Abstract
During embryonic development, regulation of gene expression is key to creating the many subtypes of cells that an organism needs throughout its lifetime. Recent work has shown that maternal genetics and environmental factors have lifelong consequences on diverse processes ranging from immune function to stress responses. The RE1-silencing transcription factor (Rest) is a transcriptional repressor that interacts with chromatin-modifying complexes to repress transcription of neural-specific genes during early development. Here we show that in zebrafish, maternally supplied rest regulates expression of target genes during larval development and has lifelong impacts on behavior. Larvae deprived of maternal rest are hyperactive and show atypical spatial preferences. Adult male fish deprived of maternal rest present with atypical spatial preferences in a novel environment assay. Transcriptome sequencing revealed 158 genes that are repressed by maternal rest in blastula stage embryos. Furthermore, we found that maternal rest is required for target gene repression until at least 6 dpf. Importantly, disruption of the RE1 sites in either snap25a or snap25b resulted in behaviors that recapitulate the hyperactivity phenotype caused by absence of maternal rest. Both maternal rest mutants and snap25a RE1 site mutants have altered primary motor neuron architecture that may account for the enhanced locomotor activity. These results demonstrate that maternal rest represses snap25a/b to modulate larval behavior and that early Rest activity has lifelong behavioral impacts.
SIGNIFICANCE STATEMENT Maternal factors deposited in the oocyte have well-established roles during embryonic development. We show that, in zebrafish, maternal rest (RE1-silencing transcription factor) regulates expression of target genes during larval development and has lifelong impacts on behavior. The Rest transcriptional repressor interacts with chromatin-modifying complexes to limit transcription of neural genes. We identify several synaptic genes that are repressed by maternal Rest and demonstrate that snap25a/b are key targets of maternal rest that modulate larval locomotor activity. These results reveal that zygotic rest is unable to compensate for deficits in maternally supplied rest and uncovers novel temporal requirements for Rest activity, which has implications for the broad roles of Rest-mediated repression during neural development and in disease states.
Keywords: locomotor behavior, maternal effect, Rest/Nrsf, snap25, zebrafish
Introduction
Precise regulation of gene expression is key to proper nervous system function and is influenced by both genetic and environmental factors. Central to the mechanisms of gene regulation are chromatin modifications, which include alterations of the acetylation and methylation status of chromatin by transcriptional activators and repressors. Changes to chromatin landscapes may have both immediate and lifelong consequences and are caused by environmental effects, including poor maternal care (Weaver et al., 2004), prenatal stress (St-Cyr and McGowan, 2015; Vangeel et al., 2015), smoking (Ivorra et al., 2015), and gestational diabetes (Petropoulos et al., 2015).
Maternal mRNAs encoding transcription factors and chromatin effectors are deposited in oocytes before fertilization and modulate developmental gene expression in many species. For example, depletion of maternal Drosophila Piwi alters heterochromatin formation (Gu and Elgin, 2013); knockdown of VegT in Xenopus alters embryonic cell fate and patterning (Zhang et al., 1998); loss of maternal runx2b dorsalizes zebrafish embryos (Flores et al., 2008); and deletion of maternal BRG1, arrests mouse development at early cleavage stages and reduces zygotic genome activation (Bultman et al., 2006). These findings suggest a broad role for maternal mRNAs in modulating chromatin landscapes in early embryos.
The RE1-Silencing Transcription factor (Rest)/Neuron Restrictive Silencing Factor (Nrsf) recruits cofactors to modify chromatin structure to silence neural-specific genes in non-neural tissues (Chong et al., 1995; Schoenherr and Anderson, 1995) and to modulate transcription within the developing nervous system (Ballas et al., 2005). Rest regulates hundreds of neural-specific genes via interactions with a conserved ∼23 bp DNA element, the RE1 site (Lunyak et al., 2002; Mortazavi et al., 2006). The N-terminal domain of Rest interacts with Sin3 family members to recruit repressor complexes that include MeCP2 and HDAC1/2 (Naruse et al., 1999; Grzenda et al., 2009). The Rest C-terminal domain interacts with CoRest family members, which associate with HDAC 1/2, LSD1 and H3K9 methyltranferase G9a, among other factors (Ballas et al., 2001; Lunyak et al., 2002; Roopra et al., 2004).
We previously showed that zebrafish rest is broadly expressed in the developing nervous system (Gates et al., 2010) but is not essential for neurogenesis (Kok et al., 2012). Rather, Rest acts to fine-tune neural gene expression (Kok et al., 2012) and consequently modulate both larval and adult behaviors (Moravec et al., 2015). Zebrafish rest mRNA is provided as a maternal transcript (Gates et al., 2010) that is essential for proper regulation of gene expression in the blastula (Kok et al., 2012). In addition, maternally supplied rest also modulates later migration of facial branchiomotor neurons (Love and Prince, 2015). An early function of REST has also been demonstrated in rodents, where maternal deprivation decreases REST levels (Uchida et al., 2010; Rodenas-Ruano et al., 2012). Subsequent misregulation of NMDA receptor gene expression leads to changes in synaptic plasticity (Rodenas-Ruano et al., 2012). Conversely, increased maternal care augments REST levels, which correlates with decreased expression of a stress hormone, corticotropin-releasing hormone (Korosi et al., 2010).
In this study, we demonstrate that in zebrafish maternal rest modulates zygotic gene expression until at least 6 days post fertilization (dpf) and that depletion of maternal rest results in behavioral changes in larvae, including hyperactivity and atypical spatial preferences. Strikingly, behavioral anomalies persist into adulthood in animals that lack maternal rest. Affected adult males, but not females, engage in abnormal swimming behaviors, including atypical wall preference combined with frequent vertical swimming and sharper turning angles. Importantly, disruption of the RE1 site of either of two target genes, snap25a or snap25b, recapitulates the larval hyperactivity phenotype. This finding implicates snap25 paralogs as key targets of Rest in controlling larval behavior. Consistent with the role of Snap25 in axon growth, we investigated the architecture of the primary motor neurons in the mutants and observed increased branching in primary motor neurons in embryos that lack maternal rest and in snap25a RE-1 site mutants. Together, these results demonstrate that maternally supplied rest influences embryonic and larval gene expression and lifelong behavior.
Materials and Methods
Fish maintenance.
Zebrafish embryos were obtained from natural crosses and maintained at 28.5°C under 13/11 h light/dark cycle. Adult fish were fed twice daily with a combination of artemia and flake food. The restsbu29 mutation was maintained as previously described (Moravec et al., 2015). All rest mutants came from intercrossing rest heterozygotes to control for effects caused by maintaining mutant inbred stocks. Larval assays were performed at 6 dpf on multiple clutches derived from different parents to minimize genetic background effects.
Housing and genotyping.
Housing and genotyping were described previously (Kok et al., 2012; Moravec et al., 2015) with a slight modification. Adult fish were raised in groups of 8–10 in 1.8 L tanks, moved into unisex tanks at 4 months, and transferred to individual 1 L tanks 2 weeks before the behavioral assays.
Behavioral testing apparatus and paradigms.
The Novel Environment and Visual-Motor-Behavioral Assays and the testing apparatus were previously described (Moravec et al., 2015). Assays of adults, of both sexes, were conducted at 6 months. All behavioral assays were performed between 1:00 and 5:00 P.M. and approved by the Stony Brook University Institutional Animal Care and Use Committee.
Deep sequencing.
Total RNA was extracted from pools of 10 embryos from each of the four groups (MrestSBU29/+, Zrest,MZrest, and wild-type [WT]), and 2 pools per group were sent to the New York Genome Center for sequencing. Samples underwent a Tru Seq V2 library prep and sequenced on a Hi Seq 2000 by 2× 50 bp paired end reads. The reads were aligned to Danio_rerio.Zv9.74 from Ensembl. Significance was defined as p < 0.05 after a correction of multiple testing hypothesis using the Benjamini and Hochberg procedure.
Expression studies.
Total RNA was extracted from pools of five embryos using Trizol (Invitrogen), and cDNA was synthesized by using Super Script II reverse transcriptase (Invitrogen). Quantitative PCR (qPCR) was performed with a Light Cycler 480 (Roche) using Quanta SYBR Green (Quanta Bioscience). Transcript levels from each sample were normalized to β-actin. Each experiment consisted of three pools of embryos run in duplicate. Primer pairs are listed below or were described previously (Kok et al., 2012): npas4a forward, GGGCTCAAGCACTTCTCAAC, reverse, AGATAGCCCACTGCTTCCTG; amph forward, CCAGAGGAAGAGACCAGTTCA, reverse, CTTCTCCTGGTTGGGTCTCA; syt4 forward, TGGAGAAATCCCAGGACAAG, reverse, GACAGACCATGTGCCTCCTT; scn3b forward, TGATGTATGTGCTGCTGGTG, reverse, TGTGCTTGCTCGTCAGATTT; nsfa forward, TTTGACAAGTCCAGGCAGTG, reverse, CTGAGTCGTAAGGGCTGGAG; kcns3a forward, GAGGATGACCCTCAGAACCA, reverse, GTGCCCTCAAACTTTTCCAA; cana1ba forward, ATACTGGATCGGCCCAAACT, reverse, ATACTGGATCGGCCCAAACT; syt10 forward, TGTGGTTCGCATTCTCAAAG, reverse, ACTTCTTTTTGCGCTCTGGA; grm5(1/2) forward, TGTCACTGATGGCTTCCAGA, reverse, TGGCTGCAGGTTCAGGTAGT; olfm1b forward, GGGACCTGCAGTACGTGGTA, reverse, TATTGCTTGGCGATGTTTTG; cadpsb forward, TTGTCGTGAGGTGTTCAAGC, reverse, CAAACTTGGCCATCCAAGAG; and nrxn1a forward, TAATGTGCGTGTGGAGGGTA, reverse, GGGTGACGTTTCTGAACGAT.
RNA whole-mount in situ hybridization was performed as described by Thisse et al. (1993). In cases where genotype differences could be attributed to tube-specific variations in staining, embryos were marked by tail clips and the procedure performed with both sets of embryos in the same tube. Probes were synthesized from plasmids or from 6 dpf cDNA using primer pair for amph antisense forward, ATTTGCCAAAAACGTCCAAA; reverse, GAGTAATACGACTCACTAGGGGGGCCTTTTTCAAGTCCTCT. For immunohistochemistry, embryos were fixed in 4% PFA overnight at 4°C and stored in methanol. ZNP-1(RRID:AB_10013783) staining was performed as described previously (Wei et al., 2013). Quantification of average fluorescence of the immunohistochemistry was done using ImageJ. The same three puncta were quantified on each sample and ratio to controls (WT or ZrestSBU29/+).
Disruption of RE1 sites.
RE1 sites in snap25a (TTCAGCACCCTGGACAGCGAC) and snap25b (TTCAGCACCGCGGAGAGCGCT) were disrupted using the CRISPR-CAS9 system. Guide RNA targets sites are as follows: snap25a, GCAAACGCAGTCGCTGTCCA; snap25b, GGTGCTGAAATCCACACAAC. gRNAs were generated using Ambion MegaScript T7 kit. Guide RNA (200 pg) was coinjected with 400 pg of Cas9 protein (PNA Bio) into the cell of one-cell embryos. Fish were genotyped using the following primers: snap25a RE1 site forward, ACGATGTGGGCGGTTTCT; reverse, TGGAAATTTAGCTGCAGGAG; snap25b RE1 site forward, TTGCACAGCTTTTGCATGA; reverse, TACCATGGAGGCTCGACTTT.
Statistics.
Statistical analyses were conducted as previously described (Moravec et al., 2015) using SPSS, version 21 (RRID:SCR_002865)and GraphPad software. Outliers were detected using the Grubs test and removed from analysis. Significance was defined as <0.05, and trending was defined as 0.099–0.05. Error bars indicate SE.
Results
Maternal Rest regulates gene expression at blastula stage
We previously observed that depletion of maternal rest caused derepression of a subset of target genes in blastula stage zebrafish embryos (Kok et al., 2012). To better understand the role of maternal rest in gene repression, we performed deep sequencing of blastula mRNA comparing Mrestsbu29/+ with Zrestsbu29/+ and MZrestsbu29/sbu29 with related WT controls. Mrestsbu29/+ fish are the offspring of a rest mutant female and a WT male and therefore lack maternal rest mRNA. The corresponding controls have normal maternal contribution of rest and are the offspring of a rest mutant male and a WT female (Zrestsbu29/+). MZrestsbu29/sbu29 lack both maternal and zygotic rest and are the offspring of two homozygous mutants. The corresponding control WTs were obtained from crosses of WT siblings of the mutant parents used to generate the MZrestsbu29/sbu29 offspring. Because of the temporal proximity of these embryos to the mid-blastula transition, we anticipate that most of the transcriptional changes detected will result from direct effects of maternal Rest depletion because the analysis occurred shortly after the activation of the zygotic genome.
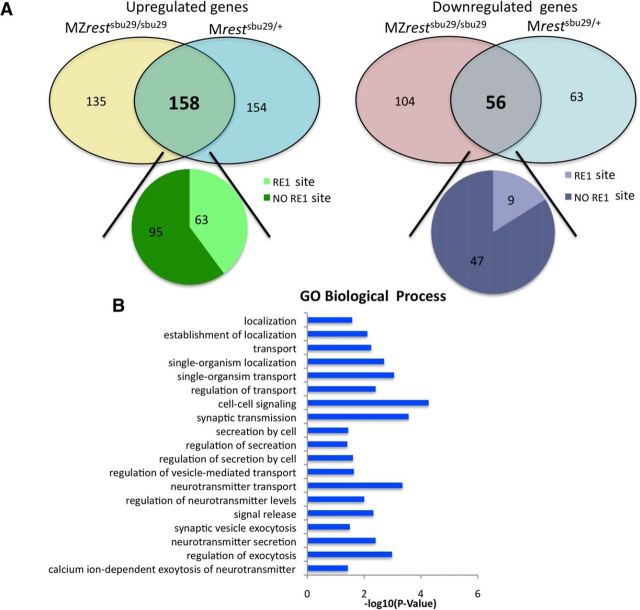
Overall, the deep sequencing identified a total of 26,000 transcripts, but only 214 were significantly misregulated in both Mrestsbu29/+ and MZrestsbu29/sbu29 RNAseqs (p < 0.05 after Benjamini and Hochberg correction) (Fig. 1A). Of these 214 genes, 158 were upregulated when maternal rest was absent. Genuine targets of maternal Rest would likely be misregulated in both Mrestsbu29/+ and MZrestsbu29/sbu29 embryos. Therefore, we focused on this set of transcripts. Because Rest is thought to influence gene expression over large chromosomal regions (Lunyak et al., 2002), we used an algorithm we previously developed (Johnson et al., 2006, 2009) to determine which of these genes had an RE1 site with 100 kb of the transcriptional start site. This analysis revealed that 63 genes (∼40%) had predicted RE1 sites (score >.91) located within 100 kb of the transcriptional start site. This set of shared upregulated genes were significantly enriched for RE1 sites (χ2 = 159.989, p < 0.0001).
Figure 1.
Transcriptome comparison of Mrestsbu29/+ and MZrestsbu29/sbu29. A, Venn diagram showing the overlap of upregulated and downregulated genes in Mrestsbu29/+ and MZrestsbu29/sbu29 blastula. The number of genes with a predicted RE1 site near them is indicated. B, GO analysis showing the significant biological processes that are enriched in the upregulated genes.
DAVID analysis of the upregulated genes revealed that 41 of the 158 misregulated genes are expressed in neural tissues, as would be expected of authentic Rest targets (Chong et al., 1995; Schoenherr and Anderson, 1995; Lunyak et al., 2002; Bruce et al., 2004). GO analysis of 158 upregulated genes indicated that their functions were enriched in exocytosis, synaptic transmissions, and cell–cell signaling (Fig. 1B). In addition, 56 significantly downregulated transcripts were identified, but only 9 had associated RE-1 sites. This set of downregulated genes was not enriched for RE-1 sites (Fig. 1A; χ2 = 1.990, p < 0.1583), although recent work has suggested that rest might act as an activator in some contexts (Kuwabara et al., 2004; Perera et al., 2015).
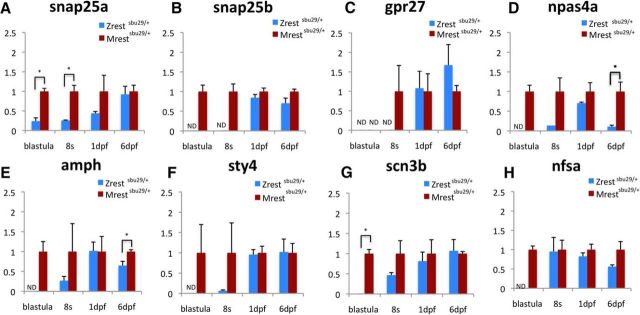
To validate the RNA-seq results, we assayed the expression of 15 upregulated RE-1-associated genes by qPCR in Mrestsbu29/+ cDNA. These genes were selected based on the significance of altered expression in the transcriptome analysis. Among them are amphiphysin, the most significantly misregulated gene, known zygotic Rest targets (snap25a, snap25b, gpr27, and syt4) (Kok et al., 2012; Love and Prince, 2015) and genes with a diversity of functions, including an ion channel (scn3b), an axon growth regulator (nsfa), and a transcription factor (npas4a) (Bruce et al., 2004).
At blastula stage, qPCR confirmed that 14 of 15 genes tested are upregulated in Mrestsbu29/+ (Fig. 2; data not shown). The remaining gene, gpr27, was not detectable by qPCR in either Mrestsbu29/+ or Zrestsbu29/+ at blastula stage. Based on these results, we conclude that identification of derepressed RE1-containing genes in the RNA-seq experiment had a low false-positive rate.
Figure 2.
RE1-containing genes are upregulated in Mrestsbu29/+ embryos. qPCR analysis showing fold differences relative to the Mrestsbu29/+ transcript levels (defined as 1). Significance: *p < 0.05 (Student's t test). All markers shown are upregulated at blastula stage in Mrestsbu29/+ embryos. snap25a (A), snap25b (B), and gpr27 (C) are upregulated at the 8 somite stage (11.5 hpf). npas4a (D) and amph (E) are upregulated at 6 dpf. ND, Not detectable.
Transcriptional effects of maternal rest depletion persist beyond blastula stages
To determine whether maternal rest is required to maintain gene expression profiles of target genes at later stages, we assayed expression of the same target genes 7.5 h later at the 8-somite stage using qPCR. Of the 15 genes we studied, three genes (snap25a, snap25b, gpr27) were significantly derepressed in Mrestsbu29/+ embryos at 8 somites (Fig. 2; data not shown). To determine whether these effects persist, we assayed expression of a set of genes, including those showing earlier derepression at 6 d and observed derepression of amph and npas4a, but no other differences were uncovered with qPCR (Fig. 2). The stage-specific effects on individual targets, such as amph and npas4a in Mrestsbu29/+ embryos, likely stem from the presence of stage-specific transcriptional activators that play significant roles in modulating transcription of these genes.
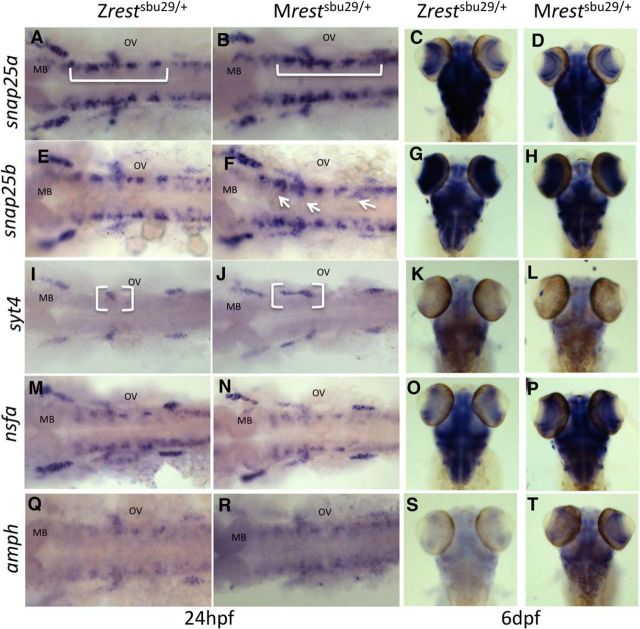
Because domain-specific differences in expression may not be detected by whole embryo qPCR, we performed RNA in situ hybridizations on 24 hpf embryos to assay gene expression in Mrestsbu29/+ embryos. It was previously shown that Rest target genes are misexpressed in the hindbrain of MZrestsbu29/sbu29 mutants at 24 hpf (Love and Prince, 2015). We observed ectopic expression of snap25a, snap25b, and syt4 in the hindbrain of Mrestsbu29/+ embryos at 24 hpf, whereas nsfa and amph expression were not altered (Fig. 3). In Mrestsbu29/+, snap25a ectopic expression spans the hindbrain and midbrain (Fig. 3A,B, bracket), whereas snap25b shows ectopic expression in hindbrain cranial ganglia (Fig. 3E,F, arrows). Syt4 has a restricted expression pattern in the hindbrain compared with snap25a and snap25b, but the domain located rostral to the otic vesicle is broadly expressed in the Mrestsbu29/+compared with Zrestsbu29/+ (Fig. 3I,J, white brackets). No spatial differences were observed in expression of nsfa or amph (Fig. 3M,N,Q,R). At 6 dpf, these genes are exclusively expressed in the brain (Fig. 3). We observed that Mrestsbu29/+ increased expression of nsfa and amph in 6 dpf Mrestsbu29/+ embryos (Fig. 3O,P,S,T) but no differences in expression of snap25a, snap25b, or syt4 at this stage (Fig. 3C,D,G,H,K,L).
Figure 3.
Rest target genes are inappropriately expressed in Mrestsbu29/+ embryos. RNA Whole-mount in situ hybridization at 24 h and 6 dpf for Rest target genes for Mrestsbu29/+ and Zrestsbu29/+. Ectopic expression (white bracket or arrow) is observed with probes for snap25a (A,B), snap25b (E,F), and syt4 (I,J) but not nfsa (M,N) or amph (Q,R) in the hindbrain of Mrestsbu29/+ embryos at 24 hpf. Increase expression in Mrestsbu29/+ observed at 6 dpf in with nsfa (O,P) and amph (S,T) but not snap25a (C,D), snap25b, or syt4 probes. OV, Otic vesicle; MB, midbrain.
Depletion of maternal rest modulates larval locomotion
In addition to derepression of rest target genes, disruption of zygotic rest results in hypolocomotion at 6 dpf (Moravec et al., 2015). To determine whether maternal rest modulates larval behavior during development, we monitored locomotor activity during spontaneous and evoked swimming behaviors in embryos lacking maternal rest mRNA at 6 dpf. Larvae were placed in 24 well plates, one animal per well, and locomotor activity was analyzed using the Zebrabox imaging system (Viewpoint).
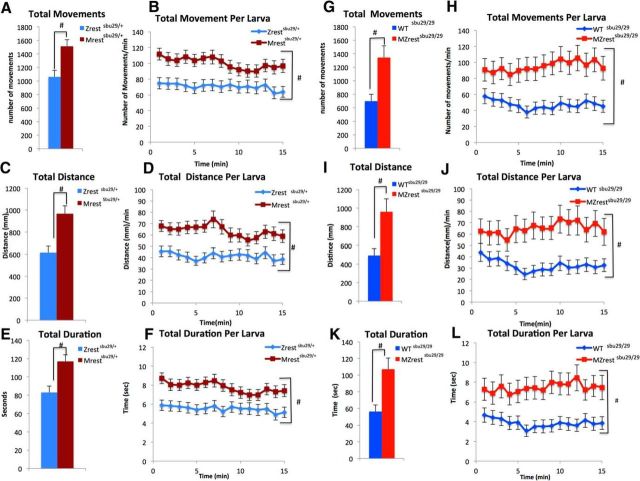
Spontaneous movements of Mrestsbu29/+, Zrestsbu29/+, MZrestsbu29/sbu29, and related WT control larvae were analyzed at 6 dpf in the light. Comparison of Mrestsbu29/+ and Zrestsbu29/+ locomotion revealed that Mrestsbu29/+ larvae move significantly more (n = 71, average of 1511 movements) than Zrestsbu29/+ (n = 72, average of 1061.57 movements) controls (Fig. 4A; p = 0.0013) over 15 min. A repeated-measures ANOVA evaluated movements over 1 min time intervals and identified a significant main effect of genotype. On average, the Zrestsbu29/+ controls traveled 70 movements/min, whereas the Mrestsbu29/+ larvae traveled 100 movements/min (Fig. 4B; Table 1). The requirement for maternal rest in modulating larval locomotor behavior was also apparent from comparisons of MZrestsbu29/sbu29 mutants (N = 48) and related WT controls (N = 72). In this assay, the MZrestsbu29/sbu29 mutants significantly surpassed the related WT controls in the number of movements, duration of movements, and distance traveled (Fig. 4G,H; Table 1). Both genotypes of maternal rest depleted larvae also show a significant increased activity in additional parameters of movement, including distance traveled and duration of movements (Fig. 4C–F,I,L; Table 1). Overall, these data revealed that the loss of maternal rest results in larval hyperactivity.
Figure 4.
Larvae lacking maternal rest are hyperactive at 6 dpf. A–F, Mrestsbu29/+ (N = 71) exceed Zrestsbu29/+ (n = 72) in total movements (A,B), total distance (C,D), and total duration (E,F) over 15 min. G–L, Similarly, MZrestsbu29/sbu29 (n = 48) exceed related WT controls (n = 72) in total movements (G,H), total distance (I,J), and total duration (K,L) over 15 min. All graphs represent mean; error bars indicate SE. Significance: Student's t test for the entire testing periods and two-way repeated-measures ANOVAs, with genotype serving as the independent factor and time serving as the repeated measure for the 1 min analysis. #Genotype (p < 0.05).
Table 1.
Two-way ANOVA with repeated-measures design to compare genotypes in 1 min intervals during spontaneous movements
| Variable | Genotype |
Time |
Time × genotype |
|||
|---|---|---|---|---|---|---|
| F | p | F | p | F | p | |
| Mrest versus Zrest | ||||||
| Total distance | 19.936154 | 0.000017 | 1.669738 | 0.103 | 0.915994 | 0.50099 |
| Total duration | 12.167352 | 0.000652 | 2.002607 | 0.036533 | 0.711894 | 0.69668 |
| Total movements | 13.437356 | 0.00035 | 2.072364 | 0.029648 | 0.836665 | 0.58133 |
| MZrest versus WT | ||||||
| Total distance | 11.085346 | 0.001179 | 0.449008 | 0.829216 | 0.88972 | 0.494282 |
| Total duration | 11.380426 | 0.001015 | 0.514421 | 0.782803 | 0.902606 | 0.48616 |
| Total movements | 16.616386 | 0.000085 | 0.639221 | 0.670649 | 1.007349 | 0.412699 |
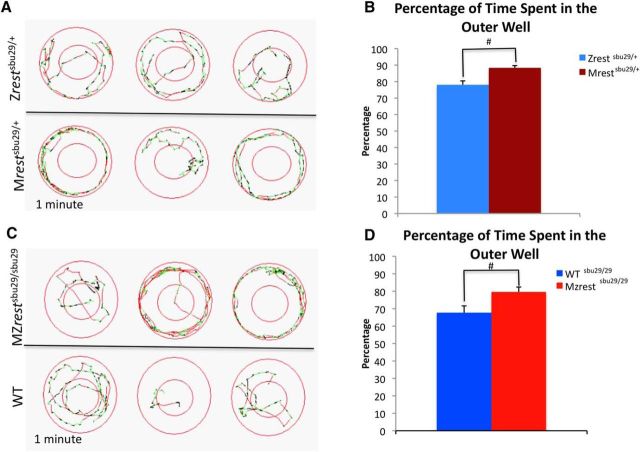
Wall preference for the four groups of larvae were assessed by calculating the percentage of time the larvae spent in both the center and the peripheral divisions of the circular wells (enter well diameter, 150 mm; center well diameter, 62 mm). Comparison of Mrestsbu29/+ versus Zrestsbu29/+ and MZrestsbu29/sbu29 versus related WT controls demonstrated that the larvae lacking maternal rest displayed a preference to be located at the periphery of the well (Fig. 5). We also examined evoked responses to a light change, but no significant differences were observed in the absence of maternal rest (data not shown). The hyperactivity and atypical spatial preference behavior that are observed in the larvae lacking maternal rest differ from that of the zygotic rest mutant (Moravec et al., 2015).
Figure 5.
Larvae lacking maternal rest show an atypical spatial preference at 6 dpf. A, C, Representative locomotion diagrams of movement in 1 min. Mrestsbu29/+ (A) and MZrestsbu29/sbu29 (C) larvae display a preference for the outer well. Green represents small velocity movements. Red represents large velocity movements during a spontaneous locomotion assay in the light. B, Quantification of percentage of time spent in the outer well over 15 min shows that Mrestsbu29/+ (n = 71) larvae spend significantly more time in the outer well compared with Zrestsbu29/+ (n = 72) (p = 0.002). D, Quantification of percentage of time spent in the outer wall over 15 min reveals that MZrestsbu29/sbu29 (n = 48) larvae spend more time in the outer well compared with related WT controls (n = 72) (p = 0. 0307). Significance was defined using a Student's t test. #p < 0.05.
Depletion of maternal rest alters adult behavior
Depletion of maternal rest in Mrestsbu29/+ or elimination of both maternal rest and zygotic rest as in MZrestsbu29/sbu29 larvae causes hyperactivity and atypical spatial preferences in spontaneous movement at 6 dpf. To determine whether depletion of maternal rest changes behavior in adults, a novel environment assay was used to measure locomotion and spatial preference at 6 months of age.
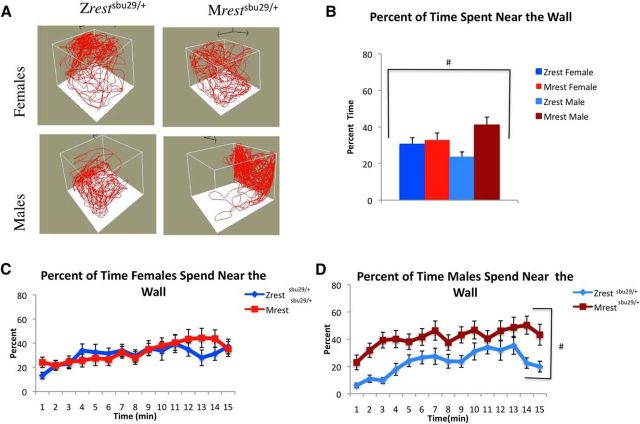
To investigate whether the effects of maternal rest on spatial preference persisted into adulthood, the amount of time that fish lacking maternal rest spent within 2.75 cm of the walls was analyzed. A comparison of Zrestsbu29/+ and Mrestsbu29/+ movement patterns revealed a strong preference of Mrestsbu29/+ males for the tank walls compared with the Zrestsbu29/+ males. No preference was observed between Mrestsbu29/+ and Zrestsbu29/+ females (Fig. 5A). A two-way ANOVA identified a significant main effect of genotype but no significant main effect of sex or sex × genotype interaction, although the sex × genotype interaction was strongly trending (Table 2). Our data showed that Mrestsbu29/+ males spend ∼40% of the interval near the edge of the tank, whereas Zrestsbu29/+ males spend ∼23% of their time near the edge of the tank. The female Mrestsbu29/+ and Zrestsbu29/+ fish spend a comparable amount of time near the edge of the tank, 32.8% and 30.6%, respectively (Fig. 6B). A within-sex analysis of time spent near the wall in 1 min intervals showed that every minute Mrestsbu29/+ males spent more time near the edge of the tank compared with Zrestsbu29/+ controls (Fig. 6D; Table 3), whereas no differences were observed when comparing females (Fig. 6C; Table 3).
Table 2.
MANOVA value from the novel environment assay to identify main effects of sex and/or genotype and significant interactions
| Variable | Sex |
Genotype |
Sex × genotype |
|||
|---|---|---|---|---|---|---|
| F | p | F | p | F | p | |
| Edge of tank | 0.011028 | 0.916645 | 6.22918 | 0.014763 | 3.472049 | 0.066329 |
Figure 6.
Mrestsbu29/+ males, but not females, showed increased wall preference in the novel environment assay. A, Locomotion diagrams for individual fish over 5 min showing the Mrestsbu29/+ male wall preference. B, During the assay, Mrestsbu29/+ (N = 21) males spent more time near the wall compared with Zrestsbu29/+ (N = 20) controls. C, D, Analysis of percentage of time spent near the walls for females (Mrestsbu29/+, N = 18; Zrestsbu29/+, N = 20) (C) and males (D) in 1 min intervals reveals that Mrestsbu29/+ males, but not females, tend to swim near the side of the tank over the entire assay. Significance was defined using a multivariate ANOVA to identify main effects of sex and/or genotype and significant interactions between the two over the testing period. A two-way repeated-measures ANOVA was also used to compare within-sex data collected in 1 min bins across the 15 min testing period. #Genotype (p < 0.05).
Table 3.
Two-way ANOVA with repeated-measures design for the novel environment assay to compare within-sex data collected in 1 min intervals across the testing period
| Edge of tank | Time |
Genotype |
Time × genotype |
|||
|---|---|---|---|---|---|---|
| F | p | F | p | F | p | |
| Sex | ||||||
| Female | 3.993977 | 0.000423 | 0.04363 | 0.835755 | 1.32888 | 0.238444 |
| Male | 5.731319 | 0.000001 | 16.835916 | 0.000223 | 1.002538 | 0.432771 |
The Mrestsbu29/+ male fish also presented with another behavioral change, erratic swimming patterns during the novel environment assay. Increased erratic swimming patterns were observed in Mrestsbu29/+ males compared with Zrestsbu29/+ males as measured by distance traveled, velocity in the vertical direction, turn angle, and location in the tank (data not shown). This behavior is similar to the movements of rest mutants of both sexes (Moravec et al., 2015).
Identification of Rest target genes that modulate locomotor behavior
To identify the Rest target genes whose misregulation produces the behavioral phenotypes we observed in the Mrestsbu29/+ and MZrestsbu29/sbu29 larvae, we deleted the RE1 elements associated with snap25a and snap25b using the CRISPR-CAS9 system. We chose these two genes because they are upregulated during embryogenesis past blastula stage (Figs. 2, 3) and have key synaptic functions. Both zebrafish snap25 paralogs have RE1 sites within the first intron, as does mammalian snap25, and Rest has been shown to frequently associate with the snap25 RE1 sites (Bruce et al., 2004).
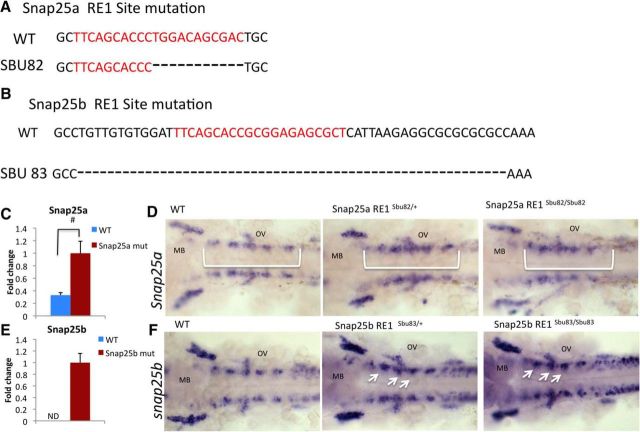
The CRISPRs were designed to recognize a portion of the RE1 site and flanking sequence to prevent cleavage events at multiple RE1 sites. RE1 sites contain two highly conserved sections (Mortazavi et al., 2006), and we aimed to delete at least one of these regions. The snap25a RE1sbu82 allele is an 11 bp deletion that removes one of these conserved regions, whereas the snap25b RE1sbu83 allele is a 53 bp deletion and removes the entire RE1 site (Fig. 7A,B).
Figure 7.
CRISPR-CAS9 targeting of RE1 sites. A, B, A sequence alignment of WT and snap25 RE1 site mutations (A) snap25asbu82 or (B) snap25bsbu83. Black represents the genomic sequence surrounding the RE1 site. Red represents RE1 site. C, E, qPCR analysis showing fold differences relative to the RE1 mutant transcript levels (defined as 1). Significance: p < 0.05 (Student's t test). D, F, RNA whole-mount in situ hybridization with (D) snap25a probe on a Snap25a RE1 sitesbu82/+ inx or (F) snap25b probe on a snap25b RE1 sitesbu83/+ inx. White bracket or white arrow indicates ectopic expression. OV, Otic vesicle; MB, midbrain. #Genotype (p < 0.05).
We first determined the effects of these RE1 site mutations on gene expression at multiple stages of development. qPCR analysis at blastula stage of snap25a in the snap25a RE1sbu82/sbu82 mutant (Fig. 7C) and snap25b in the snap25b RE1sbu83/sbu83 mutant (Fig. 7E) mirrored the upregulation of these transcripts observed in Mrestsbu29/+. RNA in situ hybridization with snap25a and snap25b probes at 24 hpf revealed ectopic expression of snap25a and snap25b in the hindbrain similar to Mrestsbu29/+ embryos. The snap25a RE1 heterozygotes and mutants both showed increase expression in the hindbrain and midbrain (as marked by the bracket) compared with sibling WTs (Fig. 7D). The snap25b RE1 heterozygous and mutants show medial ectopic expression in the hindbrain (as marked by the arrows) compared with sibling WTs (Fig. 7F).
RE-1 site mutant larvae are hyperactive
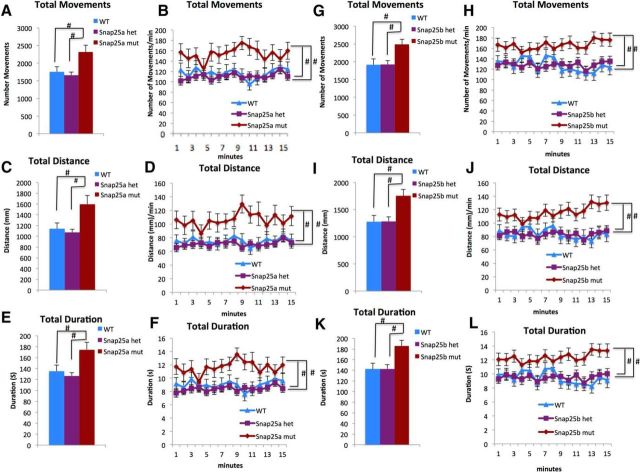
We investigated the spontaneous and light evoked movements of both the snap25a and snap25b RE-1 site mutants at 6 dpf. Remarkably, similar to the Mrestsbu29/+ and the MZrestsbu29/sbu29 larvae, the snap25a and snap25b, RE1 site mutants showed hyperactivity in spontaneous movement. Specifically, the snap25a RE1sbu82/sbu82 site mutants (n = 24) initiated significantly more swims (an average of 2316 movements), compared with sibling WTs (n = 30, an average of 1751 movements) and snap25a RE1sbu82/+ heterozygotes (n = 74, an average of 1660 movements) (Fig. 8A; Table 4). A repeated-measures ANOVA evaluated number of movements across the 1 min time bins and identified a significant main effect of genotype. The snap25a RE1sbu82/sbu82 site mutant made an average of 154 movements/min compared with the sibling WT and snap25a RE1 sbu82/+ heterozygotes who make an average of 116 movements/min and 110 movements/min, respectively (Fig. 8B; Table 5).
Figure 8.
snap25a and snap25b RE1 site mutants are hyperactive at 6 dpf. A–F, The snap25a RE1sbu82/sbu82 mutants (N = 24) exceeded sibling WT (N = 30) controls and snap25a RE1sbu82/+ heterozygotes (N = 74) in (A,B) number of movements, (C,D) distance, and (E,F) duration at 6 dpf. G–L, The snap25b RE1sbu83/sbu83 mutant (N = 44) exceeded sibling WT (N = 37) and the snap25b RE1sbu83/+ heterozygotes (N = 62) in (G,H) number of movements, (I,J) distance, and (K,L) duration at 6 dpf. Significance was defined using a one-way ANOVA over the entire test period; when the data were compared on per minute bases, the data were compared using two-way ANOVAs with repeated-measures designs, with genotype serving as the independent factor and time serving as the repeated measure. #Genotype (p < 0.05).
Table 4.
One-way ANOVA values for spontaneous movements for RE-1 mutants comparing genotypes
| Variable | One-way ANOVA |
LSD post hoc |
|||
|---|---|---|---|---|---|
| F | p | WT/Het | WT/Mut | Het/Mut | |
| Snap25a RE1sitesbu82 | |||||
| Total counts | 6.520921 | 0.002024 | 0.591579 | 0.009079 | 0.000481 |
| Total distance | 7.413463 | 0.000907 | 0.477887 | 0.013031 | 0.000469 |
| Total duration | 6.485344 | 0.00209 | 0.60111 | 0.004972 | 0.000212 |
| Snap25b RE1sitesbu83 | |||||
| Total counts | 5.724394 | 0.004077 | 0.976402 | 0.006448 | 0.002353 |
| Total distance | 6.68793 | 0.001683 | 0.992516 | 0.003439 | 0.001012 |
| Total duration | 6.259185 | 0.002491 | 0.964836 | 0.004341 | 0.00152 |
Table 5.
Two-way ANOVA with repeated-measures design to compare genotypes in 1 min intervals during spontaneous movements
| Variable | One-way ANOVA |
LSD post hoc |
|||
|---|---|---|---|---|---|
| F | p | WT/Het | WT/Mut | Het/Mut | |
| Snap25a RE1sitesbu82 | |||||
| Total counts | 6.520921 | 0.002024 | 0.591579 | 0.009079 | 0.000481 |
| Total distance | 7.413463 | 0.000907 | 0.477887 | 0.013031 | 0.000469 |
| Total duration | 6.485344 | 0.00209 | 0.60111 | 0.004972 | 0.000212 |
| Snap25b RE1sitesbu83 | |||||
| Total counts | 5.724394 | 0.004077 | 0.976402 | 0.006448 | 0.002353 |
| Total distance | 6.68793 | 0.001683 | 0.992516 | 0.003439 | 0.001012 |
| Total duration | 6.259185 | 0.002491 | 0.964836 | 0.004341 | 0.00152 |
The snap25b RE1sbu83/sbu83 mutants displayed a similar behavior to Mrestsbu29/+ larvae. These mutants engaged (n = 44) in an average of 2490 movements compared with the sibling WT (n = 37) an average of 1918 movements and snap25b RE1sbu83/+ heterozygotes (n = 62) an average of 1924 movements (Fig. 8G; Table 4). A repeated-measures ANOVA of the number of movements revealed a significant main effect of genotype. The snap25b RE1sbu83/sbu83 site mutant made an average of 166 movements/min compared with the sibling WTs and snap25b RE1sbu83/+ heterozygotes that averaged 127movements/min and 128 movements/min, respectively (Fig. 8H; Table 5). We also examined distance traveled and duration of movements and found that both the snap25a and snap2b RE1 mutants surpassed the related WTs and heterozygotes in both parameters (Fig. 8C–F,I–L; Tables 4, 5). Neither of these RE1 mutants presented with an atypical spatial preference or showed a response to a light change (data not shown). These results indicate that the rest regulation at snap25a and snap25b is sufficient to controlling locomotor behavior, but not spatial preference at 6 dpf.
Motor neurons in Mrestsbu29/+ and snap25a RE1sbu82/sbu82 site mutants have increased processes
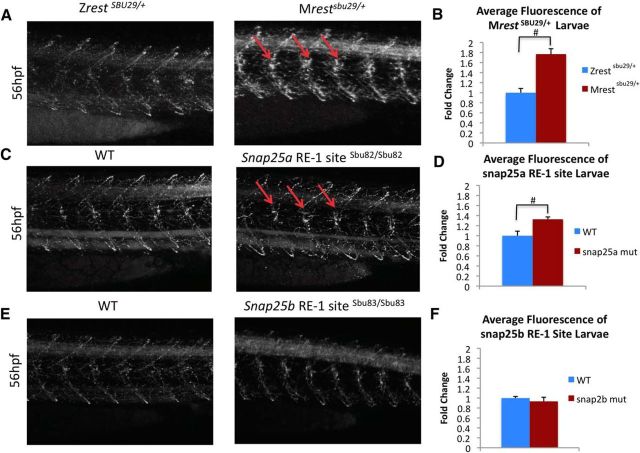
Increased expression of the zebrafish snap25 paralogs results in hyperactivity, increased branching of motor neurons, and changes to the synaptic activity at neuromuscular junctions (Wei et al., 2013). To investigate changes in the primary motor neuron architecture in the Mrestsbu29/+ and the snap25 RE1site mutants, we performed whole-mount immunostaining with Znp-1, synaptotagmin IIB(syt2b), at 56 hpf. We observed increased expression of Znp-1 in the spinal cord along with increased Znp-1 puncta associated with primary motor neurons in Mrestsbu29/+ embryos (n = 5) (marked by a red arrow) compared with ZrestSBU29/+ (n = 4) (Fig. 9A). Quantification of average fluorescence in these ZNP-1 puncta showed a significant increase in fluorescence in the Mrestsbu29/+ embryos (p = 0.0010) (Fig. 9B). We also examined the primary motor neuron architecture of the snap RE1 site mutants and observed increased Znp-1 staining (marked by red arrows) in the snap25a RE1sbu82/sbu82 site mutant (n = 7), but not the snap25bRE1sbu83/sbu83 site mutant (n = 7) compared with WT controls (snap25aRE1 site control = 7 and snap25bRE1 site control = 7) (Fig. 9B,C,E). Quantification of the ZNP-1 puncta in the snap25a RE1sbu82/sbu82 and snap25bRE1sbu83/sbu83 site mutants revealed a significant increase of fluorescence in the snap25a RE1 mutant (p = 0.0078), but not in snap25bRE1 mutant (Fig. 9D,F). These results suggest that alterations of the neuromuscular junction in MrestSBU29/+ larvae stem from derepression of snap25a, but that regulation of snap25b expression by maternally supplied Rest is important elsewhere.
Figure 9.
Rest regulates primary motor neuron development. znp-1 immunohistochemistry on whole-mount zebrafish embryos at 56 hpf to label primary motor neurons. Confocal images were acquired from the truck using the yolk extension as a landmark (10 μm stacks). A, C, Changes in the primary motor neuron architecture are apparent in Mrestsbu29/+ and snap25a RE1 site mutant embryos compared with controls. B, D, Significant increase in fluorescence was observed in the Mrestsbu29/+ and snap25a RE1 site mutant. Significance was defined using the Student's t test, and control was set to 1. No changes are apparent in primary motor neuron architecture; fluorescence was observed in snap25b RE1 site mutant embryos (E,F). #Genotype (p < 0.05).
Discussion
Our previous work demonstrated that zebrafish rest mutants undergo largely normal neurogenesis (Kok et al., 2012) but that rest mutant larvae show locomotor defects and engage in erratic swimming as adults (Moravec et al., 2015). We now present evidence that the effects of maternally supplied rest limits expression of a subset of target genes until at least 6 dpf and that larvae lacking maternal rest are hyperactive and present with a spatial preference for outer portion of the well compared with controls. To our knowledge, this is the first example of a maternally supplied mRNA that modulates behavior. Remarkably, behavioral consequences of the deficit in the early maternal rest expression persist into adulthood, as observed by the erratic swimming behavior and atypical place preference that was apparent in adult Mrestsbu29/+ males, but not females.
Rest has been proposed to play important roles in stem and progenitor cells to control self-renewal and differentiation in the nervous system (Ballas et al., 2005; Singh et al., 2008). Although we cannot conclusively rule out the possibility that maternal rest deficit alters cell fate, we have found no evidence for major cell fate changes in any of the rest mutants. Furthermore, because the larval hyperactivity phenotype can be recapitulated by disrupting the RE1 sites in either snap25a or snap25b, we favor the model that the primary effects are on gene expression of these rest target genes. This is consistent with the observations in rodents that early Rest-mediated epigenetic effects regulate the later developmental switch in synaptic NMDA receptors (Rodenas-Ruano et al., 2012).
Rest levels in mammals are diminished by maternal deprivation and elevated by augmented maternal care (Korosi et al., 2010; Uchida et al., 2010; Rodenas-Ruano et al., 2012). Whereas zebrafish do not engage in maternal care, MrestSBU29/+ embryos face a similar early deficit in Rest activity. Our transcriptome analysis did not identify GRIN2b as a key Rest target, as has been demonstrated in the rat studies of early Rest function (Rodenas-Ruano et al., 2012). Instead, our work implicates the two snap25 paralogs as key mediators of the observed behavioral phenotypes. Nonetheless, the data in rodents and fish may point to a fundamental role for Rest in establishing chromatin landscapes that have later impacts on expression of neural genes and neuronal function.
The half-life of the protein generated from maternal rest RNA is unknown, but the maternal mRNA is degraded by about shield stage, 6 h after fertilization (unpublished results). Because Rest protein is actively degraded (Westbrook et al., 2008; Kaneko et al., 2014), it seems likely that the protein has vanished long before gene expression (Figs. 2, 3), and behavioral defects (Figs. 4, 5) are observed at 6 dpf. During this period, zygotic rest is expressed (Gates et al., 2010), yet is unable to compensate for the loss of early Rest activity. The adult behavioral analysis further suggests an early unique role for maternally supplied rest in establishing chromatin states that persist lifelong. However, our data do not exclude the possibility that the effects stem from consequences of cumulative transgenerational consequences of Rest deficiency as has been observed in C. elegans mutants for the Rest complex protein, LSD1 (Katz et al., 2009).
Transcripts regulated by maternal rest
Bioinformatic analysis indicated that we enriched for both neural-specific and RE1-containing genes in the upregulated set of genes in Mrestsbu29/+ identified by RNA-seq. Our qPCR validation of 14 RE1-containing genes demonstrated that the approach robustly identified Rest targets. The downregulated genes were not enriched for RE1 sites or for neural genes, but recent work has suggested that rest might act as an activator in some contexts (Kuwabara et al., 2004; Perera et al., 2015). However, if Rest acts as an activator at blastula stages, the number of targets is quite low. Alternatively, downregulation of some transcripts could be due to secondary effects, which are expected at a low frequency because the sequence analysis was performed less than an hour (at 4 hpf) after the mid-blastula transition (Kimmel et al., 1995).
Regulation of synaptic proteins by Rest
Many of the genes regulated by maternal rest encode synaptic proteins. Indeed, the five genes that show persistent misregulation, snap25b, snap25a, syt4, npas4a, and amph, all act on presynaptic neurons. The snap25 paralogs and Syt4 enable binding of the synaptic vesicles to the presynaptic density allowing for exocytosis of the neurotransmitters into the synaptic cleft, whereas amph promotes recycling of empty synaptic vesicles from the presynaptic density after exocytosis. Npas4a regulates the expression of inhibitory synapse genes to control the excitatory/inhibitory balance in presynaptic cells. While disrupting the snap25 RE1 sites recapitulates much of the larval locomotor observed in maternal deficient larva, it is likely that misregulation of other targets produces behavioral consequences. In particular, the atypical spatial preferences observed in Mrestsbu29/+ and MZrestsbu29/sbu29 were not apparent in the RE1 site mutants.
Regulation of behavior by Rest
Zygotic rest mutant larvae are hypoactive (Moravec et al., 2015), although we now demonstrate that fish lacking maternal rest are hyperactive and demonstrate atypical spatial preferences, spending more time near the wall. These data suggest that maternal rest plays a distinct role from zygotic rest in modulating locomotive behavior at 6 dpf.
Adult zygotic rest mutants of both sexes display atypical spatial preferences in a novel environment assay characterized by edge preferences and erratic swimming (Moravec et al., 2015). When adult Mrestsbu29/+ fish underwent the same test, only the males, but not females, presented with similar phenotypes to the zygotic mutants. The observation that depletion of a maternal RNA effects behavior in a sex-specific manor is unusual and suggests that life-long effects on the epigenetic genome may be strongly influenced by sex hormones.
Changes to the architecture of primary motor neurons
Mrestsbu29/+ embryos display increased expression of Syt2b in trunk motor neurons compared with Zrestsbu29/+. This suggests a possible molecular mechanism for the hyperactivity observed in the Mrestsbu29/+ larvae (Fig. 4) as decreased locomotion has been linked to changes in axon formation and elongation of the motor neurons (Granato et al., 1996).
We also investigated the primary motor neuron architecture in the snap25 RE1 sites mutants because they are also hyperactive (Fig. 7), and increased expression of Snap25 is linked to both axon growth (Wei et al., 2013; W. Wang et al., 2014) and hyperactivity (Wei et al., 2013). We observed increased expression of Syt2b at the neuromuscular junction of the snap25a RE1sbu82/sbu82 mutant, but not in the snap25b RE1sbu83/sbu83 mutant. This suggests that the increased number of processes associated with primary motor neurons in Mrestsbu29/+ larvae is due to derepression of snap25a in the absence of maternal rest. The behavioral phenotypes of Mrestsbu29/+ are more complex because disrupting the RE1 site of snap25b results in hyperactivity, but not overt changes of Syt2b expression in motor neurons. Enhanced Snap25b levels may alter synaptic plasticity by altering trafficking/exocytosis of synaptic vesicles while not overtly altering the complexity of motor neuron processes. Because neither snap25 RE1 site mutant displays altered spatial preferences, regulation of other target gene by Rest must be responsible for this phenotype. It is likely that some of these genes also impact swimming frequency as well.
We present the first evidence that maternal rest plays a long-term role in regulation of gene expression and behavior during development. The activity of maternally supplied rest controls expression of target genes and affects behavior not only in larvae, but in adults as well. By rendering the zebrafish snap25 paralogs impervious to Rest-mediated repression at these RE1 sites, we determined that snap25a/b are key targets of maternal rest involved in modulating primary motor neuron development and larval swimming frequency. These findings strengthen the idea that a major function of Rest is to regulate synaptic activity and plasticity (Rodenas-Ruano et al., 2012). The zebrafish rest mutant provides a unique opportunity to explore the lasting requirements for maternal factors in nervous system function. This study provides the first evidence that maternal rest is necessary for long-term regulation of both gene expression and behavior.
Footnotes
This work was supported by NYSTEM C026414 and National Institutes of Health Grant 1R03HD1066000 to H.I.S. We thank our many colleagues for experimental support and advice; Dr. Victoria Prince for the syt4 plasmid; Dr. Ian Woods for the nsfa plasmid; Dr. Shaoyu Ge and Adrian Di Antonio for imaging advice; Neal Bhattacharji and the undergraduate assistants for fish care; and Drs. Nurit Ballas and Bernadette Holdener for comments on this manuscript.
The authors declare no competing financial interests.
References
- Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, Federoff HJ, Rose DW, Rosenfeld MG, Brehm P, Mandel G. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/S0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Göttgens B, Buckley NJ. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci U S A. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 2006;20:1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramírez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Flores MV, Lam EY, Crosier KE, Crosier PS. Osteogenic transcription factor Runx2 is a maternal determinant of dorsoventral patterning in zebrafish. Nat Cell Biol. 2008;10:346–352. doi: 10.1038/ncb1697. [DOI] [PubMed] [Google Scholar]
- Gates KP, Mentzer L, Karlstrom RO, Sirotkin HI. The transcriptional repressor REST/NRSF modulates hedgehog signaling. Dev Biol. 2010;340:293–305. doi: 10.1016/j.ydbio.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato M, van Eeden FJ, Schach U, Trowe T, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Nüsslein-Volhard C. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development. 1996;123:399–413. doi: 10.1242/dev.123.1.399. [DOI] [PubMed] [Google Scholar]
- Grzenda A, Lomberk G, Zhang JS, Urrutia R. Sin3: master scaffold and transcriptional corepressor. Biochim Biophys Acta. 2009;1789:443–450. doi: 10.1016/j.bbagrm.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T, Elgin SC. Maternal depletion of Piwi, a component of the RNAi system, impacts heterochromatin formation in Drosophila. PLoS Genet. 2013;9:e1003780. doi: 10.1371/journal.pgen.1003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivorra C, Fraga MF, Bayón GF, Fernández AF, Garcia-Vicent C, Chaves FJ, Redon J, Lurbe E. DNA methylation patterns in newborns exposed to tobacco in utero. J Transl Med. 2015;13:25. doi: 10.1186/s12967-015-0384-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Gamblin RJ, Ooi L, Bruce AW, Donaldson IJ, Westhead DR, Wood IC, Jackson RM, Buckley NJ. Identification of the REST regulon reveals extensive transposable element-mediated binding site duplication. Nucleic Acids Res. 2006;34:3862–3877. doi: 10.1093/nar/gkl525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Samuel J, Ng CK, Jauch R, Stanton LW, Wood IC. Evolution of the vertebrate gene regulatory network controlled by the transcriptional repressor REST. Mol Biol Evol. 2009;26:1491–1507. doi: 10.1093/molbev/msp058. [DOI] [PubMed] [Google Scholar]
- Kaneko N, Hwang JY, Gertner M, Pontarelli F, Zukin RS. Casein kinase 1 suppresses activation of REST in insulted hippocampal neurons and halts ischemia-induced neuronal death. J Neurosci. 2014;34:6030–6039. doi: 10.1523/JNEUROSCI.4045-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz DJ, Edwards TM, Reinke V, Kelly WG. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell. 2009;137:308–320. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kok FO, Taibi A, Wanner SJ, Xie X, Moravec CE, Love CE, Prince VE, Mumm JS, Sirotkin HI. Zebrafish rest regulates developmental gene expression but not neurogenesis. Development. 2012;139:3838–3848. doi: 10.1242/dev.080994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korosi A, Shanabrough M, McClelland S, Liu ZW, Borok E, Gao XB, Horvath TL, Baram TZ. Early-life experience reduces excitation to stress-responsive hypothalamic neurons and reprograms the expression of corticotropin-releasing hormone. J Neurosci. 2010;30:703–713. doi: 10.1523/JNEUROSCI.4214-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116:779–793. doi: 10.1016/S0092-8674(04)00248-X. [DOI] [PubMed] [Google Scholar]
- Love CE, Prince VE. Rest represses maturation within migrating facial branchiomotor neurons. Dev Biol. 2015;401:220–235. doi: 10.1016/j.ydbio.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- Moravec CE, Li E, Maaswinkel H, Kritzer MF, Weng W, Sirotkin HI. Rest mutant zebrafish swim erratically and display atypical spatial preferences. Behav Brain Res. 2015;284:238–248. doi: 10.1016/j.bbr.2015.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Leeper Thompson EC, Garcia ST, Myers RM, Wold B. Comparative genomics modeling of the NRSF/REST repressor network: from single conserved sites to genome-wide repertoire. Genome Res. 2006;16:1208–1221. doi: 10.1101/gr.4997306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse Y, Aoki T, Kojima T, Mori N. Neural restrictive silencer factor recruits mSin3 and histone deacetylase complex to repress neuron-specific target genes. Proc Natl Acad Sci U S A. 1999;96:13691–13696. doi: 10.1073/pnas.96.24.13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera A, Eisen D, Wagner M, Laube SK, Künzel AF, Koch S, Steinbacher J, Schulze E, Splith V, Mittermeier N, Müller M, Biel M, Carell T, Michalakis S. TET3 is recruited by REST for context-specific hydroxymethylation and induction of gene expression. Cell Rep. 2015;11:283–294. doi: 10.1016/j.celrep.2015.03.020. [DOI] [PubMed] [Google Scholar]
- Petropoulos S, Guillemin C, Ergaz Z, Dimov S, Suderman M, Weinstein-Fudim L, Ornoy A, Szyf M. Gestational diabetes alters offspring DNA methylation profiles in human and rat: identification of key pathways involved in endocrine system disorders, insulin signaling, diabetes signaling, and ILK signaling. Endocrinology. 2015;156:2222–2238. doi: 10.1210/en.2014-1643. [DOI] [PubMed] [Google Scholar]
- Rodenas-Ruano A, Chávez AE, Cossio MJ, Castillo PE, Zukin RS. REST-dependent epigenetic remodeling promotes the developmental switch in synaptic NMDA receptors. Nat Neurosci. 2012;15:1382–1390. doi: 10.1038/nn.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell. 2004;14:727–738. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ. Silencing is golden: negative regulation in the control of neuronal gene transcription. Curr Opin Neurobiol. 1995;5:566–571. doi: 10.1016/0959-4388(95)80060-3. [DOI] [PubMed] [Google Scholar]
- Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Cyr S, McGowan PO. Programming of stress-related behavior and epigenetic neural gene regulation in mice offspring through maternal exposure to predator odor. Front Behav Neurosci. 2015;9:145. doi: 10.3389/fnbeh.2015.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Funato H, Hobara T, Otsuki K, Yamagata H, McEwen BS, Watanabe Y. Early life stress enhances behavioral vulnerability to stress through the activation of REST4-mediated gene transcription in the medial prefrontal cortex of rodents. J Neurosci. 2010;30:15007–15018. doi: 10.1523/JNEUROSCI.1436-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangeel EB, Izzi B, Hompes T, Vansteelandt K, Lambrechts D, Freson K, Claes S. DNA methylation in imprinted genes IGF2 and GNASXL is associated with prenatal maternal stress. Genes Brain Behav. 2015;14:573–582. doi: 10.1111/gbb.12249. [DOI] [PubMed] [Google Scholar]
- Wang W, Wang F, Liu J, Zhao W, Zhao Q, He M, Qian BJ, Xu Y, Liu R, Liu SJ, Liu W, Liu J, Zhou XF, Wang TH. SNAP25 ameliorates sensory deficit in rats with spinal cord transection. Mol Neurobiol. 2014;50:290–304. doi: 10.1007/s12035-014-8642-8. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Wei C, Thatcher EJ, Olena AF, Cha DJ, Perdigoto AL, Marshall AF, Carter BD, Broadie K, Patton JG. miR-153 regulates SNAP-25, synaptic transmission, and neuronal development. PLoS One. 2013;8:e57080. doi: 10.1371/journal.pone.0057080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook TF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A, Leng Y, Maehr R, Shi Y, Harper JW, Elledge SJ. SCFβ-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008;452:370–374. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Houston DW, King ML, Payne C, Wylie C, Heasman J. The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell. 1998;94:515–524. doi: 10.1016/S0092-8674(00)81592-5. [DOI] [PubMed] [Google Scholar]