feitosa MF, Ferreira T, Lango Allen H, Weyant RJ, Wheeler E, Wood AR, Magic Estrada K, Goddard ME, Lettre G, Mangino M, Nyholt DR, Purcell S, Smith AV, Visscher PM, Yang J, McCarroll SA, Nemesh J, Voight BF, Absher D, Amin N, Aspelund T, Coin L, Glazer NL, Hayward C, Heard‐Costa NL, Hottenga JJ, Johansson A, Johnson T, Kaakinen M, Kapur K, Ketkar S, Knowles JW, Kraft P, Kraja AT, Lamina C, Leitzmann MF, McKnight B, Morris AP, Ong KK, Perry JR, Peters MJ, Polasek O, Prokopenko I, Rayner NW, Ripatti S, Rivadeneira F, Robertson NR, Sanna S, Sovio U, Surakka I, Teumer A, van Wingerden S, Vitart V, Zhao JH, Cavalcanti‐Proenca C, Chines PS, Fisher E, Kulzer JR, Lecoeur C, Narisu N, Sandholt C, Scott LJ, Silander K, Stark K, Tammesoo ML, Teslovich TM, Timpson NJ, Watanabe RM, Welch R, Chasman DI, Cooper MN, Jansson JO, Kettunen J, Lawrence RW, Pellikka N, Perola M, Vandenput L, Alavere H, Almgren P, Atwood LD, Bennett AJ, Biffar R, Bonnycastle LL, Bornstein SR, Buchanan TA, Campbell H, Day IN, Dei M, Dorr M, Elliott P, Erdos MR, Eriksson JG, Freimer NB, Fu M, Gaget S, Geus EJ, Gjesing AP, Grallert H, Grassler J, Groves CJ, Guiducci C, Hartikainen AL, Hassanali N, Havulinna AS, Herzig KH, Hicks AA, Hui J, Igl W, Jousilahti P, Jula A, Kajantie E, Kinnunen L, Kolcic I, Koskinen S, Kovacs P, Kroemer HK, Krzelj V, Kuusisto J, Kvaloy K, Laitinen J, Lantieri O, Lathrop GM, Lokki ML, Luben RN, Ludwig B, McArdle WL, McCarthy A, Morken MA, Nelis M, Neville MJ, Pare G, Parker AN, Peden JF, Pichler I, Pietilainen KH, Platou CG, Pouta A, Ridderstrale M, Samani NJ, Saramies J, Sinisalo J, Smit JH, Strawbridge RJ, Stringham Song HM, Swift AJ, Teder‐Laving M, Thomson B, Usala G, van Meurs JB, van Ommen GJ, Vatin V, Volpato CB, Wallaschofski H, Walters GB, Widen E, Wild SH, Willemsen G, Witte DR, Zgaga L, Zitting P, Beilby JP, James AL, Kahonen M, Lehtimaki T, Nieminen MS, Ohlsson C, Palmer LJ, Raitakari O, Ridker PM, Stumvoll M, Tonjes A, Viikari J, Balkau B, Ben‐Shlomo Y, Bergman RN, Boeing H, Smith GD, Ebrahim S, Froguel P, Hansen T, Hengstenberg C, Hveem K, Isomaa B, Jorgensen T, Karpe F, Khaw KT, Laakso M, Lawlor DA, Marre M, Meitinger T, Metspalu A, Midthjell K, Pedersen O, Salomaa V, Schwarz PE, Tuomi T, Tuomilehto J, Valle TT, Wareham NJ, Arnold AM, Beckmann JS, Bergmann S, Boerwinkle E, Boomsma DI, Caulfield MJ, Collins FS, Eiriksdottir G, Gudnason V, Gyllensten U, Hamsten A, Hattersley AT, Hofman A, Hu FB, Illig T, Iribarren C, Jarvelin MR, Kao WH, Kaprio J, Launer LJ, Munroe PB, Oostra B, Penninx BW, Pramstaller PP, Psaty BM, Quertermous T, Rissanen A, Rudan I, Shuldiner AR, Soranzo N, Spector TD, Syvanen AC, Uda M, Uitterlinden A, Volzke H, Vollenweider P, Wilson JF, Witteman JC, Wright AF, Abecasis GR, Boehnke M, Borecki IB, Deloukas P, Frayling TM, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, North KE, O'Connell JR, Peltonen L, Schlessinger D, Strachan DP, Hirschhorn JN, Assimes TL, Wichmann HE, Thorsteinsdottir U, van Duijn CM, Stefansson K, Cupples LA, Loos RJ, Barroso I, McCarthy MI, Fox CS, Mohlke KL, Lindgren CM (2010) Meta‐analysis identifies 13 new loci associated with waist‐hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 42, 949–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho KK, Participants GHDCW (2007) Consensus guidelines for the diagnosis and treatment of adults with GH deficiency II: a statement of the GH Research Society in association with the European Society for Pediatric Endocrinology, Lawson Wilkins Society, European Society of Endocrinology, Japan Endocrine Society, and Endocrine Society of Australia. Eur. J. Endocrinol. 157, 695–700. [DOI] [PubMed] [Google Scholar]
- Hong Y, Pedersen NL, Brismar K, Hall K, de Faire U (1996) Quantitative genetic analyses of insulin‐like growth factor I (IGF‐I), IGF‐binding protein‐1, and insulin levels in middle‐aged and elderly twins. J. Clin. Endocrinol. Metab. 81, 1791–1797. [DOI] [PubMed] [Google Scholar]
- Huether R, Dong L, Chen X, Wu G, Parker M, Wei L, Ma J, Edmonson MN, Hedlund EK, Rusch MC, Shurtleff SA, Mulder HL, Boggs K, Vadordaria B, Cheng J, Yergeau D, Song G, Becksfort J, Lemmon G, Weber C, Cai Z, Dang J, Walsh M, Gedman AL, Faber Z, Easton J, Gruber T, Kriwacki RW, Partridge JF, Ding L, Wilson RK, Mardis ER, Mullighan CG, Gilbertson RJ, Baker SJ, Zambetti G, Ellison DW, Zhang J, Downing JR (2014) The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat. Commun. 5, 3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JI, Clemmons DR (1995) Insulin‐like growth factors and their binding proteins: biological actions. Endocr. Rev. 16, 3–34. [DOI] [PubMed] [Google Scholar]
- Juul A, Scheike T, Davidsen M, Gyllenborg J, Jorgensen T (2002) Low serum insulin‐like growth factor I is associated with increased risk of ischemic heart disease: a population‐based case‐control study. Circulation 106, 939–944. [DOI] [PubMed] [Google Scholar]
- Kaplan RC, McGinn AP, Pollak MN, Kuller LH, Strickler HD, Rohan TE, Cappola AR, Xue X, Psaty BM (2007) Association of total insulin‐like growth factor‐I, insulin‐like growth factor binding protein‐1 (IGFBP‐1), and IGFBP‐3 levels with incident coronary events and ischemic stroke. J. Clin. Endocrinol. Metab. 92, 1319–1325. [DOI] [PubMed] [Google Scholar]
- Kaplan RC, Petersen AK, Chen MH, Teumer A, Glazer NL, Doring A, Lam CS, Friedrich N, Newman A, Muller M, Yang Q, Homuth G, Cappola A, Klopp N, Smith H, Ernst F, Psaty BM, Wichmann HE, Sawyer DB, Biffar R, Rotter JI, Gieger C, Sullivan LS, Volzke H, Rice K, Spyroglou A, Kroemer HK, Ida Chen YD, Manolopoulou J, Nauck M, Strickler HD, Goodarzi MO, Reincke M, Pollak MN, Bidlingmaier M, Vasan RS, Wallaschofski H (2011) A genome‐wide association study identifies novel loci associated with circulating IGF‐I and IGFBP‐3. Hum. Mol. Genet. 20, 1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002) The human genome browser at UCSC. Genome Res. 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, Ferreira T, Wood AR, Weyant RJ, Segre AV, Speliotes EK, Wheeler E, Soranzo N, Park JH, Yang J, Gudbjartsson D, Heard‐Costa NL, Randall JC, Qi L, Vernon Smith A, Magi R, Pastinen T, Liang L, Heid IM, Luan J, Thorleifsson G, Winkler TW, Goddard ME, Sin Lo K, Palmer C, Workalemahu T, Aulchenko YS, Johansson A, Zillikens MC, Feitosa MF, Esko T, Johnson T, Ketkar S, Kraft P, Mangino M, Prokopenko I, Absher D, Albrecht E, Ernst F, Glazer NL, Hayward C, Hottenga JJ, Jacobs KB, Knowles JW, Kutalik Z, Monda KL, Polasek O, Preuss M, Rayner NW, Robertson NR, Steinthorsdottir V, Tyrer JP, Voight BF, Wiklund F, Xu J, Zhao JH, Nyholt DR, Pellikka N, Perola M, Perry JR, Surakka I, Tammesoo ML, Altmaier EL, Amin N, Aspelund T, Bhangale T, Boucher G, Chasman DI, Chen C, Coin L, Cooper MN, Dixon AL, Gibson Q, Grundberg E, Hao K, Juhani Junttila M, Kaplan LM, Kettunen J, Konig IR, Kwan T, Lawrence RW, Levinson DF, Lorentzon M, McKnight B, Morris AP, Muller M, Suh Ngwa J, Purcell S, Rafelt S, Salem RM, Salvi E, Sanna S, Shi J, Sovio U, Thompson JR, Turchin MC, Vandenput L, Verlaan DJ, Vitart V, White CC, Ziegler A, Almgren P, Balmforth AJ, Campbell H, Citterio L, De Grandi A, Dominiczak A, Duan J, Elliott P, Elosua R, Eriksson JG, Freimer NB, Geus EJ, Glorioso N, Haiqing S, Hartikainen AL, Havulinna AS, Hicks AA, Hui J, Igl W, Illig T, Jula A, Kajantie E, Kilpelainen TO, Koiranen M, Kolcic I, Koskinen S, Kovacs P, Laitinen J, Liu J, Lokki ML, Marusic A, Maschio A, Meitinger T, Mulas A, Pare G, Parker AN, Peden JF, Petersmann A, Pichler I, Pietilainen KH, Pouta A, Ridderstrale M, Rotter JI, Sambrook JG, Sanders AR, Schmidt CO, Sinisalo J, Smit JH, Stringham HM, Bragi Walters G, Widen E, Wild SH, Willemsen G, Zagato L, Zgaga L, Zitting P, Alavere H, Farrall M, McArdle WL, Nelis M, Peters MJ, Ripatti S, van Meurs JB, Aben KK, Ardlie KG, Beckmann JS, Beilby JP, Bergman RN, Bergmann S, Collins FS, Cusi D, den Heijer M, Eiriksdottir G, Gejman PV, Hall AS, Hamsten A, Huikuri HV, Iribarren C, Kahonen M, Kaprio J, Kathiresan S, Kiemeney L, Kocher T, Launer LJ, Lehtimaki T, Melander O, Mosley TH Jr, Musk AW, Nieminen MS, O'Donnell CJ, Ohlsson C, Oostra B, Palmer LJ, Raitakari O, Ridker PM, Rioux JD, Rissanen A, Rivolta C, Schunkert H, Shuldiner AR, Siscovick DS, Stumvoll M, Tonjes A, Tuomilehto J, van Ommen GJ, Viikari J, Heath AC, Martin NG, Montgomery GW, Province MA, Kayser M, Arnold AM, Atwood LD, Boerwinkle E, Chanock SJ, Deloukas P, Gieger C, Gronberg H, Hall P, Hattersley AT, Hengstenberg C, Hoffman W, Lathrop GM, Salomaa V, Schreiber S, Uda M, Waterworth D, Wright AF, Assimes TL, Barroso I, Hofman A, Mohlke KL, Boomsma DI, Caulfield MJ, Cupples LA, Erdmann J, Fox CS, Gudnason V, Gyllensten U, Harris TB, Hayes RB, Jarvelin MR, Mooser V, Munroe PB, Ouwehand WH, Penninx BW, Pramstaller PP, Quertermous T, Rudan I, Samani NJ, Spector TD, Volzke H, Watkins H, Wilson JF, Groop LC, Haritunians T, Hu FB, Kaplan RC, Metspalu A, North KE, Schlessinger D, Wareham NJ, Hunter DJ, O'Connell JR, Strachan DP, Wichmann HE, Borecki IB, van Duijn CM, Schadt EE, Thorsteinsdottir U, Peltonen L, Uitterlinden AG, Visscher PM, Chatterjee N, Loos RJ, Boehnke M, McCarthy MI, Ingelsson E, Lindgren CM, Abecasis GR, Stefansson K, Frayling TM, Hirschhorn JN (2010) Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roith D (1997) Seminars in medicine of the Beth Israel Deaconess Medical Center. Insulin‐like growth factors. N. Engl. J. Med. 336, 633–640. [DOI] [PubMed] [Google Scholar]
- Lee PD, Giudice LC, Conover CA, Powell DR (1997) Insulin‐like growth factor binding protein‐1: recent findings and new directions. Proc. Soc. Exp. Biol. Med. 216, 319–357. [DOI] [PubMed] [Google Scholar]
- Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Investigators A , Hayward NK, Montgomery GW, Visscher PM, Martin NG, Macgregor S (2010) A versatile gene‐based test for genome‐wide association studies. Am. J. Hum. Genet. 87, 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melmed S, Colao A, Barkan A, Molitch M, Grossman AB, Kleinberg D, Clemmons D, Chanson P, Laws E, Schlechte J, Vance ML, Ho K, Giustina A, Acromegaly Consensus G (2009) Guidelines for acromegaly management: an update. J. Clin. Endocrinol. Metab. 94, 1509–1517. [DOI] [PubMed] [Google Scholar]
- Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, Prokopenko I, Kang HM, Dina C, Esko T, Fraser RM, Kanoni S, Kumar A, Lagou V, Langenberg C, Luan J, Lindgren CM, Muller‐Nurasyid M, Pechlivanis S, Rayner NW, Scott LJ, Wiltshire S, Yengo L, Kinnunen L, Rossin EJ, Raychaudhuri S, Johnson AD, Dimas AS, Loos RJ, Vedantam S, Chen H, Florez JC, Fox C, Liu CT, Rybin D, Couper DJ, Kao WH, Li M, Cornelis MC, Kraft P, Sun Q, van Dam RM, Stringham HM, Chines PS, Fischer K, Fontanillas P, Holmen OL, Hunt SE, Jackson AU, Kong A, Lawrence R, Meyer J, Perry JR, Platou CG, Potter S, Rehnberg E, Robertson N, Sivapalaratnam S, Stancakova A, Stirrups K, Thorleifsson G, Tikkanen E, Wood AR, Almgren P, Atalay M, Benediktsson R, Bonnycastle LL, Burtt N, Carey J, Charpentier G, Crenshaw AT, Doney AS, Dorkhan M, Edkins S, Emilsson V, Eury E, Forsen T, Gertow K, Gigante B, Grant GB, Groves CJ, Guiducci C, Herder C, Hreidarsson AB, Hui J, James A, Jonsson A, Rathmann W, Klopp N, Kravic J, Krjutskov K, Langford C, Leander K, Lindholm E, Lobbens S, Mannisto S, Mirza G, Muhleisen TW, Musk B, Parkin M, Rallidis L, Saramies J, Sennblad B, Shah S, Sigurethsson G, Silveira A, Steinbach G, Thorand B, Trakalo J, Veglia F, Wennauer R, Winckler W, Zabaneh D, Campbell H, van Duijn C, Uitterlinden AG, Hofman A, Sijbrands E, Abecasis GR, Owen KR, Zeggini E, Trip MD, Forouhi NG, Syvanen AC, Eriksson JG, Peltonen L, Nothen MM, Balkau B, Palmer CN, Lyssenko V, Tuomi T, Isomaa B, Hunter DJ, Qi L, Wellcome Trust Case Control C , Meta‐Analyses of G , Insulin‐related traits Consortium I , Genetic Investigation of ATC , Asian Genetic Epidemiology Network‐Type 2 Diabetes C , South Asian Type 2 Diabetes C , Shuldiner AR, Roden M, Barroso I, Wilsgaard T, Beilby J, Hovingh K, Price JF, Wilson JF, Rauramaa R, Lakka TA, Lind L, Dedoussis G, Njolstad I, Pedersen NL, Khaw KT, Wareham NJ, Keinanen‐Kiukaanniemi SM, Saaristo TE, Korpi‐Hyovalti E, Saltevo J, Laakso M, Kuusisto J, Metspalu A, Collins FS, Mohlke KL, Bergman RN, Tuomilehto J, Boehm BO, Gieger C, Hveem K, Cauchi S, Froguel P, Baldassarre D, Tremoli E, Humphries SE, Saleheen D, Danesh J, Ingelsson E, Ripatti S, Salomaa V, Erbel R, Jockel KH, Moebus S, Peters A, Illig T, de FU, Hamsten A, Morris AD, Donnelly PJ, Frayling TM, Hattersley AT, Boerwinkle E, Melander O, Kathiresan S, Nilsson PM, Deloukas P, Thorsteinsdottir U, Groop LC, Stefansson K, Hu F, Pankow JS, Dupuis J, Meigs JB, Altshuler D, Boehnke M, McCarthy MI, Replication DIG , Meta‐analysis C (2012) Large‐scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44, 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ (2010) LocusZoom: regional visualization of genome‐wide association scan results. Bioinformatics 26, 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd‐Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole‐genome association and population‐based linkage analyses. Am. J. Hum. Genet. 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpathak SN, He M, Sun Q, Kaplan RC, Muzumdar R, Rohan TE, Gunter MJ, Pollak M, Kim M, Pessin JE, Beasley J, Wylie‐Rosett J, Hu FB, Strickler HD (2012) Insulin‐like growth factor axis and risk of type 2 diabetes in women. Diabetes 61, 2248–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renehan AG, Zwahlen M, Minder C, O'Dwyer ST, Shalet SM, Egger M (2004) Insulin‐like growth factor (IGF)‐I, IGF binding protein‐3, and cancer risk: systematic review and meta‐regression analysis. Lancet 363, 1346–1353. [DOI] [PubMed] [Google Scholar]
- Saxena R, Hivert MF, Langenberg C, Tanaka T, Pankow JS, Vollenweider P, Lyssenko V, Bouatia‐Naji N, Dupuis J, Jackson AU, Kao WH, Li M, Glazer NL, Manning AK, Luan J, Stringham HM, Prokopenko I, Johnson T, Grarup N, Boesgaard TW, Lecoeur C, Shrader P, O'Connell J, Ingelsson E, Couper DJ, Rice K, Song K, Andreasen CH, Dina C, Kottgen A, Le Bacquer O, Pattou F, Taneera J, Steinthorsdottir V, Rybin D, Ardlie K, Sampson M, Qi L, van Hoek M, Weedon MN, Aulchenko YS, Voight BF, Grallert H, Balkau B, Bergman RN, Bielinski SJ, Bonnefond A, Bonnycastle LL, Borch‐Johnsen K, Bottcher Y, Brunner E, Buchanan TA, Bumpstead SJ, Cavalcanti‐Proenca C, Charpentier G, Chen YD, Chines PS, Collins FS, Cornelis M, G JC, Delplanque J, Doney A, Egan JM, Erdos MR, Firmann M, Forouhi NG, Fox CS, Goodarzi MO, Graessler J, Hingorani A, Isomaa B, Jorgensen T, Kivimaki M, Kovacs P, Krohn K, Kumari M, Lauritzen T, Levy‐Marchal C, Mayor V, McAteer JB, Meyre D, Mitchell BD, Mohlke KL, Morken MA, Narisu N, Palmer CN, Pakyz R, Pascoe L, Payne F, Pearson D, Rathmann W, Sandbaek A, Sayer AA, Scott LJ, Sharp SJ, Sijbrands E, Singleton A, Siscovick DS, Smith NL, Sparso T, Swift AJ, Syddall H, Thorleifsson G, Tonjes A, Tuomi T, Tuomilehto J, Valle TT, Waeber G, Walley A, Waterworth DM, Zeggini E, Zhao JH, consortium G, investigators M, Illig T, Wichmann HE, Wilson JF, van Duijn C, Hu FB, Morris AD, Frayling TM, Hattersley AT, Thorsteinsdottir U, Stefansson K, Nilsson P, Syvanen AC, Shuldiner AR, Walker M, Bornstein SR, Schwarz P, Williams GH, Nathan DM, Kuusisto J, Laakso M, Cooper C, Marmot M, Ferrucci L, Mooser V, Stumvoll M, Loos RJ, Altshuler D, Psaty BM, Rotter JI, Boerwinkle E, Hansen T, Pedersen O, Florez JC, McCarthy MI, Boehnke M, Barroso I, Sladek R, Froguel P, Meigs JB, Groop L, Wareham NJ, Watanabe RM (2010) Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat. Genet. 42, 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher FR, Cheng I, Freedman ML, Mucci L, Allen NE, Pollak MN, Hayes RB, Stram DO, Canzian F, Henderson BE, Hunter DJ, Virtamo J, Manjer J, Gaziano JM, Kolonel LN, Tjonneland A, Albanes D, Calle EE, Giovannucci E, Crawford ED, Haiman CA, Kraft P, Willett WC, Thun MJ, Le Marchand L, Kaaks R, Feigelson HS, Bueno‐de‐Mesquita HB, Palli D, Riboli E, Lund E, Amiano P, Andriole G, Dunning AM, Trichopoulos D, Stampfer MJ, Key TJ, Ma J (2010) A comprehensive analysis of common IGF1, IGFBP1 and IGFBP3 genetic variation with prospective IGF‐I and IGFBP‐3 blood levels and prostate cancer risk among Caucasians. Hum. Mol. Genet. 19, 3089–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Cardiogenics , Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein SE, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJ, Khaw KT, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Muhleisen TW, Muhlestein JB, Munzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nothen MM, Olivieri O, Patel RS, Patterson CC, Peters A, Peyvandi F, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Rosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schafer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith A, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stirrups K, Stoll M, Tang WH, Tennstedt S, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, Wells GA, Wichmann HE, Wild PS, Willenborg C, Witteman JC, Wright BJ, Ye S, Zeller T, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, Marz W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thompson JR, Stefansson K, Roberts R, Thorsteinsdottir U, O'Donnell CJ, McPherson R, Erdmann J, Consortium CA , Samani NJ (2011) Large‐scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 43, 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurmann C, Heim K, Schillert A, Blankenberg S, Carstensen M, Dorr M, Endlich K, Felix SB, Gieger C, Grallert H, Herder C, Hoffmann W, Homuth G, Illig T, Kruppa J, Meitinger T, Muller C, Nauck M, Peters A, Rettig R, Roden M, Strauch K, Volker U, Volzke H, Wahl S, Wallaschofski H, Wild PS, Zeller T, Teumer A, Prokisch H, Ziegler A (2012) Analyzing illumina gene expression microarray data from different tissues: methodological aspects of data analysis in the metaxpress consortium. PLoS ONE 7, e50938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP, Walter K, Menni C, Chen L, Vasquez L, Valdes AM, Hyde CL, Wang V, Ziemek D, Roberts P, Xi L, Grundberg E, Multiple Tissue Human Expression Resource C , Waldenberger M, Richards JB, Mohney RP, Milburn MV, John SL, Trimmer J, Theis FJ, Overington JP, Suhre K, Brosnan MJ, Gieger C, Kastenmuller G, Spector TD, Soranzo N (2014) An atlas of genetic influences on human blood metabolites. Nat. Genet. 46, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soranzo N, Sanna S, Wheeler E, Gieger C, Radke D, Dupuis J, Bouatia‐Naji N, Langenberg C, Prokopenko I, Stolerman E, Sandhu MS, Heeney MM, Devaney JM, Reilly MP, Ricketts SL, Stewart AF, Voight BF, Willenborg C, Wright B, Altshuler D, Arking D, Balkau B, Barnes D, Boerwinkle E, Bohm B, Bonnefond A, Bonnycastle LL, Boomsma DI, Bornstein SR, Bottcher Y, Bumpstead S, Burnett‐Miller MS, Campbell H, Cao A, Chambers J, Clark R, Collins FS, Coresh J, de Geus EJ, Dei M, Deloukas P, Doring A, Egan JM, Elosua R, Ferrucci L, Forouhi N, Fox CS, Franklin C, Franzosi MG, Gallina S, Goel A, Graessler J, Grallert H, Greinacher A, Hadley D, Hall A, Hamsten A, Hayward C, Heath S, Herder C, Homuth G, Hottenga JJ, Hunter‐Merrill R, Illig T, Jackson AU, Jula A, Kleber M, Knouff CW, Kong A, Kooner J, Kottgen A, Kovacs P, Krohn K, Kuhnel B, Kuusisto J, Laakso M, Lathrop M, Lecoeur C, Li M, Li M, Loos RJ, Luan J, Lyssenko V, Magi R, Magnusson PK, Malarstig A, Mangino M, Martinez‐Larrad MT, Marz W, McArdle WL, McPherson R, Meisinger C, Meitinger T, Melander O, Mohlke KL, Mooser VE, Morken MA, Narisu N, Nathan DM, Nauck M, O'Donnell C, Oexle K, Olla N, Pankow JS, Payne F, Peden JF, Pedersen NL, Peltonen L, Perola M, Polasek O, Porcu E, Rader DJ, Rathmann W, Ripatti S, Rocheleau G, Roden M, Rudan I, Salomaa V, Saxena R, Schlessinger D, Schunkert H, Schwarz P, Seedorf U, Selvin E, Serrano‐Rios M, Shrader P, Silveira A, Siscovick D, Song K, Spector TD, Stefansson K, Steinthorsdottir V, Strachan DP, Strawbridge R, Stumvoll M, Surakka I, Swift AJ, Tanaka T, Teumer A, Thorleifsson G, Thorsteinsdottir U, Tonjes A, Usala G, Vitart V, Volzke H, Wallaschofski H, Waterworth DM, Watkins H, Wichmann HE, Wild SH, Willemsen G, Williams GH, Wilson JF, Winkelmann J, Wright AF, WTCCC , Zabena C, Zhao JH, Epstein SE, Erdmann J, Hakonarson HH, Kathiresan S, Khaw KT, Roberts R, Samani NJ, Fleming MD, Sladek R, Abecasis G, Boehnke M, Froguel P, Groop L, McCarthy MI, Kao WH, Florez JC, Uda M, Wareham NJ, Barroso I, Meigs JB (2010) Common variants at 10 genomic loci influence hemoglobin A(1)(C) levels via glycemic and nonglycemic pathways. Diabetes 59, 3229–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souren NY, Paulussen AD, Loos RJ, Gielen M, Beunen G, Fagard R, Derom C, Vlietinck R, Zeegers MP (2007) Anthropometry, carbohydrate and lipid metabolism in the East Flanders Prospective Twin Survey: heritabilities. Diabetologia 50, 2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Magi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segre AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpelainen TO, Yang J, Bouatia‐Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard‐Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben‐Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, Busonero F, Campbell H, Cappuccio FP, Cavalcanti‐Proenca C, Chen YD, Chen CM, Chines PS, Clarke R, Coin L, Connell J, Day IN, den Heijer M, Duan J, Ebrahim S, Elliott P, Elosua R, Eiriksdottir G, Erdos MR, Eriksson JG, Facheris MF, Felix SB, Fischer‐Posovszky P, Folsom AR, Friedrich N, Freimer NB, Fu M, Gaget S, Gejman PV, Geus EJ, Gieger C, Gjesing AP, Goel A, Goyette P, Grallert H, Grassler J, Greenawalt DM, Groves CJ, Gudnason V, Guiducci C, Hartikainen AL, Hassanali N, Hall AS, Havulinna AS, Hayward C, Heath AC, Hengstenberg C, Hicks AA, Hinney A, Hofman A, Homuth G, Hui J, Igl W, Iribarren C, Isomaa B, Jacobs KB, Jarick I, Jewell E, John U, Jorgensen T, Jousilahti P, Jula A, Kaakinen M, Kajantie E, Kaplan LM, Kathiresan S, Kettunen J, Kinnunen L, Knowles JW, Kolcic I, Konig IR, Koskinen S, Kovacs P, Kuusisto J, Kraft P, Kvaloy K, Laitinen J, Lantieri O, Lanzani C, Launer LJ, Lecoeur C, Lehtimaki T, Lettre G, Liu J, Lokki ML, Lorentzon M, Luben RN, Ludwig B, MAGIC , Manunta P, Marek D, Marre M, Martin NG, McArdle WL, McCarthy A, McKnight B, Meitinger T, Melander O, Meyre D, Midthjell K, Montgomery GW, Morken MA, Morris AP, Mulic R, Ngwa JS, Nelis M, Neville MJ, Nyholt DR, O'Donnell CJ, O'Rahilly S, Ong KK, Oostra B, Pare G, Parker AN, Perola M, Pichler I, Pietilainen KH, Platou CG, Polasek O, Pouta A, Rafelt S, Raitakari O, Rayner NW, Ridderstrale M, Rief W, Ruokonen A, Robertson NR, Rzehak P, Salomaa V, Sanders AR, Sandhu MS, Sanna S, Saramies J, Savolainen MJ, Scherag S, Schipf S, Schreiber S, Schunkert H, Silander K, Sinisalo J, Siscovick DS, Smit JH, Soranzo N, Sovio U, Stephens J, Surakka I, Swift AJ, Tammesoo ML, Tardif JC, Teder‐Laving M, Teslovich TM, Thompson JR, Thomson B, Tonjes A, Tuomi T, van Meurs JB, van Ommen GJ, Vatin V, Viikari J, Visvikis‐Siest S, Vitart V, Vogel CI, Voight BF, Waite LL, Wallaschofski H, Walters GB, Widen E, Wiegand S, Wild SH, Willemsen G, Witte DR, Witteman JC, Xu J, Zhang Q, Zgaga L, Ziegler A, Zitting P, Beilby JP, Farooqi IS, Hebebrand J, Huikuri HV, James AL, Kahonen M, Levinson DF, Macciardi F, Nieminen MS, Ohlsson C, Palmer LJ, Ridker PM, Stumvoll M, Beckmann JS, Boeing H, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Collins FS, Cupples LA, Smith GD, Erdmann J, Froguel P, Gronberg H, Gyllensten U, Hall P, Hansen T, Harris TB, Hattersley AT, Hayes RB, Heinrich J, Hu FB, Hveem K, Illig T, Jarvelin MR, Kaprio J, Karpe F, Khaw KT, Kiemeney LA, Krude H, Laakso M, Lawlor DA, Metspalu A, Munroe PB, Ouwehand WH, Pedersen O, Penninx BW, Peters A, Pramstaller PP, Quertermous T, Reinehr T, Rissanen A, Rudan I, Samani NJ, Schwarz PE, Shuldiner AR, Spector TD, Tuomilehto J, Uda M, Uitterlinden A, Valle TT, Wabitsch M, Waeber G, Wareham NJ, Watkins H, Procardis C, Wilson JF, Wright AF, Zillikens MC, Chatterjee N, McCarroll SA, Purcell S, Schadt EE, Visscher PM, Assimes TL, Borecki IB, Deloukas P, Fox CS, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, Mohlke KL, O'Connell JR, Peltonen L, Schlessinger D, Strachan DP, van Duijn CM, Wichmann HE, Frayling TM, Thorsteinsdottir U, Abecasis GR, Barroso I, Boehnke M, Stefansson K, North KE, McCarthy MI, Hirschhorn JN, Ingelsson E, Loos RJ (2010) Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 42, 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y, Atzmon G, Cho MO, Hwang D, Liu B, Leahy DJ, Barzilai N, Cohen P (2008) Functionally significant insulin‐like growth factor I receptor mutations in centenarians. Proc. Natl. Acad. Sci. U S A. 105, 3438–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wagele B, Altmaier E, CardioGram DP, Erdmann J, Grundberg E, Hammond CJ, de Angelis MH, Kastenmuller G, Kottgen A, Kronenberg F, Mangino M, Meisinger C, Meitinger T, Mewes HW, Milburn MV, Prehn C, Raffler J, Ried JS, Romisch‐Margl W, Samani NJ, Small KS, Wichmann HE, Zhai G, Illig T, Spector TD, Adamski J, Soranzo N, Gieger C (2011) Human metabolic individuality in biomedical and pharmaceutical research. Nature 477, 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg SM, Baxter RC (1998) Insulin‐like growth factor (IGF)‐binding protein 5 forms an alternative ternary complex with IGFs and the acid‐labile subunit. J. Biol. Chem. 273, 6074–6079. [DOI] [PubMed] [Google Scholar]
- Vasan RS, Sullivan LM, D'Agostino RB, Roubenoff R, Harris T, Sawyer DB, Levy D, Wilson PW (2003) Serum insulin‐like growth factor I and risk for heart failure in elderly individuals without a previous myocardial infarction: the Framingham Heart Study. Ann. Intern. Med. 139, 642–648. [DOI] [PubMed] [Google Scholar]
- Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G, McCulloch LJ, Ferreira T, Grallert H, Amin N, Wu G, Willer CJ, Raychaudhuri S, McCarroll SA, Langenberg C, Hofmann OM, Dupuis J, Qi L, Segre AV, van Hoek M, Navarro P, Ardlie K, Balkau B, Benediktsson R, Bennett AJ, Blagieva R, Boerwinkle E, Bonnycastle LL, Bengtsson Bostrom K, Bravenboer B, Bumpstead S, Burtt NP, Charpentier G, Chines PS, Cornelis M, Couper DJ, Crawford G, Doney AS, Elliott KS, Elliott AL, Erdos MR, Fox CS, Franklin CS, Ganser M, Gieger C, Grarup N, Green T, Griffin S, Groves CJ, Guiducci C, Hadjadj S, Hassanali N, Herder C, Isomaa B, Jackson AU, Johnson PR, Jorgensen T, Kao WH, Klopp N, Kong A, Kraft P, Kuusisto J, Lauritzen T, Li M, Lieverse A, Lindgren CM, Lyssenko V, Marre M, Meitinger T, Midthjell K, Morken MA, Narisu N, Nilsson P, Owen KR, Payne F, Perry JR, Petersen AK, Platou C, Proenca C, Prokopenko I, Rathmann W, Rayner NW, Robertson NR, Rocheleau G, Roden M, Sampson MJ, Saxena R, Shields BM, Shrader P, Sigurdsson G, Sparso T, Strassburger K, Stringham HM, Sun Q, Swift AJ, Thorand B, Tichet J, Tuomi T, van Dam RM, van Haeften TW, van Herpt T, van Vliet‐Ostaptchouk JV, Walters GB, Weedon MN, Wijmenga C, Witteman J, Bergman RN, Cauchi S, Collins FS, Gloyn AL, Gyllensten U, Hansen T, Hide WA, Hitman GA, Hofman A, Hunter DJ, Hveem K, Laakso M, Mohlke KL, Morris AD, Palmer CN, Pramstaller PP, Rudan I, Sijbrands E, Stein LD, Tuomilehto J, Uitterlinden A, Walker M, Wareham NJ, Watanabe RM, Abecasis GR, Boehm BO, Campbell H, Daly MJ, Hattersley AT, Hu FB, Meigs JB, Pankow JS, Pedersen O, Wichmann HE, Barroso I, Florez JC, Frayling TM, Groop L, Sladek R, Thorsteinsdottir U, Wilson JF, Illig T, Froguel P, van Duijn DC, Stefansson K, Altshuler D, Boehnke M, McCarthy MI, Investigators M , Consortium G (2010) Twelve type 2 diabetes susceptibility loci identified through large‐scale association analysis. Nat. Genet. 42, 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, Christiansen MW, Fairfax BP, Schramm K, Powell JE, Zhernakova A, Zhernakova DV, Veldink JH, Van den Berg LH, Karjalainen J, Withoff S, Uitterlinden AG, Hofman A, Rivadeneira F, t Hoen PA, Reinmaa E, Fischer K, Nelis M, Milani L, Melzer D, Ferrucci L, Singleton AB, Hernandez DG, Nalls MA, Homuth G, Nauck M, Radke D, Volker U, Perola M, Salomaa V, Brody J, Suchy‐Dicey A, Gharib SA, Enquobahrie DA, Lumley T, Montgomery GW, Makino S, Prokisch H, Herder C, Roden M, Grallert H, Meitinger T, Strauch K, Li Y, Jansen RC, Visscher PM, Knight JC, Psaty BM, Ripatti S, Teumer A, Frayling TM, Metspalu A, van Meurs JB, Franke L (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD (2008) FOXO3A genotype is strongly associated with human longevity. Proc. Natl. Acad. Sci. U S A. 105, 13987–13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR (2010) METAL: fast and efficient meta‐analysis of genomewide association scans. Bioinformatics 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome‐wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv E, Hu D (2011) Genetic variation in insulin/IGF‐1 signaling pathways and longevity. Ageing Res. Rev. 10, 201–204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Materials and methods.
Fig. S1 Manhattan plots of men and women strata.
Fig. S2 QQ plots of meta‐analysis results.
Fig. S3 Regional association plots for IGF‐I traits.
Fig. S4 Regional association plots for IGFBP‐3 traits.
Fig. S5 Results of bivariate analysis of IGF‐I and IGFBP‐3.
Fig. S6 Flow chart of the study's design.
Table S1 Characteristics of study cohorts.
Table S2 Assay and genotyping information of study cohorts.
Table S3 GWAS results with P‐value <10−6 in stage 1.
Table S4 Genome‐wide significant loci of bivariate analysis.
Table S5 Sex interaction results of the 12 genome‐wide significant SNPs.
Table S6 Results of eQTL lookup in the Muther dataset.
Table S7 Results of gene expression analysis of the genome‐wide significant loci with IGF‐I and IGFBP‐3 in the SHIP‐TREND cohort.
Table S8 Results of pQTL analysis in the SHIP‐TREND cohort.
Table S9 Lookup of genome‐wide significant lead SNPs in IGF‐I/IGFBP‐3 correlated traits.
Table S10 Lookup of Top IGF‐I associated SNPs in associations with survival beyond age 90 years old.
Table S11 Lookup of Top IGFBP‐3 associated SNPs in associations with survival beyond age 90 years old.
Table S12 Enrichment of putative regulatory elements among IGF‐1 and IGFBP‐3 associated loci.
Data S1 Design and funding of participating cohort studies.


