Abstract
Genome-wide association studies have found SNPs at 17q22 to be associated with breast cancer risk. To identify potential causal variants related to breast cancer risk, we performed a high resolution fine-mapping analysis that involved genotyping 517 SNPs using a custom Illumina iSelect array (iCOGS) followed by imputation of genotypes for 3,134 SNPs in more than 89,000 participants of European ancestry from the Breast Cancer Association Consortium (BCAC). We identified 28 highly correlated common variants, in a 53 Kb region spanning two introns of the STXBP4 gene, that are strong candidates for driving breast cancer risk (lead SNP rs2787486 (OR = 0.92; CI 0.90–0.94; P = 8.96 × 10−15)) and are correlated with two previously reported risk-associated variants at this locus, SNPs rs6504950 (OR = 0.94, P = 2.04 × 10−09, r2 = 0.73 with lead SNP) and rs1156287 (OR = 0.93, P = 3.41 × 10−11, r2 = 0.83 with lead SNP). Analyses indicate only one causal SNP in the region and several enhancer elements targeting STXBP4 are located within the 53 kb association signal. Expression studies in breast tumor tissues found SNP rs2787486 to be associated with increased STXBP4 expression, suggesting this may be a target gene of this locus.
Breast cancer is one of the most common epithelial malignancies in women1,2. Ahmed et al.3 carried out a multi-stage genome wide association study (GWAS) for breast cancer susceptibility involving studies from the Cancer Genetic Markers of Susceptibility (CGEMS) and Breast Cancer Association Consortium (BCAC) and reported strong evidence for a susceptibility locus at 17q22 with single nucleotide polymorphism (SNP) rs6504950, OR = 0.95, 95% confidence interval (CI) 0.92–0.97, P = 1.4 × 10−8. Turnbull et al.4 found confirmatory evidence for association with SNPs at the same locus; they reported a breast cancer risk association with SNP rs1156287 (OR = 0.91; 95% CI 0.85–0.97; P = 5.8 × 10−3), which lies 20 kb from originally reported SNP rs6504950 (r2 = 0.91). Using data from the National Cancer Institute's Breast and Prostate Cancer Cohort Consortium (BPC3), Campa et al.5 also confirmed the association with rs6504950 (OR = 0.92; 95% CI 0.88–0.97; P = 5.83 × 10−4). Broeks et al.6 further investigated this association with respect to tumor estrogen receptor (ER) status, and reported that rs6504950 had stronger association with ER positive (ER+; OR = 0.93; 95% CI 0.90–0.95; P = 7.2 × 10−7) than ER negative disease (ER−; OR = 1.00; 95% CI 0.95–1.05; P = 0.94). Tang et al.7 conducted a meta-analysis which further confirmed the association with SNP rs6504950 (OR = 0.93; 95% CI 0.87–0.99). SNP rs6504950 lies in an intron of STXBP4 (Syntax binding protein 4) and two other genes are found within 200 kb including COX11 (cytochrome C assembly protein 11) and TOM1L1 (target of myb1-like1). As part of the Collaborative Oncological Gene-Environment Study (COGS), we conducted a comprehensive fine-scale mapping of this 17q22 breast cancer susceptibility locus using 517 SNPs chosen to give dense coverage across this locus. These were genotyped on a custom-designed Illumina iSelect genotyping array (iCOGS) in 50 studies participating in BCAC. We used these data to define the variants most strongly associated with risk, and combined these data with additional in-silico and functional data in an attempt to determine the most likely causal variants.
Material and Methods
Genetic Mapping
Tagging strategy for the fine-scale mapping
We defined the region for fine-mapping by identifying the flanking SNPs with minor allele frequency (MAF) > 2% and detectable correlation (r2 > 0.1) with rs6504950, based on the 1000 genomes project European population (March 2010 Pilot version 60 CEU project data). From this 468 kb interval we selected all SNPs correlated with rs6504950 at r2 > 0.1, plus a set of SNPs designed to tag all remaining SNPs with r2 > 0.9. We thus aimed to genotype 525 SNPs, between chromosome 17 positions 52,816,899 and 53,284,506 (NCBI build 37 assembly), that had an Illumina designability score (DS) > 0.9. Of these, 517 were successfully genotyped on the array and passed QC filters.
iCOGS genotyping and imputation
Case and control samples were drawn from studies participating in the BCAC, of which 41 (total: 46450 cases/42600 controls) were predominantly of European ancestry and nine (6269 cases/6624 controls) of Asian ancestry. We performed iCOGS genotyping in four centres, as part of the Collaborative Oncological Gene-Environment Study (COGS). All BCAC studies had local human ethical approvals as described previously8. We then used the genotype data from 517 SNPs that passed quality control to impute genotypes, among European subjects, at all additional known variants in the interval, using IMPUTE version 2.0 (IMPUTE2; without pre-phasing) and the 1000 genome project multi-population data (March 2012 version) as a reference panel9,10. IMPUTE2 was run with default parameters and "effective size" of the population Ne = 20,000. Using an imputation–r2 > 0.3 in Europeans, we successfully imputed 3,134 SNPs (MAF ≥ 1%).
Statistical Analysis
For each SNP, we estimated the per-allele log-odds ratio (OR) and standard error using logistic regression, including principal components and per-study fixed-effects to capture study-specific differences as previously described8. For the analyses of European subjects, we included the first six principal components as covariates, together with a seventh component derived specifically for one study (LMBC) for which there was substantial inflation not accounted for by the components derived from the analysis of all studies (this component was set to zero for all other studies). For the analysis of Asian subjects, we included two principal components8. We estimated per allele ORs under the assumption of a log-additive mode of inheritance, i.e. SNPs were coded according to the number of minor alleles 0, 1 or 2. We estimated main effects by subtype specific status (ER +/−) using case-control logistic regression and restricting the case sample to a specific subtype. We evaluated heterogeneity of association across tumour subtypes in a case-only analysis, treating subtype status as a dependent variable. We derived the P values by means of a likelihood-ratio test (one degree of freedom). Tests were two-sided. We carried out analyses separately among women of European and of Asian ancestry, defined by multiple dimensional scaling as previously described8. We performed multiple logistic regression analyses to identify SNPs independently associated with each phenotype. To identify the most parsimonious model, we included all SNPs with a P < 10−4 and MAF ≥ 2% in the single SNP analysis in forward selection regression analyses, utilizing the step function in R11 with penalty term set to 2012; we also used a joint analysis of all SNPs using a Bayesian-inspired penalised maximum likelihood approach (HyperLasso)13. To correctly account for uncertainty in the data resulting from the imputation process, we conducted analysis by regressing on the allele dosage for each genotype. For HyperLasso we utilized the most probable genotype as input, based on the posterior probability from the imputation algorithm (set to missing if all posterior probabilities were <0.9).
Bioinformatic analyses
We combined multiple sources of in silico annotation from public databases to help identify potential functional SNPs. To investigate functional elements enriched across the previously defined fine-mapped region, more specifically in the region encompassing the strongest candidate causal SNPs, we analysed chromatin biofeatures data from the Encyclopedia of DNA Elements (ENCODE) Project14 namely: Chromatin State Segmentation by Hidden Markov Models (chromHMM), DNase I hypersensitivity sites (DHS) and histone modifications of epigenetic markers H3K4, H3K9, and H3K27 in Human Mammary Epithelial Cells (HMEC) and MCF7 breast cancer cells. To identify putative target genes, we examined chromatin interactions between distal and proximal regulatory transcription-factor binding sites and gene promoters, using Chromatin Interaction Analysis by Paired End Tag (ChiA-PET) in MCF7 cells. This detects genome-wide interactions associated with CCCTC-binding factor (CTCF) and DNA polymerase II (Pol2) – both involved in transcriptional regulation15. Putative regulatory elements were determined using data from ENCODE, Roadmap Epigenomics16, the “Predicting Specific Tissue Interactions of Genes and Enhancers” (PreSTIGE)17 algorithm, Hnisz18 and FANTOM. Intersections between candidate causal variants and regulatory elements were identified using Galaxy, and visualised in the UCSC Genome Browser. We used the ENCODE RNAseq data to evaluate the expression of exons across the 17q22 locus in HMEC and MCF7 cell lines. The alignment files for HMEC (4 biological replicates) and MCF7 (19 biological replicates) were downloaded from ENCODE and the read count in the defined region was extracted and normalized in reads per million (RPM).
Allele specific expression (ASE) analysis
ASE analysis was performed using The Cancer Genome Atlas (TCGA) breast cancer data as described previously19. SNP rs2787481, genotyped on the Affymetrix SNP Array 6.0 was used as a representative SNP for the candidate causal variants (r2 = 0.90 with rs2787486). SNP rs2787481 genotype calls and the corresponding confidence scores were retrieved using level 2 TCGA SNP array Birdseed data downloaded from TCGA portal. Genotypes with confidence scores equal to or above 0.1 were excluded.
We utilised RNA-sequencing data from 742 breast cancer samples from women of Caucasian ancestry. The corresponding RNA-sequencing BAM files and metadata are available from the Cancer Genomics Hub (CGHub). Markers used to assess relative allelic expression were exonic SNPs located in KIF2B, TOM1L1, COX11, STXBP4, HLF, MMD, TMEM100, PCTP, and ANKFN1 extracted from dbSNP human Build 142. Homozygote marker SNPs, those with low coverage (less than 15x) and those within overlapping regions of the target genes, were removed. RNA-sequencing read counts on SNP sites for reference and alternative alleles were computed. The major allele fraction (μ), representing allelic imbalance for each marker SNP, was computed and an average of allelic imbalances for each gene was calculated for individual tumour samples. Marker SNPs with extreme μ values (μ > 0.75) were not included in the analysis. Level 3 SNP array data were downloaded from TCGA portal and GISTIC version 2.0.16 was used to identify copy number variations (CNVs) for each sample. Samples with low or high CNV levels, as presented in the gene-based GISTIC module report, were excluded from the analysis of the corresponding gene.
Allelic imbalance for the target transcripts was compared between rs2787481 heterozygote (CT) and homozygote (CC and TT) samples using Levene's Test for equality of variances. KIF2B, TMEM100, and ANKFN1 were excluded from the statistical analyses as they did not have enough informative marker SNPs left after applying the filtering criteria.
Local gene expression by SNP (eQTL) association analysis
We examined the association of all genotyped or imputed SNPs with expression of nine genes (KIF2B, TOM1L1, COX11, STXBP4, HLF, MMD, TMEM100, PCTP, and ANKFN1) in the 1 Mb region on either side of the fine-mapping interval, using data from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) study. METABRIC comprises normal tissues adjacent to tumours from breast cancer patients genetically confirmed to be of European ancestry20. The samples (n = 135) were assayed for expression with the Illumina HT-12 v3 microarray. Matched germline SNP genotypes were derived using the Affymetrix SNP 6.0 array. Genotyping quality control and imputation for the METABRIC data are described in Guo et al.21. Association between genotype and expression was tested by linear regression with FDR control as implemented in the MatrixEQTL22 package in R11.
Four additional SNP-expression data sets were available and analysed separately: (1) NB116 consists of 116 Caucasian normal breast samples (the majority of Norwegian descent) with n = 10 tumour-adjacent normal biopsies. (2) BC241 consists of 241 Caucasian tumor (all stages) samples (the majority of Norwegian origin). These were both genotyped on the “iCOGS” SNP array, and gene expression levels were measured with Agilent 44 K23. (3) NB93 consists of 93 Caucasian adjacent normal breast samples from TCGA. Birdseed processed germline genotype data from the Affy6 SNP array were obtained from the TCGA dbGaP data portal24. (4) BC765 consists of 765 Caucasian breast tumour samples from TCGA24. Gene expression levels were assayed by RNA sequencing, RSEM (RNAseq by Expectation-Maximization25) normalized per gene or isoform, as obtained from the TCGA consortium24. Unexpressed and minimally expressed genes/isoforms whose sum in expression level was less than ten were excluded, and the data log2 transformed prior to analysis. The influence of SNPs on local gene expression (transcripts within 1 MB from the most strongly associated SNP) was assessed using a linear regression model, as implemented in the R11 library eMAP26. An additive effect was assumed by modelling the patient’s number of copies of the rare allele, i.e. 0, 1 or 2 for a given genotype. Correction for multiple testing was performed using the false discovery rate (FDR) as implemented in the p.adjust function in R.
eQTL data from the Genotype-Tissue Expression (GTEx) project27 were downloaded from the v6 release.
Results
A total of 517 SNPs at chromosome 17 positions 52,816,899 to 53,284,506 (NCI build 37 assembly) were successfully genotyped using the iCOGs chip. Genotypes of other common variants across the region were imputed in the European studies using known genotypes in combination with a reference panel from the 1000 Genomes Project. 3,134 SNPs and insertion/deletion (indel) polymorphisms were reliably imputed (imputation r2 score > 0.3, MAF ≥ 0.01) and included in further analysis together with the 517 genotyped SNPs. In the European studies 139 genotyped or imputed SNPs were associated with overall risk of breast cancer (P values < 10−7) (Fig. 1). This set included SNPs rs6504950 and rs1156287 (r2 = 0.84), both previously reported3,4 to be associated with breast cancer risk among Europeans (Supplementary Table 1).
Figure 1. Association Results for Overall Breast Cancer Risk.
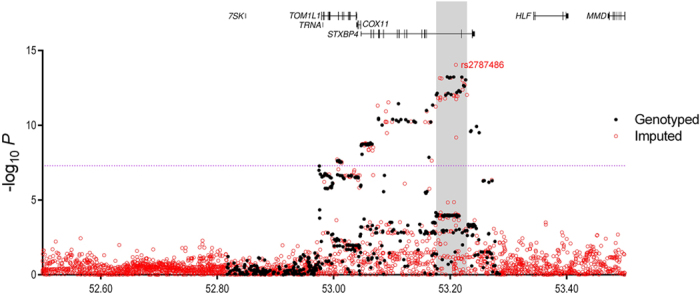
Directly genotyped SNPs are shown as filled black circles, and imputed SNPs (r2 > 0.3, MAF > 0.02) are shown as open red circles, plotted as the negative log of the P value against relative position across the locus. A schematic of the gene structures is shown. Signals, encompassing all SNPs with a likelihood ratio of <1:100 compared with the most significant SNP, are labelled and are shown as grey regions. The dashed purple line represents genome-wide significance (P < 5 × 10−08).
Among the European ancestry studies the strongest association detected was with imputed SNP rs2787486 (OR [minor/major allele] = 0.92 [C/A]; 95% CI 0.90–0.94; P = 8.96 × 10−15), located in an intron of STXBP4 and strongly correlated with both previously reported GWAS hits (r2 = 0.83 with rs1156287; r2 = 0.73 with rs6504950). The strongest genotyped SNP association was rs244353 (OR = 0.92; 95% CI 0.90–0.94); P = 5.75 × 10−14) which lies ~15 kb from rs2787486 and is correlated with it (r2 = 0.99). A regression model suggests that both rs2787486 (P = 0.02) and rs244353 (P = 0.11) are detecting the same risk association. To dissect further the observed associations all SNPs displaying evidence for association (P < 10−4 and MAF ≥ 0.02) with overall breast cancer risk (228 SNPs, Supplementary Table 1) in European studies were included in a forward stepwise regression model. This analysis identified a single association signal marked by top imputed variant rs2787486 (that is, no further SNPs were associated after adjustment for rs2787486). We also utilized penalized logistic regression models (based on the normal exponential gamma probability density) implemented in HyperLasso13, including all typed and imputed variants with an specified lambda of 0.05 and a penalty of 491 for overall risk (based on the sample size and a type I error of 0.001)28.
In this analysis, the best fitting model also included just one SNP, rs2787486.
On the assumption of a single causal variant, we calculated the likelihood ratio of each SNP relative to rs2787486 with respect to overall risk and SNPs with a relative likelihood ratio of <1:100 were excluded from further consideration29. After this exclusion process 28 SNPs (17 genotyped and 11 imputed), spanning 52.3 Kb (positions 53,176,211 to 53,228,543), remained as candidate causal variants (Table 1, Supplementary Table 2). These SNPs have very similar allele frequencies and are strongly correlated with SNP rs2787486. The two SNPs first reported to be associated with breast cancer were both excluded from this set of 28 candidate causal variants by likelihood ratio tests relative to SNP rs2787486 (likelihood ratios: 1:177439 for rs6504950 and 1: 3271 for rs1156287). Caswell et al.30 subsequently identified marker rs11658717 as a potential causal candidate, but this variant is ranked 49th and has a likelihood ratio of 1: 3875 relative to lead SNP rs2787486 - hence this has also been ruled out as potential casual candidate by our analysis.
Table 1. Association result of independent signal among European and the set of highly correlated common variants.
| Population | European | Candidate Causal SNPs | Positionb | r2c | RLc | Infod | P-valuee | OR (95% CI)e | |
|---|---|---|---|---|---|---|---|---|---|
| rs2787497§ | 53176211 | 0.89 | 0.01 | 1 | 9.27E-13 | 0.93 (0.91, 0.95) | |||
| Independent Signal | rs2787486* | rs8082622§ | 53177567 | 0.90 | 0.01 | 1 | 7.06E-13 | 0.93 (0.91, 0.94) | |
| rs2529510§§ | 53182046 | 0.84 | 0.13 | 0.91 | 6.92E-14 | 0.92 (0.90, 0.94) | |||
| Major/Minor (MAF) | A/C (0.28) | rs244373§§ | 53184949 | 0.99 | 0.13 | 0.98 | 7.30E-14 | 0.92 (0.90, 0.94) | |
| rs244358§ | 53192231 | 0.89 | 0.01 | 1 | 9.68E-13 | 0.93 (0.91, 0.95) | |||
| Overall Risk a | P-value | 8.96E-15 | rs187242§§ | 53192946 | 0.98 | 0.01 | 0.98 | 8.17E-13 | 0.93 (0.91, 0.95) |
| OR (95% CI) | 0.92 (0.90, 0.94) | rs244353§ | 53194769 | 0.99 | 0.16 | 1 | 5.75E-14 | 0.92 (0.90, 0.94) | |
| rs244348§ | 53196291 | 0.90 | 0.01 | 1 | 7.08E-13 | 0.93 (0.91, 0.94) | |||
| ER+ a | P-value | 1.39E-14 | rs244342§ | 53198407 | 0.99 | 0.14 | 1 | 6.73E-14 | 0.92 (0.90, 0.94) |
| OR (95% CI) | 0.91 (0.88, 0.93) | rs244338§ | 53200418 | 0.99 | 0.15 | 1 | 6.21E-14 | 0.92 (0.90, 0.94) | |
| rs244337§§ | 53201124 | 0.99 | 0.14 | 0.99 | 6.60E-14 | 0.92 (0.90, 0.94) | |||
| ER− a | P-value | 0.0177 | rs244336§ | 53201502 | 0.90 | 0.01 | 1 | 8.80E-13 | 0.93 (0.91, 0.95) |
| OR (95% CI) | 0.95 (0.91, 0.99) | chr17:53205761:D§§ | 53205761 | 0.90 | 0.01 | 0.99 | 9.42E-13 | 0.93 (0.91, 0.95) | |
| rs2628321§ | 53205917 | 0.99 | 0.16 | 1 | 5.88E-14 | 0.92 (0.90, 0.94) | |||
| rs2787486§§* | 53209774 | 0.96 | 8.96E-15 | 0.92 (0.90, 0.94) | |||||
| rs2787481§ | 53211110 | 0.90 | 0.01 | 1 | 6.44E-13 | 0.92 (0.91, 0.94) | |||
| rs244315§ | 53214654 | 0.90 | 0.02 | 1 | 4.72E-13 | 0.92 (0.91, 0.94) | |||
| rs244317§ | 53216985 | 0.90 | 0.02 | 1 | 5.79E-13 | 0.92 (0.91, 0.94) | |||
| rs2628316§ | 53219837 | 0.99 | 0.15 | 1 | 6.00E-14 | 0.92 (0.90, 0.94) | |||
| chr17:53221365:I§§ | 53221365 | 0.99 | 0.10 | 0.98 | 9.68E-14 | 0.92 (0.90, 0.94) | |||
| rs244318§§ | 53221368 | 0.97 | 0.02 | 0.98 | 4.19E-13 | 0.93 (0.91, 0.95) | |||
| rs244319§ | 53222874 | 0.98 | 0.04 | 1 | 2.19E-13 | 0.93 (0.91, 0.94) | |||
| rs244320§ | 53222920 | 0.98 | 0.04 | 1 | 2.19E-13 | 0.93 (0.91, 0.94) | |||
| rs244321§§ | 53223998 | 0.98 | 0.05 | 0.99 | 1.72E-13 | 0.92 (0.91, 0.94) | |||
| rs10432032§ | 53224088 | 0.98 | 0.04 | 1 | 2.47E-13 | 0.93 (0.91, 0.94) | |||
| rs244322§§ | 53224759 | 0.98 | 0.05 | 0.99 | 2.03E-13 | 0.93 (0.91, 0.94) | |||
| rs2628315§ | 53226622 | 0.99 | 0.10 | 1 | 9.02E-14 | 0.92 (0.90, 0.94) | |||
| rs244294§§ | 53228543 | 0.98 | 0.01 | 0.99 | 9.28E-13 | 0.93 (0.91, 0.95) |
aP-value, Odds ratio (OR) and 95% Confidence Interval (CI) for association with overall breast cancer risk, Estrogen Positive (ER+) and Estrogen Negative (ER−) disease.
bBuild 37 coordinates on chromosome 17.
cCorrelation (r2) and Relative Likelihood ratio (RL) with respect to lead SNP rs2787486
dIMPUTE2 info score.
eP-value, Odds ratio (OR) and 95% Confidence Interval (CI) for association with overall breast cancer risk.
§/§§Genotyped/Imputed marker.
*Lead SNP.
Association with breast cancer subtypes
Based on data from European studies, 66 genotyped SNPs and 72 imputed SNPs were associated with risk of ER+ breast cancer (P values 10−7 to 10−14). The most strongly associated SNP for overall breast cancer (rs2787486) was also the most strongly associated for ER+ disease (OR = 0.91 (0.88–0.93), P = 1.39 × 10−14), but was more weakly associated with ER− disease (OR = 0.95 (0.91–0.99), P = 1.77 × 10−02,  = 0.017). The most strongly associated SNP for ER− disease was c17_pos53079506 (OR = 1.19 (1.07–1.33),
P = 0.0017,
= 0.017). The most strongly associated SNP for ER− disease was c17_pos53079506 (OR = 1.19 (1.07–1.33),
P = 0.0017, 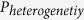 = 0.015) located ~130 kb from rs2787486.
= 0.015) located ~130 kb from rs2787486.
To determine whether there were additional subtype-specific association signals, we included all SNPs displaying evidence for association with ER+ disease (345 SNPs, P < 10−4 and MAF ≥ 2%) in a separate forward stepwise regression model. The top associated SNP was rs2787486 (OR = 0.91 (0.88–0.93), P = 1.39 × 10−14) - the same SNP best-associated with overall risk and the same signal was localized by the HyperLasso search with penalty term set to 424. We calculated the likelihood ratio of each ER+ associated SNP (r2 > 0.6) relative to rs2787486 and retained a list of 37 markers (17 genotyped, 20 imputed) with a likelihood ratio of >1:100. This list included all 28 candidate causal SNPs for overall risk, except for imputed variant rs187242. No stepwise selection for ER- risk was performed as none of the markers fulfilled the inclusion criteria.
Overall breast cancer and subtype risk association in Asian studies
Among Asian studies, the strongest association with overall breast cancer risk was observed for genotyped SNP rs244353 (OR = 0.91 (0.85–0.93), P = 2.57 × 10−3). This SNP was one of the candidate causal SNPs in Europeans, and conferred a similar OR in both populations (Supplementary Table 3). Of the genotyped markers 299 (among Europeans) and 38 (among Asians) exhibit a marginal P-value ≤ 0.05; of which 27 were significant in both populations and 9 (Supplementary Table 4) were selected as potential candidates by relative likelihood filtering in the European population. No evidence of heterogeneity in tumour subtype OR was observed for this SNP. The strongest association with ER+ disease was with SNP rs7503456 (OR = 0.89 (0.84–0.95), P = 2.73 × 10−4), which showed no association in the European studies (OR = 1.00 (0.97–1.03), P = 0.775). For ER− disease the strongest association was found with c17_pos52831447 (OR = 1.91 (1.25–2.92), P = 3.8 × 10−3), which showed no association among the European studies (OR = 1.04 (0.98–1.09), P = 0.181).
Analyses of overlap between candidate causal variants and regulatory sites
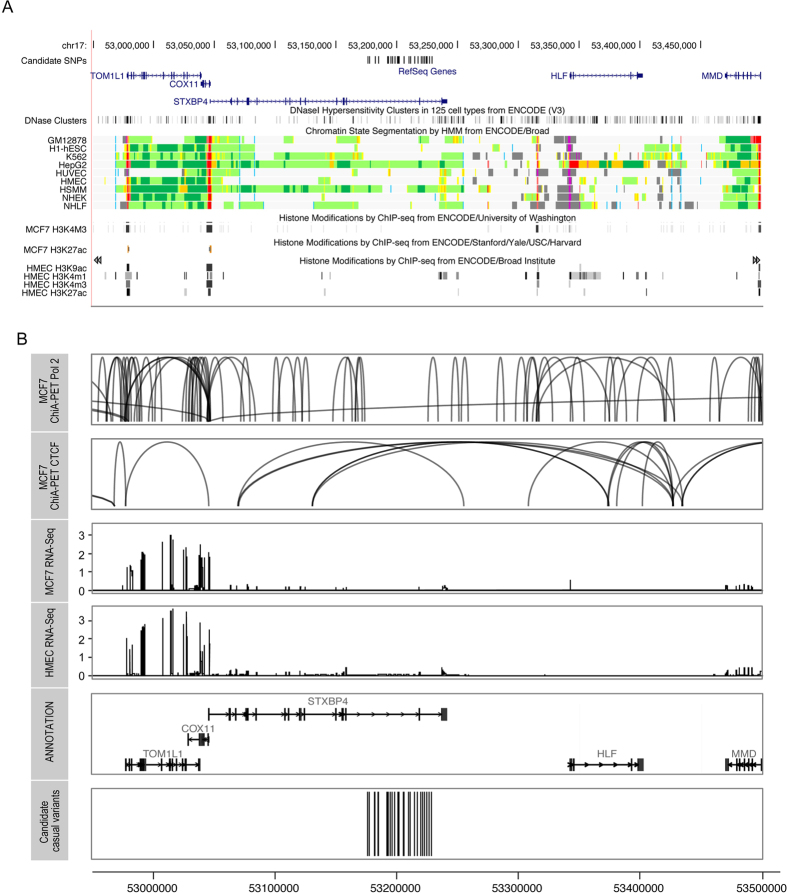
The 28 candidate causal variants (Table 1, Supplementary Table 5) fall in a 53.2 kb region spanning two introns of STXBP4 (Fig. 1). We mapped these to regulatory annotations from ENCODE. Analysis of DNase hypersensitivity clusters indicates that SNP rs244353 overlaps with a DHS in 23 cell lines, while rs2787481 and rs244317 show overlap in one and three cell lines, respectively. However, none of these overlaps were observed in mammary cells. None of the candidate causal SNPs overlapped with histone modification marks (H3K4me1, H3K4me3, H3K9ac, H3K27ac) in the mammary cells line HMEC and MCF7 breast cancer cells (Fig. 2A). We analysed enhancer-promoter interactions using Chromatin Interaction Analysis by Paired End Tag (ChiA-PET) data for CCCTC-binding factor (CTCF) and DNA polymerase II (Pol2) in MCF7 breast tumour derived cells. Although multiple chromosomal interactions were observed across the locus for both Pol2 and CTCF in MCF7 cells there was a notable dearth of such interactions in the region encompassing the strongest candidate causal variants (Fig. 2B). No interactions were observed in Hi-C data from HMEC cells in this region (data not shown).
Figure 2. In silico analysis of the 17q22 locus.
(A) Functional annotations using data from the ENCODE project. From top to bottom, epigenetic signals evaluated included DNase clusters, and ENCODE chromatin states (ChromHMM) and histone modifications in HMEC and MCF7 cell lines. The detailed colour scheme in chromatin states is described in the UCSC browser. All tracks were generated by the UCSC genome browser (hg 19). (B) Long-range chromatin interactions. From top to bottom, ChIA-PET data for Pol2 and CTCF in MCF7 cell lines. The ChIA-PET raw data available on GEO under the following accession (GSE63525.K56, GSE33664, GSE39495) were processed with the GenomicRanges package. -RNAseq data from MCF7 and HMEC cell lines. The value of the RNAseq analysis corresponds to the mean RPM value for COX11, TOM1L1 and STXBP4 from 4 HMEC and 19 MCF7 datasets, respectively. The annotation has been obtained through the Bioconductor annotation package TxDb.Hsapiens.UCSC.hg19.knownGene. The tracks have been generated using ggplot2 and ggbio library in R.
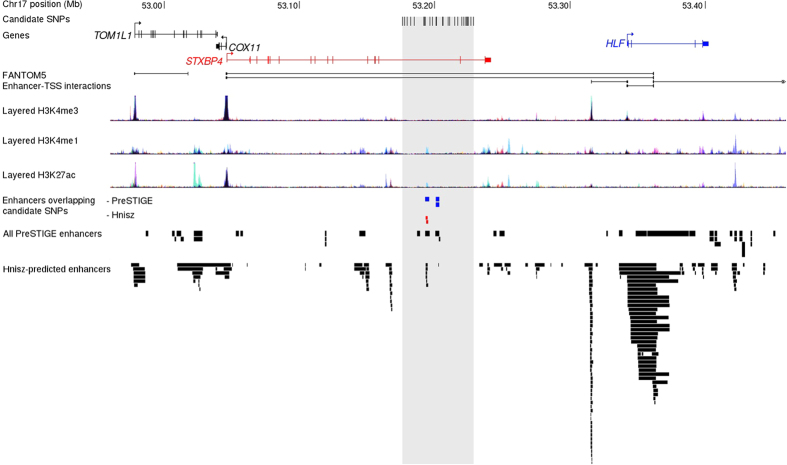
Data from Hnisz et al.18 indicates the existence of several enhancers across the region, including a small one predicted to target the STXBP4 gene (observed in both HUVEC and CD4 memory cells) that includes the candidate causal variant SNP rs244353 (Fig. 3). However, PreSTIGE17 indicates that an overlapping enhancer element (also containing rs244353) active in HepG2 cells may target the HLF gene. Another PreSTIGE element containing rs244336 and rs244337 is predicted to target HLF in colon crypt cells (Fig. 3).
Figure 3. Functional annotation of the 17q22 locus.
Positions of candidate causal variants are shown as black tick marks in relation to local genes. Epigenomic marks associated with enhancers are shown as described by FANTOM, ENCODE, PreSTIGE and Hnisz et al.18. Enhancers which overlap candidate SNPs are coded in color to match their predicted target gene. The grey shaded region area depicts the region bound by un-excluded variants.
Local Gene Expression analyses
ENCODE RNA-seq data show that COX11 and TOM1L1 are highly expressed in both MCF7 and HMEC cells lines while STXBP4 shows much lower expression levels (Fig. 2B). We performed allele specific expression (ASE) analysis using RNAseq and SNP array genotype data from TCGA19. Allelic imbalance at marginal statistical significance (P =0.032) in COX11 expression was detected with the alleles of candidate causal SNP rs2787481 (r2 = 0.90 with rs2787486 ~1.3 Kb away) but not with any other genes within 1 Mb (TOM1L1, STXBP4, HLF, MMD, PCTP) using the same SNP (Supplementary Figure 3, Supplementary Table 6).
We also examined the associations of SNPs with the expression levels of the same local genes. In the normal tissue samples (n=135) from the METABRIC study the top breast cancer candidate causal variant was also associated with COX11 expression levels (Supplementary Table 7). The most significant breast cancer associated SNP, rs2787486, was associated with differential expression of COX11 (P = 0.00019, FDR corrected P = 0.05) but not significantly associated with expression of any other genes after FDR correction. However, other SNPs across this region were more significantly associated with COX11 expression (strongest association with SNP rs138326143, P = 1.4 × 10−7, FDR corrected P = 0.003,31) suggesting that the observed change in COX11 expression in normal breast tissue is unlikely to be the main driver of breast cancer risk. By contrast, no associations with COX11 expression were observed in the TCGA breast tumour samples with the top breast cancer risk SNPs (Supplementary Figure 5). However, in TCGA multiple SNPs associate with expression of the shortest isoform of STXBP4 (uc010dcc) with the top breast cancer risk SNP, rs2787486 having a FDR corrected P = 4.0 × 10−8 (r2 = 0.06, Supplementary Figure 4). Other SNPs, including rs244317 and rs11658717 displayed more significant associations with expression of this isoform (FDR corrected P = 3.8 × 10−9, r2 = 0.07, Supplementary Figure 4) than the top breast cancer risk SNP. The minor alleles are associated with increased expression of isoform uc010dcc, and explain 7% of the variation in its expression levels. Of note Caswell et al.30 reported that the A-G base change of SNP rs11658717 mediates the use of different splice junction between exons 5 and 6 of the STXBP4 gene and thus generates the shorter uc010dcc isoform. Our expression data thus support this report but our association evidence (likelihood ratio 1:3875 relative to lead SNP rs2787486) indicates that this SNP is unlikely to be a causal variant driving breast cancer risk.
We also interrogated candidate variants in the v6 data release from the Gene-Tissue Expression (GTEx) project32. We found a significant association between the minor allele of SNP rs244353 and decreased expression of their measured STXBP4 (full length) isoform in multiple tissues including breast (n = 183; P = 1.3 × 10−6; Supplementary Figure 6, Supplementary Table 8). These different METABRIC, TCGA and GTEX findings appear contradictory of each other: SNPs rs244353 and rs244317 are highly correlated (r2 = 0.90 and yet their minor alleles are significantly associated with decreased STXBP4 expression in GTEx but increased expression of isoform (uc010dcc) in TCGA. One possible explanation is that the STXBP4 full length transcript (measured in GTEx) and the short transcript (uc010dcc, measured in TCGA) are regulated by different mechanisms33.
Discussion
In this - study, using more than 100,000 cases and controls of European and Asian ancestry participating in BCAC, we have confirmed previous reports of associations of SNPs in the 17q22 region with risk of breast cancer3,4. Moreover, we identified a set of 28 strong candidate causal variants, of which one or more is the likely driver of these reported associations. Of these, SNP rs2787486, which is correlated with previously reported candidates: rs6504950 (r2 = 0.73), rs1156287 (r2 = 0.83) and rs11658717 (r2 = 0.84)5,30; was the most strongly associated variant with overall risk (OR = 0.92 (95% CI: 0.90–0.94), P = 8.96 × 10−15). A similar magnitude of association was observed in both European and Asian women, consistent with the same causal variant mediating risk in both populations. The association was stronger for ER+ than ER− breast cancer.
All the remaining candidate causal variants lie in a 53 Kb region (positions 52,176,211 to 53,228,543) spanning two introns of the STXBP4 gene. None are predicted to alter the coding sequence of this gene and so it is most likely that the association is mediated through altering the regulation of one or more nearby genes. CHIA-Pet studies in the breast cancer MCF7 cell line reveal many chromatin interactions across the wider region (Fig. 2); however, there is a dearth of such interactions in the region encompassing the strongest candidate causal variants. Furthermore, in MCF7 or HMEC mammary cell lines there was no evidence of histone modification or open chromatin, indicative of the existence of regulatory regions, overlapping the best candidate causal variants, although such regions do exist in the wider region studied (Fig. 3). In this respect, this association signal differs from other breast cancer association signals in which strong evidence of regulatory elements in mammary cell lines has been observed34. An enhancer is predicted by FANTOM in many cell types while data from Hnisz et al.18 (Fig. 3) indicates the existence of a small enhancer region, targeting STXBP4 (observed in both HUVEC and CD4 memory cells) that overlaps with candidate causal variant rs244353. However, PreSTIGE data indicate that nearby enhancer elements may target HLF in HepG2 and colonic crypt cells (Fig. 3).
Of the candidate genes in the region, both COX11 and TOM1L1 are highly expressed in both the HMEC and MCF7 breast cancer cell lines, while STXBP4 shows much lower expression (detected by RNAseq in TCGA). In support of COX11 and TOM1L1 being the targets of this breast cancer susceptibility locus, eQTL analyses in normal breast tissue showed borderline significant associations of the risk alleles of top candidate causal SNP rs2787486 with increased expression levels of both TOM1L1 and COX11; candidate SNP rs2787481 also showed evidence of allelic imbalance in COX11 expression. COX11 encodes a cytochrome c oxidase copper chaperone – a nuclear-encoded protein component of a mitochondrial-membrane-embedded respiratory complex and TOM1L1 encodes a Target of myb1-like1 membrane trafficking protein35. Both genes are expressed in the majority of tissues examined in the Human Protein Atlas32. eQTL analysis in breast tumour tissues in TCGA find the risk allele of top candidate breast cancer risk SNP, rs2787486, to be significantly associated with increased STXBP4 expression, but not with COX11 (Supplementary Figures 4 and 5).
Furthermore, Hnisz et al.18 indicates the presence of an enhancer element that overlaps with candidate causal SNP rs244353 and potentially targets STXBP4 (observed in both HUVEC and CD4 memory cells). Consistent with this, TCGA eQTL studies in breast tumour tissues find the risk allele of top candidate breast cancer risk SNP, rs2787486, to be significantly associated with increased STXBP4 mRNA expression. The STXBP4 gene encodes Syntaxin binding protein 4, a scaffold protein, which has been shown to stabilise and prevent degradation of an isoform of p6336. P63 is, in turn, a member of the p53 tumour suppressor protein family and thus possibly a biologically more plausible candidate cancer gene than COX11 or TOM1L1.
We conclude that one or more of the 28 variants we identified is causally related to breast cancer risk, most likely through regulation of STXBP4, COX11 and TOM1L1, with the balance of the evidence favouring STXBP4 as the most important target. It remains possible, however, that the target gene(s) is more distant (>1 Mb) from the associated variants and so have not yet been considered. Further functional analyses will be required to determine the mechanism underlying this association and the downstream targets.
Additional Information
How to cite this article: Darabi, H. et al. Fine scale mapping of the 17q22 breast cancer locus using dense SNPs, genotyped within the Collaborative Oncological Gene-Environment Study (COGs). Sci. Rep. 6, 32512; doi: 10.1038/srep32512 (2016).
Supplementary Material
Acknowledgments
The authors thank all the individuals who took part in these studies and all the researchers, study staff, clinicians, and other healthcare providers, technicians, and administrative staff who have enabled this work to be carried out. In particular, they thank: COGS: Andrew Berchuck (OCAC), Rosalind A. Eeles, Ali Amin Al Olama, Zsofia Kote-Jarai, Sara Benlloch (PRACTICAL), Antonis Antoniou, Lesley McGuffog and Ken Offit (CIMBA), Joe Dennis, Andrew Lee, and Ed Dicks, Craig Luccarini and the staff of the Centre for Genetic Epidemiology Laboratory and the staff of the CNIO genotyping unit, Francois Bacot, Sylvie LaBoissière and Frederic Robidoux and the staff of the McGill University and Genome Quebec Innovation Centre, Sune F. Nielsen, Borge G. Nordestgaard, and the staff of the Copenhagen DNA laboratory, and Julie M. Cunningham, Sharon A. Windebank, Christopher A. Hilker, Jeffrey Meyer and the staff of Mayo Clinic Genotyping Core Facility; ABCFS: Maggie Angelakos, Judi Maskiell, Gillian Dite; ABCS: Sten Cornelissen, Richard van Hien, Linde Braaf, Frans Hogervorst, Senno Verhoef, Laura van 't Veer, Emiel Rutgers, C Ellen van der Schoot, Femke Atsma; ACP: The ACP study wishes to thank the participants in the Thai Breast Cancer study. Special Thanks also go to the Thai Ministry of Public Health (MOPH), doctors and nurses who helped with the data collection process. Finally, the study would like to thank Dr Prat Boonyawongviroj, the former Permanent Secretary of MOPH and Dr Pornthep Siriwanarungsan, the Department Director-Generalof Disease Control who have supported the study throughout; BBCS: Eileen Williams, Elaine Ryder-Mills, Kara Sargus; BIGGS: Niall McInerney, Gabrielle Colleran, Andrew Rowan, Angela Jones; BSUCH: Peter Bugert, Medical Faculty Mannheim; CGPS: Staff and participants of the Copenhagen General Population Study. For the excellent technical assistance: Dorthe Uldall Andersen, Maria Birna Arnadottir, Anne Bank, Dorthe Kjeldgård Hansen. The Danish Cancer Biobank is acknowledged for providing infrastructure for the collection of blood samples for the cases; CNIO-BCS: Charo Alonso, Daniel Herrero, Nuria ælvarez, Pilar Zamora, Primitiva Menendez, the Human Genotyping-CEGEN Unit (CNIO); CTS: The CTS Steering Committee includes Leslie Bernstein, James Lacey, Sophia Wang, Huiyan Ma, Yani Lu, and Jessica Clague DeHart at the Beckman Research Institute of City of Hope, Dennis Deapen, Rich Pinder, Eunjung Lee, and Fred Schumacher at the University of Southern California, Pam Horn-Ross, Peggy Reynolds, Christina Clarke Dur and David Nelson at the Cancer Prevention Institute of California, Argyrios Ziogas and Hannah Park at the University of California Irvine; ESTHER: Hartwig Ziegler, Sonja Wolf, Volker Hermann, Christa Stegmaier, Katja Butterbach; GC-HBOC: Stefanie Engert, Heide Hellebrand, Sandra Kröber; GENICA: The GENICA Network: Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart, and University of Tübingen, Germany [HB, Wing-Yee Lo, Christina Justenhoven], German Cancer Consortium (DKTK) and German Cancer Research Center (DKFZ) [HB], Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany [Yon-Dschun Ko, Christian Baisch], Institute of Pathology, University of Bonn, Germany [Hans-Peter Fischer], Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Germany, Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Bochum, Germany [TB, Beate Pesch, Sylvia Rabstein, Anne Lotz]; and Institute of Occupational Medicine and Maritime Medicine, University Medical Center Hamburg-Eppendorf, Germany [Volker Harth]; HEBCS: Kirsimari Aaltonen, Karl von Smitten, Tuomas Heikkinen, Irja Erkkilä; HMBCS: Peter Hillemanns, Hans Christiansen and Johann H. Karstens; KBCP: Eija Myöhänen, Helena Kemiläinen; kConFab/AOCS: We wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study (which has received funding from the NHMRC, the National Breast Cancer Foundation, Cancer Australia, and the National Institute of Health (USA)) for their contributions to this resource, and the many families who contribute to kConFab; LAABC: We thank all the study participants and the entire data collection team, especially Annie Fung and June Yashiki; LMBC: Gilian Peuteman, Dominiek Smeets, Thomas Van Brussel and Kathleen Corthouts; MARIE: Petra Seibold, Dieter Flesch-Janys, Judith Heinz, Nadia Obi, Alina Vrieling, Sabine Behrens, Ursula Eilber, Muhabbet Celik, Til Olchers and Stefan Nickels; MBCSG: Bernard Peissel and Jacopo Azzollini Daniela Zaffaroni of the Fondazione IRCCS Istituto Nazionale dei Tumori (INT); Bernardo Bonanni, Monica Barile and Irene Feroce of the Istituto Europeo di Oncologia (IEO) and the personnel of the Cogentech Cancer Genetic Test Laboratory; MTLGEBCS: We would like to thank Martine Tranchant (CHU de Québec Research Center), Marie-France Valois, Annie Turgeon and Lea Heguy (McGill University Health Center, Royal Victoria Hospital; McGill University) for DNA extraction, sample management and skillful technical assistance. J.S. is Chairholder of the Canada Research Chair in Oncogenetics; MYBRCA: Phuah Sze Yee, Peter Kang, Kang In Nee, Kavitta Sivanandan, Shivaani Mariapun, Yoon Sook-Yee, Daphne Lee, Teh Yew Ching and Nur Aishah Mohd Taib for DNA Extraction and patient recruitment; NBCS: The following are NBCS Collaborators: Dr. Kristine K.Sahlberg, PhD (Department of Research, Vestre Viken Hospital, Drammen, Norway and Department of Cancer Genetics, Institute for Cancer Research, Oslo University Hospital-Radiumhospitalet, Oslo, Norway), Dr. Lars Ottestad, MD (Department of Cancer Genetics, Institute for Cancer Research, Oslo University Hospital-Radiumhospitalet, Oslo, Norway), Prof. Em. Rolf Kåresen, MD (Institute of Clinical Medicine, University of Oslo, Oslo, Norway and Department of Breast- and Endocrine Surgery, Division of Surgery, Cancer and Transplantation, Oslo University Hospital, Oslo, Norway), Dr. Anita Langerød, PhD (Department of Cancer Genetics, Institute for Cancer Research, Oslo University Hospital-Radiumhospitalet, Oslo, Norway), Dr. Ellen Schlichting, MD (Section for Breast- and Endocrine Surgery, Department of Cancer, Division of Surgery, Cancer and Transplantation Medicine, Oslo University Hospital, Oslo, Norway), Dr. Marit Muri Holmen, MD (Department of Radiology and Nuclear Medicine, Oslo University Hospital, Oslo, Norway), Prof. Toril Sauer, MD (Department of Pathology at Akershus University hospital, Lørenskog, Norway and Institute of Clinical Medicine, Faculty of Medicine, University of Oslo, Oslo, Norway), Dr. Vilde Haakensen, MD (Department of Cancer Genetics, Institute for Cancer Research, Oslo University Hospital-Radiumhospitalet, Oslo, Norway), Dr. Olav Engebråten, MD (Department of Tumor Biology, Institute for Cancer Research, Oslo University Hospital, Oslo, Norway, Department of Oncology, Division of Surgery and Cancer and Transplantation Medicine, Oslo University Hospital, Oslo, Norway and Institute for Clinical Medicine, Faculty of Medicine, University of Oslo, Oslo, Norway), Prof. Bjørn Naume, MD (Department of Oncology, Division of Surgery and Cancer and Transplantation Medicine, Oslo University Hospital-Radiumhospitalet, Oslo, Norway and K.G. Jebsen Centre for Breast Cancer, Institute for Clinical Medicine, University of Oslo, Oslo, Norway.), Dr. Cecile E. Kiserud, MD (National Advisory Unit on Late Effects after Cancer Treatment, Department of Oncology, Oslo University Hospital, Oslo, Norway and Department of Oncology, Oslo University Hospital, Oslo, Norway), Dr. Kristin V. Reinertsen, MD (National Advisory Unit on Late Effects after Cancer Treatment, Department of Oncology, Oslo University Hospital, Oslo, Norway and Department of Oncology, Oslo University Hospital, Oslo, Norway), Assoc. Prof. Åslaug Helland, MD (Department of Genetics, Institute for Cancer Research and Department of Oncology, Oslo University Hospital Radiumhospitalet, Oslo, Norway), Dr. Margit Riis, MD (Dept of Breast- and Endocrine Surgery, Oslo University Hospital, Ullevål, Oslo, Norway), Dr. Ida Bukholm, MD (Department of Breast-Endocrine Surgery, Akershus University Hospital, Oslo, Norway and Department of Oncology, Division of Cancer Medicine, Surgery and Transplantation, Oslo University Hospital, Oslo, Norway), Prof. Per Eystein Lønning, MD (Section of Oncology, Institute of Medicine, University of Bergen and Department of Oncology, Haukeland University Hospital, Bergen, Norway), OSBREAC (Oslo Breast Cancer Research Consortium), Prof. Anne-Lise Børresen-Dale, PhD (Department of Cancer Genetics, Institute for Cancer Research, Oslo University Hospital-Radiumhospitalet, Oslo, Norway and Institute of Clinical Medicine, Faculty of Medicine, University of Oslo, Norway) and Grethe I. Grenaker Alnæs, M.Sc. (Department of Cancer Genetics, Institute for Cancer Research, Oslo University Hospital-Radiumhospitalet, Oslo, Norway); NBHS: We thank study participants and research staff for their contributions and commitment to this study; OBCS: We thank Arja Jukkola-Vuorinen, Mervi Grip, Saila Kauppila, Meeri Otsukka and Kari Mononen for their contributions to this study; OFBCR: Teresa Selander, Nayana Weerasooriya; ORIGO: We thank E. Krol-Warmerdam, and J. Blom for patient accrual, administering questionnaires, and managing clinical information. The LUMC survival data were retrieved from the Leiden hospital-based cancer registry system (ONCDOC) with the help of Dr. J. Molenaar; PBCS: Louise Brinton, Mark Sherman, Neonila Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao, Michael Stagner; pKARMA: The Swedish Medical Research Counsel; RBCS: Petra Bos, Jannet Blom, Ellen Crepin, Elisabeth Huijskens, Annette Heemskerk, the Erasmus MC Family Cancer Clinic; SASBAC: The Swedish Medical Research Counsel; SBCGS: We thank study participants and research staff for their contributions and commitment to this study; SBCS: Sue Higham, Helen Cramp, Ian Brock, Sabapathy Balasubramanian and Dan Connley; SEARCH: The SEARCH and EPIC teams; SGBCC: We thank the participants and research coordinator Kimberley Chua; SKKDKFZS: We thank all study participants, clinicians, family doctors, researchers and technicians for their contributions and commitment to this study; TNBCC:Robert Pilarski and Charles Shapiro were instrumental in the formation of the OSU Breast Cancer Tissue Bank. We thank the Human Genetics Sample Bank for processing of samples and providing OSU Columbus area control samples. UKBGS: We thank Breast Cancer Now and the Institute of Cancer Research for support and funding of the Breakthrough Generations Study, and the study participants, study staff, and the doctors, nurses and other health care providers and health information sources who have contributed to the study. We acknowledge NHS funding to the Royal Marsden/ICR NIHR Biomedical Research Centre. The authors would also like to acknowledge Dr Katherine A. Hoadley for normalization and sharing of all of TCGA BRCA RNAseq gene expression data.
The work conducted for this project is supported by BCAC: BCAC is funded by Cancer Research UK [C1287/A10118, C1287/A12014] and by the European Community ‹s Seventh Framework Programme under grant agreement number 223175 (grant number HEALTH-F2-2009-223175); COGS: Funding for the iCOGS infrastructure came from: the European Community's Seventh Framework Programme under grant agreement n› 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692, C8197/A16565), the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112 - the GAME-ON initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund; ABCFS: The Australian Breast Cancer Family Study (ABCFS) was supported by grant UM1 CA164920 from the National Cancer Institute (USA). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the USA Government or the BCFR. The ABCFS was also supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation (Australia) and the Victorian Breast Cancer Research Consortium. J.L.H. is a National Health and Medical Research Council (NHMRC) Senior Principal Research Fellow. M.C.S. is a NHMRC Senior Research Fellow; ABCS: The ABCS study was supported by the Dutch Cancer Society [grants NKI 2007-3839; 2009 4363]; BBMRI-NL, which is a Research Infrastructure financed by the Dutch government (NWO 184.021.007); and the Dutch National Genomics Initiative; ACP: The ACP study is funded by the Breast Cancer Research Trust, UK; BBCC: The work of the BBCC was partly funded by ELAN-Fond of the University Hospital of Erlangen; BBCS: The BBCS is funded by Cancer Research UK and Breast Cancer Now and acknowledges NHS funding to the NIHR Biomedical Research Centre, and the National Cancer Research Network (NCRN); BIGGS: ES is supported by NIHR Comprehensive Biomedical Research Centre, Guy's & St. Thomas' NHS Foundation Trust in partnership with King's College London, United Kingdom. IT is supported by the Oxford Biomedical Research Centre; BSUCH: The BSUCH study was supported by the Dietmar-Hopp Foundation, the Helmholtz Society and the German Cancer Research Center (DKFZ); CECILE: The CECILE study was funded by Fondation de France, Institut National du Cancer (INCa), Ligue Nationale contre le Cancer, Ligue contre le Cancer Grand Ouest, Agence Nationale de Sécurité Sanitaire (ANSES), Agence Nationale de la Recherche (ANR); CGPS: The CGPS was supported by the Chief Physician Johan Boserup and Lise Boserup Fund, the Danish Medical Research Council and Herlev Hospital; CNIO-BCS: he CNIO-BCS was supported by the Instituto de Salud Carlos III, the Red Temática de Investigación Cooperativa en Cáncer and grants from the Asociación Española Contra el Cáncer and the Fondo de Investigación Sanitario (PI11/00923 and PI12/00070); CTS: The CTS was initially supported by the California Breast Cancer Act of 1993 and the California Breast Cancer Research Fund (contract 97-10500) and is currently funded through the National Institutes of Health (R01 CA77398). Collection of cancer incidence data was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885. HAC receives support from the Lon V Smith Foundation (LVS39420); ESTHER: The ESTHER study was supported by a grant from the Baden Württemberg Ministry of Science, Research and Arts. Additional cases were recruited in the context of the VERDI study, which was supported by a grant from the German Cancer Aid (Deutsche Krebshilfe); GC-HBOC: The GC-HBOC (German Consortium of Hereditary Breast and Ovarian Cancer) is supported by the German Cancer Aid (grant no 110837, coordinator: Rita K. Schmutzler); GENICA: The GENICA was funded by the Federal Ministry of Education and Research (BMBF) Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0 and 01KW0114, the Robert Bosch Foundation, Stuttgart, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, the Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Bochum, as well as the Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany; HEBCS: The HEBCS was financially supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (266528), the Finnish Cancer Society, The Nordic Cancer Union and the Sigrid Juselius Foundation; HERPACC: The HERPACC was supported by MEXT Kakenhi (No. 170150181 and 26253041) from the Ministry of Education, Science, Sports, Culture and Technology of Japan, by a Grant-in-Aid for the Third Term Comprehensive 10-Year Strategy for Cancer Control from Ministry Health, Labour and Welfare of Japan, by Health and Labour Sciences Research Grants for Research on Applying Health Technology from Ministry Health, Labour and Welfare of Japan, by National Cancer Center Research and Development Fund, and "Practical Research for Innovative Cancer Control (15ck0106177h0001)" from Japan Agency for Medical Research and development, AMED, and Cancer Bio Bank Aichi; HMBCS: The HMBCS was supported by a grant from the Friends of Hannover Medical School and by the Rudolf Bartling Foundation; KARBAC: Financial support for KARBAC was provided through the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet, the Swedish Cancer Society, The Gustav V Jubilee foundation and Bert von Kantzows foundation; KBCP: The KBCP was financially supported by the special Government Funding (EVO) of Kuopio University Hospital grants, Cancer Fund of North Savo, the Finnish Cancer Organizations, and by the strategic funding of the University of Eastern Finland; kConFab/AOCS: kConFab is supported by a grant from the National Breast Cancer Foundation, and previously by the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. Financial support for the AOCS was provided by the United States Army Medical Research and Materiel Command [DAMD17-01-1-0729], Cancer Council Victoria, Queensland Cancer Fund, Cancer Council New South Wales, Cancer Council South Australia, The Cancer Foundation of Western Australia, Cancer Council Tasmania and the National Health and Medical Research Council of Australia (NHMRC; 400413, 400281, 199600). G.C.T. and P.W. are supported by the NHMRC. RB was a Cancer Institute NSW Clinical Research Fellow; LAABC: LAABC is supported by grants (1RB-0287, 3PB-0102, 5PB-0018, 10PB-0098) from the California Breast Cancer Research Program. Incident breast cancer cases were collected by the USC Cancer Surveillance Program (CSP) which is supported under subcontract by the California Department of Health. The CSP is also part of the National Cancer Institute's Division of Cancer Prevention and Control Surveillance, Epidemiology, and End Results Program, under contract number N01CN25403; LMBC: LMBC is supported by˜the 'Stichting tegen Kanker' (232–2008 and 196–2010). Diether Lambrechts is supported by the FWO and the KULPFV/10/016-SymBioSysII; MARIE: The MARIE study was supported by the Deutsche Krebshilfe e.V. [70-2892-BR I, 106332, 108253, 108419], the Hamburg Cancer Society, the German Cancer Research Center (DKFZ) and the Federal Ministry of Education and Research (BMBF) Germany [01KH0402]; MBCSG: MBCSG is supported by grants from the Italian Association for Cancer Research (AIRC) and by funds from the Italian citizens who allocated the 5/1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects “5 × 1000”); MCBCS: The MCBCS was supported by the NIH grants CA128978, CA116167, CA176785 an NIH Specialized Program of Research Excellence (SPORE) in Breast Cancer [CA116201], and the Breast Cancer Research Foundation and a generous gift from the David F. and Margaret T. Grohne Family Foundation and the Ting Tsung and Wei Fong Chao Foundation; MCCS: MCCS cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 209057, 251553 and 504711 and by infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry (VCR) and the Australian Institute of Health and Welfare (AIHW), including the National Death Index and the Australian Cancer Database; MEC: The MEC was support by NIH grants CA63464, CA54281, CA098758 and CA132839; MTLGEBCS: The work of MTLGEBCS was supported by the Quebec Breast Cancer Foundation, the Canadian Institutes of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” program – grant # CRN-87521 and the Ministry of Economic Development, Innovation and Export Trade – grant # PSR-SIIRI-701; MYBRCA: MYBRCA is funded by research grants from the Malaysian Ministry of Science, Technology and Innovation (MOSTI), Malaysian Ministry of Higher Education (UM.C/HlR/MOHE/06) and Cancer Research Initiatives Foundation (CARIF). Additional controls were recruited by the Singapore Eye Research Institute, which was supported by a grant from the Biomedical Research Council (BMRC08/1/35/19/550), Singapore and the National medical Research Council, Singapore (NMRC/CG/SERI/2010); NBCS: The NBCS has received funding from the K.G. Jebsen Centre for Breast Cancer Research; the Research Council of Norway grant 193387/V50 (to A-L Børresen-Dale and V.N. Kristensen) and grant 193387/H10 (to A-L Børresen-Dale and V.N. Kristensen), South Eastern Norway Health Authority (grant 39346 to A-L Børresen-Dale) and the Norwegian Cancer Society (to A-L Børresen-Dale and V.N. Kristensen); NBHS: The NBHS was supported by NIH grant R01CA100374. Biological sample preparation was conducted the Survey and Biospecimen Shared Resource, which is supported by P30 CA68485; OBCS: The OBCS was supported by research grants from the Finnish Cancer Foundation, the Academy of Finland (grant number 250083, 122715 and Center of Excellence grant number 251314), the Finnish Cancer Foundation, the Sigrid Juselius Foundation, the University of Oulu, the University of Oulu Support Foundation and the special Governmental EVO funds for Oulu University Hospital-based research activities; OFBCR: The Ontario Familial Breast Cancer Registry (OFBCR) was supported by grant UM1 CA164920 from the National Cancer Institute (USA). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the USA Government or the BCFR; ORIGO: The ORIGO study was supported by the Dutch Cancer Society (RUL 1997–1505) and the Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL CP16); PBCS: The PBCS was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA; pKARMA: The pKARMA study was supported by Märit and Hans Rausings Initiative Against Breast Cancer; RBCS:The RBCS was funded by the Dutch Cancer Society (DDHK 2004-3124, DDHK 2009-4318); SASBAC: The SASBAC study was supported by funding from the Agency for Science, Technology and Research of Singapore (A*STAR), the US National Institute of Health (NIH) and the Susan G. Komen Breast Cancer Foundation; SBCGS: The SBCGS was supported primarily by NIH grants R01CA64277, R01CA148667, and R37CA70867. Biological sample preparation was conducted the Survey and Biospecimen Shared Resource, which is supported by P30 CA68485. The scientific development and funding of this project were, in part, supported by the Genetic Associations and Mechanisms in Oncology (GAME-ON) Network U19 CA148065; SBCS: The SBCS was supported by Yorkshire Cancer Research S295, S299, S305PA and Sheffield Experimental Cancer Medicine Centre; SCCS: The SCCS is supported by a grant from the National Institutes of Health (R01 CA092447). Data on SCCS cancer cases used in this publication were provided by the Alabama Statewide Cancer Registry; Kentucky Cancer Registry, Lexington, KY; Tennessee Department of Health, Office of Cancer Surveillance; Florida Cancer Data System; North Carolina Central Cancer Registry, North Carolina Division of Public Health; Georgia Comprehensive Cancer Registry; Louisiana Tumor Registry; Mississippi Cancer Registry; South Carolina Central Cancer Registry; Virginia Department of Health, Virginia Cancer Registry; Arkansas Department of Health, Cancer Registry, 4815 W. Markham, Little Rock, AR 72205. The Arkansas Central Cancer Registry is fully funded by a grant from National Program of Cancer Registries, Centers for Disease Control and Prevention (CDC). Data on SCCS cancer cases from Mississippi were collected by the Mississippi Cancer Registry which participates in the National Program of Cancer Registries (NPCR) of the Centers for Disease Control and Prevention (CDC). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the CDC or the Mississippi Cancer Registry; SEARCH: SEARCH is funded by a programme grant from Cancer Research UK [C490/A10124] and supported by the UK National Institute for Health Research Biomedical Research Centre at the University of Cambridge; SEBCS: SEBCS was supported by the BRL (Basic Research Laboratory) program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2012-0000347); SGBCC: SGBCC is funded by the NUS start-up Grant, National University Cancer Institute Singapore (NCIS) Centre Grant and the NMRC Clinician Scientist Award. Additional controls were recruited by the Singapore Consortium of Cohort Studies-Multi-ethnic cohort (SCCS-MEC), which was funded by the Biomedical Research Council, grant number: 05/1/21/19/425; SKKDKFZS: SKKDKFZS is supported by the DKFZ; SZBCS: The SZBCS was supported by Grant PBZ_KBN_122/P05/2004; TBCS: The TBCS was funded by The National Cancer Institute Thailand; TNBCC: The TNBCC was supported by: a Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), a grant from the Breast Cancer Research Foundation, a generous gift from the David F. and Margaret T. Grohne Family Foundation, the Stefanie Spielman Breast Cancer fund and the OSU Comprehensive Cancer Center, the Hellenic Cooperative Oncology Group research grant (HR R_BG/04) and the Greek General Secretary for Research and Technology (GSRT) Program, Research Excellence II, the European Union (European Social Fund – ESF), and Greek national funds through the Operational Program "Education and Lifelong Learning" of the National Strategic Reference Framework (NSRF)- ARISTEIA; TWBCS: The TWBCS is supported by the Taiwan Biobank project of the Institute of Biomedical Sciences, Academia Sinica, Taiwan; UKBGS: The UKBGS is funded by Breast Cancer Now and the Institute of Cancer Research (ICR), London. ICR acknowledges NHS funding to the NIHR Biomedical Research Centre.
Footnotes
Author Contributions Analysed the data: H.D., J. Beesley, A.D., S. Kar, S.N., M.M.M. and A.M.D. J.Beesley prepared Figure 1,3 and Supplementary Figure 1, 2 and 6. A.D. prepared Figure 2. S.N. prepared Supplementary Figures 4 and 5. M.M.M. prepared Supplementary Figure 3. Wrote the manuscript: H.D., J.Beesley, A.D., S. Kar, S.N., M.M.M. and A.M.D. Provided critical review of the manuscript: H.D., J. Beesley, A.D., S. Kar, S.N., M.M.M., D.F.E., W.Z., S.L.E., J.D.F., G.C.T. and A.M.D. Approved the final version of the manuscript and were involved in sample and data collection for each of the involved studies together with other listed staff (see Acknowledgements): H.D., J. Beesley, A.D., S.Kar, S.N., M.M.M., P.S., K. Michailidou, M.G., H.F.W., M.K.B., Q.W., J.D., M.R.A., I.L.A., H.A.C., V.A., M.W.B., J. Benitez, N.V.B., S.E.B., H. Brauch, H. Brenner, A.B., T.B., B.B., J.C.C., J.Y.C., D.M.C., F.J.C., A.C., S.S.C., K.Z., P.D., T.D., D.F.E., P.A.F., J.F., O.F., H.F., E.G., M.G.C., G.G.G., M.S.G., A.G.N., P.G., C.A.H., E.H., U.H., M.H., A.H., J.L.H., H.I., A.J., N.J., D.K., S.Khan, V.M.K., M.K., V.K., D.L., L.L.M., S.C.L., A. Lindblom, A.Lophatananon, J.L., A. Mannermaa, S. Manoukian, S. Margolin, K. Matsuo, R.M., J.M., A.Meindl, R.L.M., K. Muir, S.L.N., H.N., C.O., N.O., P.P., G.P., K.P., A.R., S.S., E.J.S., M.K.S., R.K.S., C.S., M.S., C.Y.S., X.O.S., M.C.S., D.O.S., H.S., A.W., S.H.T., D.C.T., I.T., D.T., T.T., C.M.V., D.V., R.W., A.H.W., P.E.W., C.H.Y., W.Z., P.D.P.P., P.H., S.L.E., J.S., J.D.F., G.C.T. and A.M.D.
References
- Ferlay J. et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127, 2893–2917, 10.1002/ijc.25516 (2010). [DOI] [PubMed] [Google Scholar]
- Ferlay J. et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136, E359–386, 10.1002/ijc.29210 (2015). [DOI] [PubMed] [Google Scholar]
- Ahmed S. et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet 41, 585–590, 10.1038/ng.354 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C. et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet 42, 504–507, 10.1038/ng.586 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campa D. et al. Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium. J Natl Cancer Inst 103, 1252–1263, 10.1093/jnci/djr265 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeks A. et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: findings from the Breast Cancer Association Consortium. Hum Mol Genet 20, 3289–3303, 10.1093/hmg/ddr228 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L. et al. Association of STXBP4/COX11 rs6504950 (G > A) polymorphism with breast cancer risk: evidence from 17,960 cases and 22,713 controls. Arch Med Res 43, 383–388, 10.1016/j.arcmed.2012.07.008 (2012). [DOI] [PubMed] [Google Scholar]
- Michailidou K. et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet 45, 353–361, 361e351-352, 10.1038/ng.2563 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B. N., Donnelly P. & Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics 5, e1000529, 10.1371/journal.pgen.1000529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B., Marchini J. & Stephens M. Genotype imputation with thousands of genomes. G3 1, 457–470, 10.1534/g3.111.001198 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team R. C. R: A Language and Environment for Statistical Computing, https://www.R-project.org/ (2016).
- French J. D. et al. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. American journal of human genetics 92, 489–503, 10.1016/j.ajhg.2013.01.002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggart C. J., Whittaker J. C., De Iorio M. & Balding D. J. Simultaneous analysis of all SNPs in genome-wide and re-sequencing association studies. PLoS genetics 4, e1000130, 10.1371/journal.pgen.1000130 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium E. P. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9, e1001046, 10.1371/journal.pbio.1001046 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680, 10.1016/j.cell.2014.11.021 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics C. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330, 10.1038/nature14248 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradin O. et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res 24, 1–13, 10.1101/gr.164079.113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947, 10.1016/j.cell.2013.09.053 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. et al. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell 152, 633–641, 10.1016/j.cell.2012.12.034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352, 10.1038/nature10983 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q. et al. Identification of novel genetic markers of breast cancer survival. J Natl Cancer Inst 107, 10.1093/jnci/djv081 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin A. A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28, 1353–1358, 10.1093/bioinformatics/bts163 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haakensen V. D. et al. Gene expression profiles of breast biopsies from healthy women identify a group with claudin-low features. BMC Med Genomics 4, 77, 10.1186/1755-8794-4-77 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70, 10.1038/nature11412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Ruotti V., Stewart R. M., Thomson J. A. & Dewey C. N. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 26, 493–500, 10.1093/bioinformatics/btp692 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W. R Package eMAP (http://www.bios.unc.edu/~weisun/software.htm) (2010).
- Consortium G. T. The Genotype-Tissue Expression (GTEx) project. Nat Genet 45, 580–585, 10.1038/ng.2653 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W. Y. et al. Identification and characterization of novel associations in the CASP8/ALS2CR12 region on chromosome 2 with breast cancer risk. Hum Mol Genet 24, 285–298, 10.1093/hmg/ddu431 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer A. V., Cox A. & Walters K. Comparing the efficacy of SNP filtering methods for identifying a single causal SNP in a known association region. Annals of human genetics 78, 50–61, 10.1111/ahg.12043 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell J. L. et al. Multiple breast cancer risk variants are associated with differential transcript isoform expression in tumors. Hum Mol Genet 24, 7421–7431, 10.1093/hmg/ddv432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela M. J. & Chanock S. J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557, 10.1093/bioinformatics/btv402 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele M. et al. Human genomics. The human transcriptome across tissues and individuals. Science 348, 660–665, 10.1126/science.aaa0355 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J. et al. The human genome browser at UCSC. Genome Res 12, 996–1006, 10.1101/gr.229102. Article published online before print in May 2002 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darabi H. et al. Polymorphisms in a Putative Enhancer at the 10q21.2 Breast Cancer Risk Locus Regulate NRBF2 Expression. American journal of human genetics 97, 22–34, 10.1016/j.ajhg.2015.05.002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C. et al. TOM1L1 drives membrane delivery of MT1-MMP to promote ERBB2-induced breast cancer cell invasion. Nat Commun 7, 10765, 10.1038/ncomms10765 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Peart M. J. & Prives C. Stxbp4 regulates DeltaNp63 stability by suppression of RACK1-dependent degradation. Mol Cell Biol 29, 3953–3963, 10.1128/MCB.00449-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.