Abstract
Human kallikrein-related peptidases (KLKs) are a group of 15 secreted serine proteases encoded by the largest contiguous cluster of protease genes in the human genome. KLKs are involved in coordination of numerous physiological functions including regulation of blood pressure, neuronal plasticity, skin desquamation, and semen liquefaction, and thus represent promising diagnostic and therapeutic targets. Until now, quantification of KLKs in biological and clinical samples was accomplished by enzyme-linked immunosorbent assays (ELISA). Here, we developed multiplex targeted mass spectrometry assays for the simultaneous quantification of all 15 KLKs. Proteotypic peptides for each KLK were carefully selected based on experimental data and multiplexed in single assays. Performance of assays was evaluated using three different mass spectrometry platforms including triple quadrupole, quadrupole-ion trap, and quadrupole-orbitrap instruments. Heavy isotope-labeled synthetic peptides with a quantifying tag were used for absolute quantification of KLKs in sweat, cervico-vaginal fluid, seminal plasma, and blood serum, with limits of detection ranging from 5 to 500 ng/ml. Analytical performance of assays was evaluated by measuring endogenous KLKs in relevant biological fluids, and results were compared with selected ELISAs. The multiplex targeted proteomic assays were demonstrated to be accurate, reproducible, sensitive, and specific alternatives to antibody-based assays. Finally, KLK4, a highly prostate-specific protein and a speculated biomarker of prostate cancer, was unambiguously detected and quantified by immunoenrichment-SRM assay in seminal plasma and blood serum samples from individuals with confirmed prostate cancer and negative biopsy. Mass spectrometry revealed exclusively the presence of a secreted isoform and thus unequivocally resolved earlier disputes about KLK4 identity in seminal plasma. Measurements of KLK4 in either 41 seminal plasma or 58 blood serum samples revealed no statistically significant differences between patients with confirmed prostate cancer and negative biopsy. The presented multiplex targeted proteomic assays are an alternative analytical tool to study the biological and pathological roles of human KLKs.
The human tissue kallikrein-related peptidases (KLKs) 1 are a large family of 15 closely related serine proteases with trypsin- or chymotrypsin-like activities. Kallikreins exhibit important similarities both at the gene and protein level (1). The genes are localized to chromosome 19q13.4, forming the largest contiguous cluster of proteases within the human genome (2). The enzymes are initially secreted as inactive zymogens and subsequently activated by removal of a short N-terminal pro-sequence (3). KLKs are expressed in many tissues, including steroid hormone-producing or hormone-dependent tissues and are responsible for the coordination of various physiological functions including regulation of blood pressure (4), neuronal plasticity (5), semen liquefaction (6), skin desquamation (7) and inflammation (8), and thus represent attractive diagnostic and therapeutic targets (9). Aberrant levels of some KLKs were observed in tissues, blood serum and proximal fluids of cancer patients, particularly in cases of adenocarcinomas derived from steroid hormone-regulated tissues, and were correlated with the course of disease. For instance, concurrent up-regulation of multiple KLKs has been observed in ovarian carcinoma (10). It has also been shown for breast, prostate, and testicular cancers that KLKs may facilitate neoplastic progression through promotion of cell proliferation and metastasis (11). Based on this evidence, kallikrein-related peptidases have been extensively studied for their potential as biomarkers of various malignancies (12–16).
High-quality ELISAs are now available for the majority of KLKs, except KLKs 1, 9, 12, and 15. Because of their specificity, sensitivity, accuracy, high throughput, and relative simplicity, ELISAs have been used for decades to measure KLKs in biological and clinical samples (17). Rapid evolution of biological mass spectrometry provided powerful alternatives to immunoassays for unambiguous identification and accurate quantification of proteins in clinical samples (18, 19). Selected reaction monitoring (SRM) and parallel reaction monitoring (PRM) assays have high specificity, dynamic range of six orders of magnitude, and limits of detection (LOD) and quantification (LOQ) in the low ng/ml level. Unlike multiplex ELISA, which allows for the detection of up to 25 analytes in a single assay (20), multiplex SRM assays facilitate measurements of tens to hundreds of proteins in a single run without compromising assay sensitivity and accuracy (21–24).
Here, we present multiplatform targeted mass spectrometry assays for the simultaneous quantification of all 15 KLKs with limits of quantification in the low ng/ml range in clinical samples. The analytical performance of the SRM assays was evaluated by measuring absolute levels of endogenous KLKs in sweat, cervico-vaginal fluid, seminal plasma, and blood serum samples and by comparison to the performance of selected ELISAs. Finally, using the whole set of SRM, immuno-SRM and sandwich immunoassays, we investigated KLK4 and unequivocally resolved earlier disputes about its isoform identity in seminal plasma and its levels in prostate cancer.
EXPERIMENTAL PROCEDURES
Materials and Reagents
Iodoacetamide, dithiothreitol, acetonitrile, formic acid, and sequencing grade modified trypsin were purchased from Sigma-Aldrich (Oakville, ON, Canada). RapiGest SF surfactant was purchased from Waters (Milford, MA). Quantified & Stable Isotope Labeled Peptides (SpikeTides™_TQL) were obtained from JPT Peptide Technologies GmbH (Berlin, Germany). The recombinant KLK4 was purified through a two-step purification protocol and was used as an immunogen for the production of monoclonal antibodies in mice, as previously described (25). Clone 10F4.1G6 was used as a capture antibody for the analysis of seminal plasma and blood serum samples with immunoenrichment-SRM and ELISA. Rabbit polyclonal anti-KLK4 antibodies were used as detection antibodies.
Biological Samples
All biological samples were collected from individuals of different ethnic background with an informed consent. Sample collection was approved by the institutional review boards of Mount Sinai Hospital (approval #08–117-E and, #16–0137-E) and University Health Network (# 09–0830-AE). Semen samples were collected by masturbation into sterile collection cups. Following liquefaction for 1 h at room temperature, semen samples were centrifuged at 16,000 rpm for 30 min at 4 °C three times to separate seminal plasma (SP) from cells and cellular components, and were stored at −80 °C until further use. Ten SP samples were obtained from healthy men (median age 35 years old) prior to vasectomy, 21 SPs from men with biopsy-confirmed prostate cancer (median age 62) and 20 SPs from men with negative biopsy outcome (median age 62). In addition, blood serum samples were obtained from 36 men with biopsy-confirmed prostate cancer (serum PSA > 4 ng/ml, median age 63), 22 men with negative biopsy (serum PSA > 4 ng/ml, median age 61), and 3 healthy men (serum PSA < 1 ng/ml, median age 36). Five cervico-vaginal fluid (CVF) samples were collected from nonpregnant women (median age 30) in flexible cups that are worn internally, around the cervix. Ten sweat (SW) samples were collected from five men and five women (median age 28) after physical exercise in wood fiber wipes (KimWipes, Kimberly-Clark, Mississauga, ON, Canada). A 20 ml syringe was used to squeeze the fluid out of the wipes.
Sample Preparation and Trypsin Digestion
All samples were centrifuged at 16,000 × g for 30 min to ensure complete removal of cells and cellular debris. Before trypsin digestion, the total protein content of each sample was measured using the Pierce BCA protein assay (Thermo Scientific, Mississauga, ON, Canada). Twenty micrograms of total protein per sample were subjected to proteomic sample preparation. To prepare for digestion, denaturation of proteins and reduction of disulfide bonds were achieved by treatment of samples with 0.1% RapiGest SF and 10 mm dithiothreitol at 65 °C for 15 min. After reduction, the samples were alkylated with 20 mm iodoacetamide in the dark at room temperature for 40 min. Heavy isotope-labeled peptides with a quantitation tag (500 fmoles) were added to all samples prior to trypsin digestion. Digestion (1:20, trypsin/total protein) was performed overnight at 37 °C. RapiGest SF was cleaved with 1% trifluoroacetic acid and removed with centrifugation at 16,000 × g for 15 min. C18 Bond Elute OMIX tips (10 μl; Agilent Technologies, Mississauga, ON, Canada) were used for desalting and concentration of tryptic peptides (final solution in 5% acetonitrile and 0.1% formic acid).
Chromatographic and Mass Spectrometry Conditions
Shotgun and PRM experiments were performed on a Q Exactive™ Plus Hybrid Quadrupole-Orbitrap™ Mass Spectrometer (Thermo Scientific). The tryptic peptides were loaded on a 3 cm long C18 (5 μm) trap precolumn (i.d. 200 μm) via EASY-nLC 1000 pump (Thermo Scientific) at 8 μl/min. Mobile phases included 0.1% formic acid in water (buffer A) and 0.1% formic acid in acetonitrile (buffer B). Peptides were separated with a 15 cm long C18 (3 μm) analytical column (i.d. 75 μm) with a 8 μm tip (New Objective, Woburn, MA) with a 22 min gradient elution at 350 nL/min flow rate. A five-step gradient was used: 1% to 14% of buffer B for 1 min, 14% to 40% for 11 min, 40% to 65% for 2 min, 65% to 100% for 1 min, and 100% for 7 min. Shotgun experiments included one full MS1 scan followed by 12 data-dependent MS2 scans. In-source collision induced dissociation (CID) was set to 0.0 eV, and the ion transfer capillary temperature was 275 °C. MS transitions in the orbitrap were acquired with 70,000 resolving power at 200 m/z. Automatic Gain Control (AGC) for MS1 acquisition was set to 3 × 106 with a maximum injection time of 100 ms. AGC target for the data dependent MS2 scans acquisition was set to 105 with a maximum injection time of 50 ms and the normalized collision energy of 27. The scan range was 250 to 1500 m/z, microscans were set to 1 for both MS1 and MS2, charge state screening was enabled and unassigned, and +1 charge states were excluded from MS2 activation. The performance settings for the PRM assays were the following: in-source CID was set to 3.0 eV, MS transitions in the orbitrap were acquired with 17,500 resolving power at 200 m/z, AGC target was set to 3 × 106 with a maximum injection time of 100 ms, the isolation window was set to 1.0 m/z, the normalized collision energy was set to 23 and scan times were set to 100 ms.
The development of SRM assays and the analysis of clinical samples were performed using a TSQ QuantivaTM triple quadrupole mass spectrometer (Thermo Scientific), as previously described (26). Polarity was set to positive, ion transfer tube temperature was 300°C, and the CID argon pressure was 1.5 mTorr. The resolution settings of the first and third quadrupoles were 0.2 and 0.7 Da FWHM, respectively (comparable to the “high” and “unit” resolution of the QTRAP 6500 quadrupole-ion trap mass spectrometer). High resolution in the first quadrupole facilitated exclusion of potentially interfering ions, thus improving selectivity in the complex biological matrices. Scan times were set to 10 ms. Optimized SRM method was also tested in a QTRAP 6500 quadrupole-ion trap mass spectrometer (AB Sciex, Concord, ON, Canada). Declustering and entrance potentials were set to 150 and 10 V, respectively. Resolutions for both the first quadrupole and the ion trap were set to “unit,” and scan times were 15 ms.
Selection of Proteotypic Peptides
To facilitate identification of proteotypic peptides, we aimed at identifying five to seven peptides that would result in intense and reproducible MS1 signal. For the selection of top peptides, we used tryptic digests of 15 recombinant KLKs previously expressed in our lab in prokaryotic (27), mammalian (HEK293) (28–30) or yeast (Pichia pastoris) (13, 15, 31) expression systems. The LC-MS/MS raw data were analyzed using the Proteome Discoverer™ software (Thermo Scientific, version 1.4.1.14) with SEQUEST search algorithm and the human UniProtKB/Swiss-Prot database (HUMAN_sProt-07092014; 20,209 entries). The mass tolerances for precursor and fragment ions were set to 7 ppm and 0.02 Da, respectively. The Proteome Discoverer decoy database and Percolator algorithm were used to compute the number of false positive protein identifications based on the Posterior Error Probabilities. The false discovery rate based on Q-values was set to 1%. The following parameters were set for the search: (1) digestion enzyme: trypsin; (2) maximum allowance for miss-cleavages: 1; (3) oxidation of methionine and tryptophan, as well as N-terminal peptide acetylation were set as variable modifications, whereas cysteine carbamidomethylation was set as a static modification. Unmodified peptides as well as peptides with proline were preferred, if possible. Peptides with abundant modifications based on three analytical replicates that included the digestion step (MS1 area of forms with methionine oxidation, glutamine and asparagine deamidation, N-terminal protein acetylation, or missed cleavage sites >10% of total MS1 area) were selected for further investigation. All peptides were searched with the protein Basic Local Alignment Tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to ensure that the selected peptides were unique for each KLK. Raw mass spectrometry data and Proteome Discoverer output files with peptide and protein identifications were deposited to the ProteomeXchange Consortium via the PRIDE partner repository (http://www.ebi.ac.uk/pride/archive/login) with the data set identifier PXD003324 and the following credentials: Username: reviewer73059@ebi.ac.uk; Password: b4Ge5SYs.
Development of PRM and SRM Assays
To facilitate accurate protein quantification, we aimed at experimental testing of three peptides for each KLK. First, in silico digestion of all proteins was performed using the Skyline Targeted Proteomics Environment v3.1.0.7382 (MacCoss Lab Software, Seattle, WA). The top three peptides for each protein were selected based on our full MS data dependent MS/MS identification data and were subsequently confirmed with the SRM atlas (www.srmatlas.org). Because of the presence of N terminus cysteine in one of the selected peptides for KLK5, the pyro-carbamidomethyl modified form of this peptide was also selected for quantification. In the second step of method development, the selected peptides were included into each of 15 unscheduled survey PRM assays designed for Q Exactive Plus (supplemental Table S1). Comparison of LC retention time for shotgun and PRM gradients was used as another indication of identity of selected peptides (32). The ten most intense and selective transitions per peptide were selected. In the third step, 10 transitions per selected peptide were included into each of 15 unscheduled targeted mass spectrometry methods on QTRAP 6500 quadrupole-ion trap and TSQ QuantivaTM triple quadrupole mass spectrometers. Based on the relative intensities, the most intense peptides and the three most intense and reproducible transitions per peptide were selected for SRM assays (supplemental Tables S2 and S3). Calibration curves for all 45 peptides (derived by digestion of 15 recombinant KLKs) were built, and a single peptide per each KLK was selected based on its limit of detection, quantification, linearity and coefficient of variation (supplemental Table S4). The limit of detection was defined as the lowest analyte concentration distinguished from the background (S/N≥3). The limit of quantification was defined as the lowest concentration measured with CV≤20% within the linear range of the calibration curve.
In order to exclude possible interferences, ensure correct identity of each peak and facilitate absolute quantification of each protein, heavy isotope-labeled peptide internal standards with a quantifying tag were synthesized for all 15 KLKs. Following peptide synthesis, the quantifying tag (serine-alanine-[3-nitro]tyrosine-glycine) ensured accurate peptide quantification using UV absorption at 350 nm and was readily cleaved by trypsin, thus accounting for digestion efficiency. The area of the endogenous peptide was compared with that of the heavy peptide, and their ratio was used to calculate the absolute concentration of the endogenous light peptide. In the fourth step of method development, 32 heavy and light peptides and 96 transitions were included into multiplex unscheduled PRM or SRM assays. Selectivity of transitions and possible interferences in each sample matrix were assessed. Finally, 15 KLKs, 32 peptides, and 96 transitions where scheduled within 2 min intervals in a single multiplex scheduled SRM assay (supplemental Figs. S1–S3). Scan times were optimized to ensure acquisition of at least 20 points per peak. Precursor-to-product transitions were provided in the supplemental Tables S5–7. SRM and PRM raw mass spectrometry data were deposited to the Peptide Atlas repository (http://www.peptideatlas.org/PASS/PASS00777) with the dataset identifier PASS00777 and the following credentials: Username: PASS00777; Password: VU2858zr.
Protein Quantification by SRM
Peptides were separated on a C18 analytical column with a 22 min gradient elution and subsequently detected by TSQ QuantivaTM mass spectrometer with a nanoelectrospray ionization source. The total method duration, including sample pickup and loading, pre-column and column equilibration was 48 min, thus resulting in a throughput of 30 runs per day. The performance of the nanoLC-MS platforms was assessed at the beginning of each day and every six runs thereafter using injections of 10 μl of 1 fmol/μl bovine serum albumin (BSA) solution. Thus, the throughput was 12 clinical samples per day in duplicates (24 injections). Repeatability of the method and the system stability were estimated by the coefficient of variation (CV%). Carryover ranged between 0.2–3.1% and was estimated by blank injections. Linearity of the assay was studied by spiking increasing amounts of the heavy labeled peptides into pools of SP, CVF, and SW. The area ratios of heavy labeled peptides to the corresponding endogenous light peptides were plotted against the spiked internal standard concentrations. Sixteen heavy isotope-labeled peptide standards were mixed and diluted to a final concentration of 100 fmol/μl. Five microliters of the internal standard mixture were spiked to each sample before the digestion and each digest was analyzed in duplicate. Because all measurements were executed within the linear ranges of calibration curves, the absolute concentration of each endogenous protein was calculated using the heavy-to-light peptide ratio and the concentration of the spiked-in heavy peptide.
Quantification of Kallikrein 4 by Immunoenrichment-SRM
For the analysis of SP and blood serum samples with immunoenrichment-SRM, 500 ng of monoclonal anti-KLK4 antibody (clone 10F4.1G6) were diluted in the coating buffer (50 mm Tris-HCl, pH 7.8) in each well of a 96-well white polystyrene microliter plate and incubated overnight. The plate was then washed three times with phosphate-buffered saline (PBS). Ten μl of recombinant protein or biological samples were added to each well and were further diluted up to 100 μl with assay buffer (6% BSA in 50 mm Tris-HCl at pH 7.8). The plate was incubated for 2 h with continuous shaking and was finally washed three times with PBS and three times with 50 mm ammonium bicarbonate buffer. Heavy isotope-labeled peptide with a quantitation tag (500 fmoles) was added to all samples, and trypsin (0.25 ng per well) was used for digestion. Calibration curves were built with a recombinant KLK4 within a range 0.05 ng/ml - 50 ng/ml. To differentiate between the two isoforms of KLK4, we targeted one peptide common for both secreted Q9Y5K2–1 and intracellular Q9Y5K2–2 isoforms and two peptides unique for the secreted isoform. The common peptide for both isoforms was used for protein quantification. All samples were analyzed in duplicate.
Quantification of Kallikreins in Biological Samples with ELISA
Concentrations of kallikreins 2, 4, 8, and 13 in SP, CVF, SW and blood serum samples were measured using in-house ELISAs (17, 25). The assays were standardized using recombinant proteins produced in-house in yeast or mammalian expression systems.
Validation of Assay on Quadrupole-orbitrap, Quandrupole-ion Trap, and Triple Quadrupole Platforms
The analytical performance of a multiplex assay of 15 KLKs using three different mass spectrometry platforms (Q Exactive Plus, QTRAP 6500 and TSQ Quantiva) was tested by spiking increasing amounts of the heavy isotope-labeled peptides in SP, CVF, and SW pools and plotting calibration curves of the heavy labeled peptide area against the spiked internal standard concentrations. Completeness of trypsin digestion was assessed by SRM measurement of peptides with a cleavable tag (supplemental Table S8). The coefficient of variation of peptide retention times for all KLKs in 10 SP samples ranged between 0.7% and 1.7%, allowing the scheduling of the method within 2 min intervals. Linearity of assay was validated by spiking increasing amounts of heavy peptides (0.01–20,000 fmol/μl for SP and CVF and 0.01–2,000 fmol/μg for SW) into pools of 10 SP, 5 CVF, and 10 SW samples. Samples with each concentration level were analyzed in triplicate, and the area ratios of internal standards to the endogenous peptides were plotted against internal standard concentrations.
Data analysis
Raw files of shotgun data-dependent acquisition and PRM experiments generated by Q Exactive Plus, raw files of SRM assays generated by TSQ Quantiva and raw .wiff files of SRM assays generated by QTRAP 6500 were analyzed using Skyline Software (v3.1.0.7382; MacCoss Lab), and the .csv files with peptide areas were extracted. Peak integration and peak areas were manually verified and normalized by the respective internal standards to account for variations of sample preparation and MS analysis.
Experimental Design and Statistical Rationale
For the SRM and PRM development, we pooled 5 CVF, 10 SW, or 10 SP samples, in order to reduce inter-individual biological variation. Three process replicates were performed for each pool, and each process replicate was analyzed by mass spectrometry in triplicate. For the individual sample analysis by SRM and PRM, one process and two technical replicates were analyzed. All ELISA and immunoenrichment-SRM assays were performed with process duplicates. GraphPad Prism (v5.03; Graphpad Software, San Diego, CA) was used to generate scatter dot plots, perform statistical analysis and calculate the area under the Receiver Operating Characteristic curve (ROC AUC) and diagnostic sensitivity and specificity. Statistically significant differences for two or three groups of clinical samples were determined using the non-parametric two-tailed Mann-Whitney U or Kruskal-Wallis tests, respectively. p values <0.05 were considered significant.
RESULTS
Development of Targeted Mass Spectrometry Assays
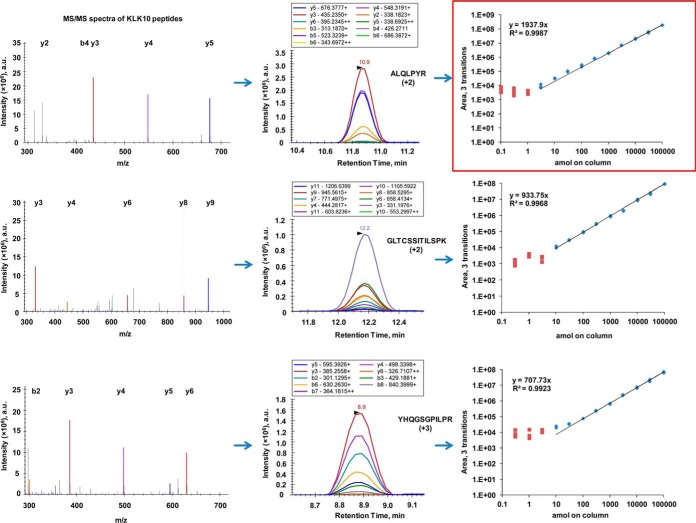
Our method development was based on selection of proteotypic peptides using recombinant proteins and experimental LC-MS/MS, LC-PRM, and LC-SRM data. Shotgun LC-MS/MS revealed sequence coverage 26% to 92% for all 15 human recombinant kallikreins. The top 3 to 4 peptides identified by our shotgun data were chosen for further investigation. The selection of peptides was based on the following criteria: (1) peptides were 7 to 20 amino acids long, (2) contained no methionine, if possible, and (3) contained no N-terminal cysteine or glutamine, if possible. Using PRM mode of the quadrupole-orbitrap mass spectrometer, we monitored simultaneously all possible products of each selected peptide. Because patterns of higher-energy collisional dissociation (HCD) and CID fragmentation are similar (33), the top ten transitions per peptide were selected using Q Exactive Plus data and subsequently tested on the triple quadrupole mass spectrometer. Finally, one proteotypic peptide per protein was used for quantification (supplemental Table S4). The pyro-carbamidomethyl modified peptide of KLK5, as well as the unmodified peptide, were used in the final assay. Multiplex PRM and SRM assays for all 15 KLKs and for three different mass spectrometry platforms (Q Exactive Plus, TSQ Quantiva, and QTRAP 6500) were developed (supplemental Tables S5–7). Superposition of peaks of the heavy isotope-labeled standards and light endogenous peptides, identical peak shapes and the same order of peak heights for heavy and light transitions confirmed the correct identity of each proteotypic peptide and absence of interferences. A workflow of the SRM assay development for kallikrein-10 is presented at Fig. 1.
Fig. 1.
Development of SRM assay for quantification of KLK10. Human recombinant KLK10 was digested by trypsin and subjected to LC-MS/MS, LC-PRM and LC-SRM analyses. Top three peptides were selected based on LC-MS/MS data-dependent acquisition. Top 10 transitions were selected based on LC-PRM experiments. LC-SRM calibration curves for the top three peptides allowed for the selection of a top proteotypic peptide ALQLPYR for quantification of KLK10. Square and diamond shapes denote concentrations below and above LODs, respectively.
According to the UniProt database, our multiplex PRM and SRM assays with a single proteotypic peptide per protein could measure 32 of total 41 alternative splicing isoforms of all 15 KLKs. Missing isoforms included isoform 4 of KLK2, isoform 4 of KLK3, isoform 3 of KLK6, isoform 2 of KLK7, isoform 4 of KLK8, isoform 2 of KLK9, isoform 3 of KLK12, and isoforms 2 and 4 of KLK15. Interestingly, the isoform-specific transcript expression analysis using the Genotype Tissue Expression Portal (www.gtexportal.org) revealed that the relative expression of these missing isoforms in human tissues was negligible (<6% of the major isoform abundance; except isoform 3 of KLK12 with 59%). Assays with three peptides per each KLK (supplemental Tables S1–3) allowed for measurement of all 41 alternative splicing isoforms.
Comparison of PRM and SRM Assays
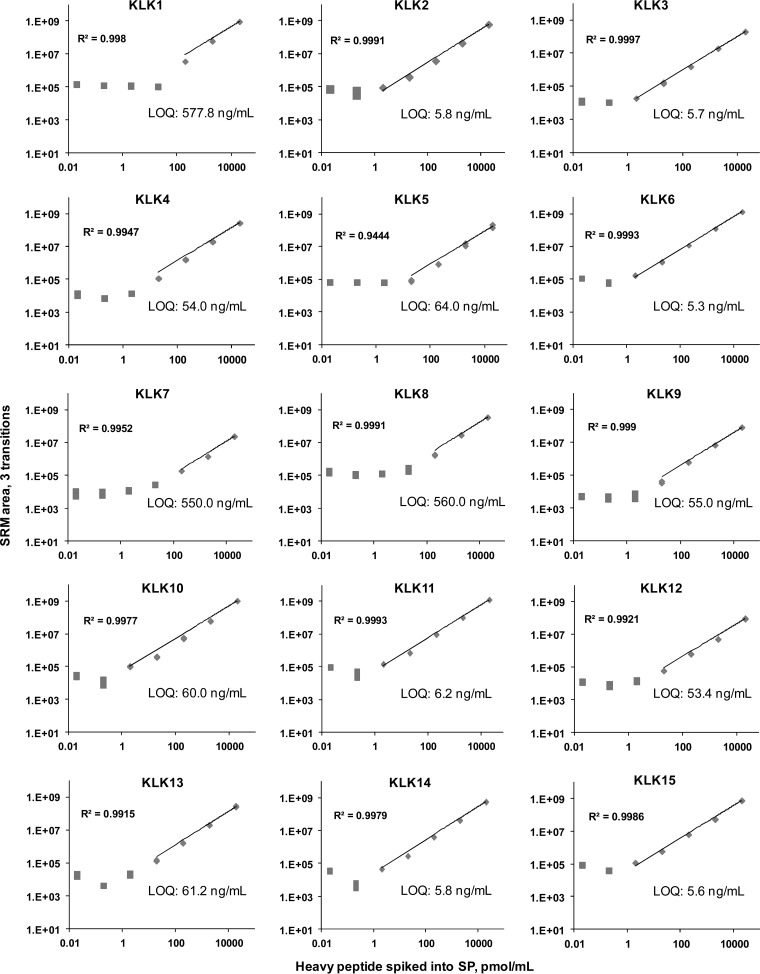
We compared the performance of our final PRM and SRM assays on three different mass spectrometry platforms (quadrupole-orbitrap, quadrupole-ion trap, and triple quadrupole). Calibration curves for the heavy peptides in pooled SP, CVF and SW samples presented linear ranges of 2 to 5 orders of magnitude and linear regression coefficients (R2) greater than 0.99. Limits of detection ranged from 5 to 600 ng/ml (Fig. 2,supplemental Table S9 and supplemental Figs. S4–S5). Comparison of inter-platform calibration curves revealed similar sensitivities and linear ranges for SRM assays executed on quadrupole-ion trap and triple quadrupole instruments. Multiplex PRM assay revealed slightly lower sensitivity because of much higher scan times (100 versus 10 ms) and thus fewer scans per each chromatography peak.
Fig. 2.
SRM method validation in SP using the triple quadrupole mass spectrometer. Each sample was analyzed in triplicates, and LOQs were defined as the lowest concentration measured with CV ≤ 20% within the linear range of the calibration curve. Because of different performances of proteotypic peptides, LOQs ranged from 5.3 to 578 ng/ml. CVs at the LOQ level were between 0.4% and 18.3%. Square and diamond shapes denote concentrations below and above LODs, respectively.
SRM Analysis of Cervico-vaginal Fluid Samples
The total protein amount in the studied CVF samples ranged from 1.6 to 3.0 mg/ml. KLKs 5, 6, 7, 8, 10, 11, and 13 were quantified at low μg/ml levels in at least one of five CVF samples (Table I). KLKs 10 and 13 were quantified in all CVF samples, whereas KLK12 was measured near the detection limit only in two CVF samples (supplemental Fig. S6). Endogenous levels of KLK5 ranged from 0.1 to 1.0 μg/ml. KLK6 concentration ranged from 0.1–3.8 μg/ml, KLK7 from 4.5 to 29.0 μg/ml, KLK10 from 0.4 to 7.4 μg/ml, KLK11 from 0.3 to 1.2 μg/ml, and KLK13 from 0.5 to 2.4 μg/ml. KLK8 was found in 2 out of 5 samples at the 0.4 μg/ml concentration. Interestingly, detection and quantification of KLKs 5 and 8 in CVF by mass spectrometry is reported for the first time.
Table I. Analysis of five CVF samples by SRM. Analysis revealed expression of seven different kallikreins in CVF, with KLK10 and KLK13 being the most abundant. Quantification of KLK5 and KLK8 in CVF by mass spectrometry is reported here for the first time. The samples were analyzed in duplicates and the coefficient of variation of the calculated concentration ranged from 0.2% to 12.7%.
| Kallikrein | Protein concentration (μg/ml) |
||||
|---|---|---|---|---|---|
| CVF1 | CVF 2 | CVF 3 | CVF 4 | CVF 5 | |
| KLK5 | – | – | 1.0 | 0.1 | |
| KLK6 | 3.8 | – | – | 0.1 | 1.1 |
| KLK7 | – | – | 29.0 | 4.5 | – |
| KLK8 | – | – | 0.4 | 0.4 | – |
| KLK10 | 0.4 | 2.8 | 7.4 | 1.7 | 4.0 |
| KLK11 | 0.3 | 0.5 | – | 1.0 | 1.2 |
| KLK13 | 0.5 | 1.3 | 2.4 | 1.0 | 0.4 |
SRM Analysis of Sweat Samples
The total protein amount in SW samples ranged from 0.05 to 0.8 mg/ml. Because total protein concentration in SW could vary substantially because of differences in secretion levels or dilution, amounts of KLKs in each SW sample were normalized per μg of total protein. Results of SRM analysis of individual samples are summarized in Table II. No significant differences in the levels of endogenous KLKs were found between male and female SW samples. To the best of our knowledge, our study reported for the first time detection of KLKs 1, 6, 9, 10, and 13 in human SW by mass spectrometry. KLK1 was detected in five samples at the lowest concentration levels, among all KLKs quantified in SW. KLKs 9 and 10 were detected with concentrations slightly above LOD (supplemental Figs. S7–S8). Kallikreins 5 and 7 were the most abundant KLKs in SW, with concentrations at the mg/g levels. In addition, KLK1 concentration ranged from 0.03 to 0.43 mg/g, KLK5 from 1.3 to 15.9 mg/g, KLK6 from 0.02 to 0.15 mg/g, KLK7 from 11.1 to 67.7 mg/g, KLK8 from 0.5 to 3.17 mg/g, KLK11 from 0.28 to 3.41 mg/g, and KLK13 from 1.68 to 7.19 mg/g.
Table II. Analysis of five female (SW1-SW5) and five male (SW6-SW10) sweat samples by SRM. Analysis revealed expression of nine different kallikreins in sweat, with KLK5 and KLK1 being the most and the least abundant, respectively. The samples were analyzed in duplicates (CVs ranged from 0.2% to 20%).
| Kallikrein | Protein concentration (mg/g) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SW1 | SW2 | SW3 | SW4 | SW5 | SW6 | SW7 | SW8 | SW9 | SW10 | |
| KLK1 | 0.11 | 0.08 | – | – | 0.03 | – | – | – | 0.16 | 0.43 |
| KLK5 | 11.7 | 15.9 | 5.7 | – | – | 3.5 | 4.9 | 1.3 | 2.8 | 5.8 |
| KLK6 | 0.11 | 0.13 | 0.15 | 0.02 | 0.12 | 0.12 | 0.10 | 0.08 | 0.06 | 0.05 |
| KLK7 | - | 67.7 | – | – | – | – | 11.1 | – | 15.8 | – |
| KLK8 | 1.62 | 1.67 | 3.00 | 0.50 | 3.17 | 3.02 | 1.05 | 0.62 | 0.96 | 1.02 |
| KLK11 | 1.92 | 1.28 | 0.74 | – | 3.41 | 0.79 | 0.29 | 0.28 | 0.42 | 0.36 |
| KLK13 | 4.33 | 7.19 | – | – | – | – | 2.85 | 1.68 | 2.32 | 2.24 |
Overall, quantification of KLK7 and KLK12 was proven to be the most challenging among all KLKs. Interestingly, sequences of proteotypic peptides selected for the quantification of KLK7 and KLK12 exhibited high similarity (WVLTAAHCK and WVLTAAHCSGSR, respectively). Difficulty of KLK7 and KLK12 detection might be the result of previously not reported post-translational or chemical modifications, the presence of proteases cleaving the these proteotypic peptides, as well as variations in salt concentration of SW samples. KLK15 was the only member of the kallikrein family that was not detected in any of the studied samples. That was expected because KLK 15 (according to the Human Protein Atlas) is not expressed at the mRNA level in the tissues that secrete proteins to CVF, SW, and SP (supplemental Table S10).
SRM Analysis of Seminal Plasma Samples
The multiplex SRM assay was first applied for the analysis of ten normal pre-vasectomy SP samples. The total protein amounts in SP ranged from 13.6 to 46.8 mg/ml. KLKs 2, 3 and 11 were found to be the most abundant in SP with concentrations ranging from 2.8 to 21.4, 199 to 1368 μg/ml, and 1.24 to 10.4 μg/ml respectively. KLKs 1, 6, and 10 were also detected, but at lower concentrations. Endogenous levels of KLK1 in SP ranged from 0.55 to 0.59 μg/ml. KLK6 concentration ranged from 0.01 to 0.04 μg/ml, whereas KLK10 ranged from 0.06 to 0.26 μg/ml. KLK13 was detected in only two samples, but was not quantified (<61.2 ng/ml). KLK4, a highly prostate-specific protein, was not detected in any of the SP samples. Results of SP analysis are summarized in Table III.
Table III. Analysis of 10 SP samples by SRM. Analysis revealed expression of seven different kallikreins in seminal plasma, with KLK3 (PSA), KLK2, and KLK11 being the most abundant. KLK4 was detected in the low ng/ml level after additional immunoenrichment. The samples were analyzed in duplicates, and CVs ranged from 0.1% to 6.4%.
| Kallikrein | SP1 | SP2 | SP3 | SP4 | SP5 | SP6 | SP7 | SP8 | SP9 | SP10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Protein concentration measured by SRM (μg/ml) | ||||||||||
| KLK1 | – | – | 0.55 | – | – | 0.59 | – | – | – | – |
| KLK2 | 8.09 | 10.2 | 9.03 | 2.79 | 7.36 | 11.3 | 15.2 | 6.41 | 21.4 | 7.79 |
| KLK3 | 579 | 430 | 917 | 199 | 398 | 786 | 1368 | 459 | 412 | 405 |
| KLK6 | 0.02 | 0.01 | 0.04 | 0.03 | 0.01 | 0.01 | 0.02 | 0.01 | 0.02 | 0.01 |
| KLK10 | 0.26 | – | 0.11 | 0.06 | 0.06 | 0.06 | 0.08 | – | 0.10 | – |
| KLK11 | 8.62 | 4.82 | 10.2 | 6.01 | 5.61 | 6.61 | 10.4 | 6.48 | 10.4 | 1.24 |
| Protein concentration measured by immunoenrichment-SRM (ng/ml) | ||||||||||
| KLK4 | 2.9 | 4.3 | 4.3 | 5.7 | 3.6 | 4.3 | 2.9 | 6.4 | 2.9 | 1.4 |
Identification and Quantification of KLK4 in Seminal Plasma
Despite relatively high levels of KLK4 mRNA in the prostate tissue (202 FPKM), concentration of KLK4 protein in SP measured by ELISA was found surprisingly low (ng/ml), thus questioning its secretion (34) and raising concerns about possible cross-reactivity of ELISA with high-abundance KLK2 and KLK3. In an attempt to investigate the presence of KLK4 in SP by orthogonal assays, we analyzed the same set of 10 normal prevasectomy samples by immunoenrichment-SRM. For the purpose of this study, we targeted two peptides for identification of the protein and one peptide for quantification, whereas all samples were analyzed in duplicate. Immunoenrichment of KLK4 resulted in a substantial improvement of assay sensitivity (LOD 0.1 ng/ml versus 50 ng/ml for SRM) and allowed the quantification of KLK4 in all 10 normal SP samples. The concentration of KLK4 in SP was found within the range of 2–7 ng/ml, which was approximately a thousand- and a million-fold lower than the levels of other prostate-specific KLK3 and KLK2, respectively.
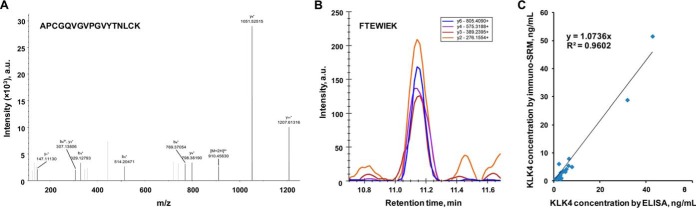
Using immunoenrichment-LC-MS/MS and immunoenrichment-SRM assays, we detected in SP three proteotypic peptides of KLK4 (Fig. 3). According to UniProt, peptide LDESVSESDTIR was shared by the secreted (Q9Y5K2–1; 254 amino acids) and intracellular (Q9Y5K2–2; 107 amino acids) isoforms, whereas peptides APCGQVGVPGVYTNLCK and FTEWIEK belonged exclusively to the secreted isoform. Comparison of peptide areas of the endogenous KLK4 in SP and the recombinant KLK4 suggested the exclusive presence of secreted isoform Q9Y5K2–1 in SP (supplemental Fig. S9). Interestingly, the isoform-specific analysis using the mRNA sequencing data of the GTEx Portal confirmed the predominant expression in prostate of the transcript isoform ENST00000324041 which encoded the secreted protein isoform Q9Y5K2–1 (219 RPKM). Expression of the transcript ENST00000431178 encoding the presumed intracellular isoform Q9Y5K2–2 was negligible (3.5 RPKM) (supplemental Fig. S10).
Fig. 3.
Identification and quantification of KLK4 in SP. A, Immunoenrichment-LC-MS/MS identification of peptide APCGQVGVPGVYTNLCK of the secreted isoform Q9Y5K2–1 of KLK4. B, Immunoenrichment-SRM identification of peptide FTEWIEK of the secreted isoform Q9Y5K2–1. C, Quantification of total KLK4 by ELISA and immunoenrichment-SRM assay in 22 SP samples. SRM assay measured peptide LDESVSESDTIR shared by secreted Q9Y5K2–1 and intracellular Q9Y5K2–2 isoforms. Ratio of intensities of secreted versus total KLK4 revealed the exclusive presence of the secreted isoform Q9Y5K2–1 in SP (see also supplemental Figs. S9–S11). Median CVs: 1.9% by immuno-SRM and 3.9% by ELISA; LOQs: 0.1 ng/ml by immuno-SRM and 0.03 ng/ml by ELISA.
Quantification of KLK4 in Prostate Cancer and Negative Biopsy SP Samples by Immunoenrichment-SRM
Upon identification of the secreted isoform of KLK4 in SP, we next questioned if immunoenrichment-SRM and ELISA measurements would provide correlated results, thus ruling out the potential cross-reactivity of KLK4 antibodies. First, 10 negative biopsy and 12 prostate cancer SP samples were analyzed in duplicates by immunoenrichment-SRM using one peptide for quantification and two additional peptides for confirmation (supplemental Fig. S11). Concentration of KLK4 in prostate cancer SP samples ranged from 1 to 51 ng/ml, whereas concentrations in negative biopsy samples ranged from 0.6 to 53 ng/ml (with CVs 1 to 20%), resulting in no statistically significant difference (p = 0.25).
Quantification of KLK4 in Prostate Cancer and Negative Biopsy SP Samples by ELISA
Because comparison of KLK4 concentrations measured in SP by immunoenrichment-SRM and ELISA showed similar trends (R2 = 0.96), a bigger set of age-matched prostate cancer (n = 21) and negative biopsy (n = 20) SP samples, as well as normal prevasectomy SP samples (n = 10), were analyzed by ELISA. Our goal was to investigate the potential of KLK4 in SP as an independent biomarker to predict the outcome of prostate cancer biopsy or as a biomarker to predict Gleason score (indicator of prostate cancer aggressiveness). All samples were analyzed in duplicates and the CV values ranged from 0.2 to 13%. Concentration of KLK4 in the negative biopsy SP samples were within a range 0.06–53 ng/ml, whereas concentrations in prostate cancer ranged from 0.05 to 46 ng/ml. Statistical analysis revealed no significant difference (p = 0.12) between groups (Fig. 4). Based on the cut-off level of 42 ng/ml derived from the current data, KLK4 levels in SP provided only 9.5% sensitivity at ≥95% specificity and ROC AUC of 0.64 (p = 0.12) for detecting prostate cancer on biopsy. In addition, comparison of low-grade (n = 7; Gleason score = 6), intermediate-grade (n = 7; Gleason score = 7, 3 + 4) and high-grade (n = 7; Gleason score = 7, 4 + 3 and ≥8) prostate cancer samples revealed no statistically significant difference (Kruskal-Wallis test p = 0.28).
Fig. 4.
Evaluation of KLK4 as a prostate cancer biomarker in SP, as measured by ELISA. A, SP samples of three groups of men were analyzed: men with biopsy-confirmed prostate cancer (n = 21, median age 62 y.o.), men with negative biopsy (n = 20, median age 62) and healthy fertile men pre-vasectomy (n = 10, median age 35). Horizontal lines represent median concentrations of proteins in each sample set. No statistically significant differences between three groups of samples were detected (Kruskal-Wallis test p = 0.12). B, ROC curve for prediction of prostate cancer on biopsy using KLK4 measured in SP (AUC = 0.64, p = 0.12).
Quantification of KLK4 in Prostate Cancer and Negative Biopsy Blood Serum Samples by Immunoenrichment-SRM and ELISA
Low diagnostic specificity of KLK3 (PSA) is attributed to the fact that KLK3 is elevated in blood of patients with negative biopsy and prostate inflammation. Thus, novel markers which better differentiate between cancer and benign disease are needed in order to complement or even replace KLK3. Even though KLK4 was previously detected by immunohistochemistry in normal and prostate cancer tissues (35), its concentration in SP, urine and blood serum of prostate cancer patients has never been previously measured.
In order to investigate KLK4 potential as a novel prostate cancer biomarker in blood, we first needed to confirm the presence of its secreted isoform in blood serum and the absence of ELISA cross-reactivity. Thus, we analyzed 36 prostate cancer and 22 negative biopsy blood serum samples by immunoenrichment-SRM. KLK4 was confidently quantified in only 17 of 58 blood serum samples (LOQ = 0.1 ng/ml). KLK4 concentration in negative biopsy serum samples was found in the range from 0.1–0.3 ng/ml, whereas concentrations in prostate cancer ranged from 0.1 to 0.7 ng/ml (CVs 3 to 21%). Statistical analysis showed no significant difference between the two sets of samples (two-tailed Mann-Whitney U test p = 0.19).
Because KLK4 levels in blood serum were below the LOD of our immunoenrichment-SRM assay (0.1 ng/ml) in most samples and because KLK4 concentrations measured by immunoenrichment-SRM and ELISA showed similar trends (R2 = 0.87), the same set of blood serum samples, as well as additional three healthy male samples, were analyzed by ELISA. Our goal was to investigate whether blood serum KLK4 could complement serum PSA≥4 ng/ml and improve its specificity to predict prostate cancer on biopsy. As a result, concentrations of KLK4 ranged from 0.03 to 0.31 ng/ml for negative biopsy and from 0.03 to 1.76 ng/ml for prostate cancer samples. Based on the cut-off level of 0.28 ng/ml, KLK4 levels provided 20% sensitivity at >95% specificity and ROC AUC 0.59 (p = 0.26) for predicting prostate cancer on biopsy. Statistical analysis revealed no significant difference (p = 0.41) between groups (Fig. 5).
Fig. 5.
Evaluation of KLK4 as a prostate cancer biomarker in blood serum, as measured by ELISA. A, Blood serum samples of three groups of men were analyzed: men with biopsy-confirmed prostate cancer (n = 36, serum PSA > 4 ng/ml, median age 63 y.o.), men with negative biopsy (n = 22, PSA > 4 ng/ml, median age 61) and healthy men (n = 3; serum PSA < 1 ng/ml, median age 36). Horizontal lines represent median concentrations of proteins in each sample set. No statistically significant differences between three groups of samples were detected (Kruskal-Wallis test p = 0.41). B, ROC curve for prediction of prostate cancer on biopsy using KLK4 measured in blood serum (AUC = 0.59, p = 0.26).
Comparison of SRM and ELISA Quantification of KLKs 2, 4, 8, and 13
In order to compare the performance, accuracy, reproducibility and sensitivity of our targeted mass spectrometry assays with established ELISAs, we measured KLK2 by SRM in SP, KLK4 by immunoenrichment-SRM in SP and blood serum, KLK8 in sweat, and KLK13 in CVF. Comparison of concentrations measured by SRM and ELISA (Fig. 3 and supplemental Fig. S12) showed similar trends. Concentrations in some samples, however, were not correlated, which may be attributed to degradation of KLKs (potentially missed by ELISA) or cross-reactivity of antibodies.
DISCUSSION
Because of its high selectivity, sensitivity (low pg/ml) and precision (<10%), ELISA remains a gold standard for the quantitative analysis of proteins in complex biological and clinical samples (36). Limitations of ELISA, however, include possible cross-reactivity within protein families, interferences from high-abundance proteins in the matrix, inability to differentiate between proteoforms and post-translational modifications, and high cost of monoclonal antibody production (37).
Recent advances in sensitivity and mass accuracy of mass analyzers, resolving power of nanoscale LC, and automation of sample preparation led to an increasing interest in mass spectrometry as a powerful alternative to ELISA (38–40). A competitive advantage of mass spectrometry was suggested for proteins that had no high-quality antibodies or had ELISAs with suspected cross-reactivities, poor reproducibility, or inconsistent responses for assay calibrators and endogenous analytes. The clear advantage of SRM assays was the very high assay selectivity. The nonscanning mode of quadrupole operation resulted in an increased sensitivity (down to ng/ml) and an increased linear response range (up to six orders of magnitude), if compared with “full scan” modes of ion traps and orbitraps. That enabled the detection and routine quantification of low-abundance proteins in highly complex mixtures, which was crucial for translational and clinical applications of mass spectrometry and proteomics (41–43).
In the present work, our goal was to develop straightforward and easy-to-use targeted mass spectrometry assays for the simultaneous quantification of all 15 tissue kallikrein-related peptidases. We adopted our assays for the three most common mass spectrometry platforms: triple quadrupole, quadrupole-ion trap and quadrupole-orbitrap mass spectrometers. Three peptides were selected for protein identification, whereas one peptide was selected for protein quantification for each KLK. Scheduling of all 15 assays within a single chromatographic gradient enabled monitoring of each peptide within its own retention time window without decreasing scan times or the number of data points across each chromatographic peak.
We reported details of the instrumental and analytical parameters for all three mass spectrometry platforms, thus allowing an easy implementation of our assay in research and clinical laboratories without additional optimization. Multiplex SRM and PRM assays exhibited a linear range of several orders of magnitude and LODs and LOQs in the low ng/ml ranges for most of KLKs, without the need for additional separation or enrichment. For the absolute quantification of each protein, heavy isotope-labeled peptide internal standards with a quantifying tag were synthesized for all 15 KLKs. Our SRM and PRM assays were validated in the top three biological fluids enriched with KLKs (SW, CVF and SP) and were successfully applied for the quantification of endogenous KLKs in sets of biological fluids from healthy individuals. Lower LOQs were obtained with the triple quadrupole mass spectrometer and it was therefore adopted for the analysis of individual samples.
Comparison of KLKs concentrations measured by SRM and ELISAs revealed clear trends, even though correlations were off in some samples. It well may be that both SRM and ELISA recognized only a fraction of the whole population of KLK proteoforms, while missing some post-translational modifications, alternatively spliced forms or complexes with protease inhibitors. For these reasons, we suggest that detailed characterization of KLKs in clinical samples may require both ELISA and SRM measurements. It should be noted that limitations of our SRM assay versus ELISA included lower sensitivity (low ng/ml versus low pg/ml) and lower throughput (12 samples or 180 analytes per sample per day, versus close to 500 samples per day for a well-established ELISA) (44).
In general, cervico-vaginal fluid (CVF) is a pool of secretions originating from the fallopian tube, endometrium, cervix, and vagina (45). KLK levels in CVF samples vary significantly between individuals and between pregnant and nonpregnant women (46) and play important roles in the desquamation of vaginal epithelial cells and the digestion or activation of antimicrobial proteins and peptides. It was also previously reported that certain KLKs played important roles in female urogenital tract malignancies such as endometrial, cervical and ovarian cancers and thus could be used as biomarkers of early diagnosis and prognosis (11). Previous studies by ELISA reported high levels of KLKs 6, 7, 8, 10, 11, 12, and 13 in CVF from nonpregnant women by ELISA (17). KLKs 6, 7, 10, 11, 12, and 13 have also been previously identified in CVF from non-pregnant women by shotgun mass spectrometry (46). Using the proposed SRM assay, we were able to detect eight kallikreins in CVF samples (KLKs 5, 6, 7, 8, 10, 11, 12, and 13), but quantify only seven of them (KLK12 levels were below LOQ). KLK levels measured in our work were in a good agreement with levels previously reported in literature. For example, KLK14 has been previously measured by ELISA within a range of 1.5–112 ng/ml (17). To the best of our knowledge, the present study reported for the first time the mass spectrometric detection of KLKs 5 and 8 in CVF.
Multiple skin kallikreins are expressed either as active or inactive pro-KLK forms in the upper stratum granulosum and stratum corneum of human epidermis and its associated appendages, such as in the inner lumen of sweat gland ducts (47). KLKs cleave substrates that are responsible for skin desquamation and antimicrobial defense. The main limitations of sweat as a clinical sample originate from the difficulty to produce enough sweat for analysis, sample dilution, evaporation during collection and lack of standardized sampling devices. Moreover, the total protein content in sweat may vary significantly between individuals, based on the amount of water consumed on the day of sample collection. Therefore, normalization of results for total protein is crucial. Kallikreins 5, 6, 7, 8, 10, 11, 13, and 14 have been previously quantified in sweat by ELISA (48) and KLKs 5, 7, 8, and 11 have been previously detected by shotgun proteomics (49–51). Using the proposed SRM assay, we were able to detect and quantify nine KLKs in sweat (KLKs 1, 5, 6, 7, 8, 9, 10, 11, and 13). Our findings were in good agreement with the previously reported KLK levels measured by ELISA. For example, concentration of endogenous KLK6 in sweat previously measured with ELISA was found in the range 1–50 μg/g of total protein (48). To the best of our knowledge, the present study reports for the first time the mass spectrometric detection of KLKs 1, 6, 9, 10, and 13 in human sweat. According to the literature, KLK5 and KLK7 are highly expressed in the stratum granulosum and transported to stratum corneum, where they form a proteolytic cascade in which KLK5 activates itself and KLK7 (52). Despite their important role in the skin desquamation, we did not detect KLKs 5 and 7 in all sweat samples. It is worth mentioning that samples which failed for endogenous peptides, also failed for the synthetic heavy labeled internal standards, even after spiking increased amounts of internal standards. We hypothesized that high proteolytic activity in some sweat samples could result in degradation of both light endogenous and heavy internal standard peptides.
Seminal plasma, a proximal fluid originating from secretions of five glands in the male urogenital system, has been previously investigated in detail in respect to KLKs and their potential role in prostate cancer and male infertility (53, 54). KLK3 (PSA) is the major component of SP and is a clinically used prostate cancer biomarker in blood serum (55). Apart from KLK3, KLKs 1, 2, 4, 5, 6, 8, 10, 11, 13, and 14 were previously detected in SP by ELISAs or Western blots (13, 15, 25). KLKs 5, 10, 11, 13, and 14 are expressed in multiple glands in the male urogenital tract, whereas KLKs 2, 3, and 4 are highly tissue-specific proteins and are expressed exclusively in prostate. KLK2 is closely related to KLK3 (sequence identity 80%) and is also highly abundant in SP. Using our SRM assay, we were able to detect and quantify six kallikreins (KLKs 1, 2, 3, 6, 10, and 11) in SP. Vegvari et al. (56) previously reported an SRM method for the analysis of KLK2 in SP using a triple quadrupole mass spectrometer. Their levels of the endogenous KLK2 in SP ranged from 0.8 to 12.3 μg/ml and were in good agreement with our findings. Although we quantified the same proteotypic peptide, we were able to improve the LOQ by more than fivefold because of the use of a more sensitive triple quadrupole mass spectrometer.
Finally, we decided to demonstrate the practical use of our SRM assays and thus focused on a rarely studied prostate-specific KLK4 and its potential use as a prostate cancer biomarker. It should be mentioned that PSA (KLK3) has been used for over two decades for prostate cancer diagnostics and that its introduction into clinic has undoubtedly revolutionized the practice of urology. Although PSA was an exceptional biomarker, it was elevated in both prostate cancer and nonmalignant prostatic diseases, which often resulted in overdiagnosis and unnecessary negative biopsies. The lack of prognostic significance of PSA (failure to predict cancer aggressiveness), often led to overtreatment and unnecessary radical prostatectomies. With all these limitations, not a single emerging biomarker could substantially outperform PSA as yet.
Recent advances in prostate cancer diagnostics included introduction of the 4Kscore test, which measured in blood serum a panel of four analytes: total PSA, free PSA, intact PSA, and KLK2. Interestingly, prostate-specific KLK2 marginally improved the overall performance of 4Kscore test from AUC 0.700 to 0.711 (57). Because the need for novel prostate cancer biomarkers remains, in this work we hypothesized that KLK4, another highly prostate-specific protein, could emerge as an independent diagnostic and prognostic biomarker in SP or a blood serum biomarker to complement serum PSA≥4 ng/ml and improve its diagnostic specificity.
KLK4 is expressed exclusively in the prostate under the control of steroid hormones (34, 58), and its expression is restricted to the luminal and basal epithelium (59, 60). KLK4 mRNA is expressed at high levels in both primary and metastatic prostate adenocarcinomas, and KLK4 protein levels were found to be significantly higher in malignant prostate carcinomas compared with benign and normal prostate tissues (61). Previously, expression of KLK4 gene was studied mostly at the mRNA level. Alternative splicing predictions suggested the existence of extracellular (62) and intracellular (58) protein isoforms. However, immunohistochemical staining confirmed the presence of the intracellular isoform, localized to the cytoplasm of normal human prostate tissue and to primary and metastatic prostate tumor tissues (63, 64), as well as to the nuclei of prostate cancer cells (34, 35). A tremendous discrepancy between high KLK4 mRNA levels in prostate tissue and low ng/ml protein levels in SP were previously discovered. Because immunohistochemistry and ELISA were the only methods to detect KLK4 expression, numerous controversial hypotheses appeared and argued that (1) low levels detected by ELISA were attributed to the cross-reactivity with high-abundance KLK2 and KLK3, whereas KLK4 mRNA was not translated into protein; (2) KLK4 protein was expressed, but quickly degraded, and (3) KLK4 was expressed but not secreted into SP.
Using immunoenrichment-SRM assay, we unambiguously confirmed for the first time the secretion of KLK4 into SP. Overall, presence or absence of KLK4 in SP was an important, but missing piece of knowledge regarding prostate-specific kallikreins. Our experimental evidence that only the secreted isoform Q9Y5K2–1 was present in SP and was measurable by immunoenrichment-SRM and ELISA encouraged us to further investigate the role of KLK4 as a potential prostate cancer biomarker. It should be noted that KLK4 concentration in SP and blood serum of patients with prostate cancer or negative biopsy has never been previously reported.
We investigated the potential of KLK4 in SP as an independent biomarker for two unmet clinical needs: (1) prediction of prostate cancer on biopsy and (2) prediction of prostate cancer aggressiveness. KLK4 levels in SP showed no statistically significant differences for both needs. We also investigated whether blood serum KLK4 could complement serum PSA and predict prostate cancer on biopsy for men with elevated serum PSA ≥ 4 ng/ml. KLK4 levels also revealed no statistically significant differences.
In conclusion, we reported multi-platform targeted mass spectrometry assays for the quantification of all the members of the tissue kallikrein-related peptidases. The present study reported for the first time the mass spectrometry detection of KLKs 5 and 8 in CVF and detection of KLKs 1, 6, 9, 10, and 13 in human sweat. We also demonstrated the presence of a secreted form of KLK4 in SP, and investigated for the first time its potential as an SP- and blood serum-based biomarker of prostate cancer. Comparison of mass spectrometry and ELISA measurements revealed that multiplex SRM and PRM assays can serve as reliable alternative tools to quantify kallikreins and study their biological and pathological roles.
Supplementary Material
Acknowledgments
We thank Keith Jarvi and Susan Lau for providing access to SP samples.
Footnotes
Author contributions: A.P.D. designed the research project, T.D.K. performed the experiments, A.S. and E.P.D. provided antibodies and recombinant kallikreins and assisted with ELISAs, and I.B. assisted with mass spectrometry. A.P.D. and T.D.K. wrote the manuscript, and all authors contributed to the revision of the manuscript.
* This work was supported in parts by grants from the Canadian Institute of Health Research (#285693) to EPD. and APD. and Prostate Cancer Canada (RS2015–01) to APD.
 This article contains supplemental material.
This article contains supplemental material.
Authors declare that they have no conflict of interest.
1 The abbreviations used are:
- KLKs
- Tissue kallikrein-related peptidases
- AUC
- Area under the curve
- CVF
- Cervicovaginal fluid
- ELISA
- Enzyme-linked immunosorbent assay
- FPKM
- Fragments per kilobase of transcript per million mapped reads
- FWHM
- Full width at half maximum
- PRM
- Parallel reaction monitoring
- ROC
- Receiver operating characteristic
- SP
- Seminal plasma
- SRM
- Selected reaction monitoring
- SW
- Sweat.
REFERENCES
- 1. Sotiropoulou G., Pampalakis G., and Diamandis E. P. (2009) Functional roles of human kallikrein-related peptidases. J. Biol. Chem. 284, 32989–32994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yousef G. M., Kopolovic A. D., Elliott M. B., and Diamandis E. P. (2003) Genomic overview of serine proteases. Biochem. Biophys. Res. Commun. 305, 28–36 [DOI] [PubMed] [Google Scholar]
- 3. Yousef G. M., and Diamandis E. P. (2001) The new human tissue kallikrein gene family: structure, function, and association to disease. Endocr. Rev. 22, 184–204 [DOI] [PubMed] [Google Scholar]
- 4. Pathak M., Wong S. S., Dreveny I., and Emsley J. (2013) Structure of plasma and tissue kallikreins. Thromb Haemost 110, 423–433 [DOI] [PubMed] [Google Scholar]
- 5. Attwood B. K., Bourgognon J. M., Patel S., Mucha M., Schiavon E., Skrzypiec A. E., Young K. W., Shiosaka S., Korostynski M., Piechota M., Przewlocki R., and Pawlak R. (2011) Neuropsin cleaves EphB2 in the amygdala to control anxiety. Nature 473, 372–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Michael I. P., Pampalakis G., Mikolajczyk S. D., Malm J., Sotiropoulou G., and Diamandis E. P. (2006) Human tissue kallikrein 5 is a member of a proteolytic cascade pathway involved in seminal clot liquefaction and potentially in prostate cancer progression. J. Biol. Chem. 281, 12743–12750 [DOI] [PubMed] [Google Scholar]
- 7. Borgono C. A., Michael I. P., Komatsu N., Jayakumar A., Kapadia R., Clayman G. L., Sotiropoulou G., and Diamandis E. P. (2007) A potential role for multiple tissue kallikrein serine proteases in epidermal desquamation. J. Biol. Chem. 282, 3640–3652 [DOI] [PubMed] [Google Scholar]
- 8. Hollenberg M. D., Oikonomopoulou K., Hansen K. K., Saifeddine M., Ramachandran R., and Diamandis E. P. (2008) Kallikreins and proteinase-mediated signaling: proteinase-activated receptors (PARs) and the pathophysiology of inflammatory diseases and cancer. Biol. Chem. 389, 643–651 [DOI] [PubMed] [Google Scholar]
- 9. Prassas I., Eissa A., Poda G., and Diamandis E. P. (2015) Unleashing the therapeutic potential of human kallikrein-related serine proteases. Nat. Rev. Drug Discov. 14, 183–202 [DOI] [PubMed] [Google Scholar]
- 10. Yousef G. M., Polymeris M. E., Yacoub G. M., Scorilas A., Soosaipillai A., Popalis C., Fracchioli S., Katsaros D., and Diamandis E. P. (2003) Parallel overexpression of seven kallikrein genes in ovarian cancer. Cancer Res. 63, 2223–2227 [PubMed] [Google Scholar]
- 11. Dorn J., Bayani J., Yousef G. M., Yang F., Magdolen V., Kiechle M., Diamandis E. P., and Schmitt M. (2013) Clinical utility of kallikrein-related peptidases (KLK) in urogenital malignancies. Thromb. Haemost. 110, 408–422 [DOI] [PubMed] [Google Scholar]
- 12. Borgono C. A., and Diamandis E. P. (2004) The emerging roles of human tissue kallikreins in cancer. Nat. Rev. Cancer 4, 876–890 [DOI] [PubMed] [Google Scholar]
- 13. Yousef G. M., Polymeris M. E., Grass L., Soosaipillai A., Chan P. C., Scorilas A., Borgono C., Harbeck N., Schmalfeldt B., Dorn J., Schmitt M., and Diamandis E. P. (2003) Human kallikrein 5: a potential novel serum biomarker for breast and ovarian cancer. Cancer Res. 63, 3958–3965 [PubMed] [Google Scholar]
- 14. Diamandis E. P., Okui A., Mitsui S., Luo L. Y., Soosaipillai A., Grass L., Nakamura T., Howarth D. J., and Yamaguchi N. (2002) Human kallikrein 11: a new biomarker of prostate and ovarian carcinoma. Cancer Res. 62, 295–300 [PubMed] [Google Scholar]
- 15. Borgono C. A., Grass L., Soosaipillai A., Yousef G. M., Petraki C. D., Howarth D. H., Fracchioli S., Katsaros D., and Diamandis E. P. (2003) Human kallikrein 14: a new potential biomarker for ovarian and breast cancer. Cancer Res. 63, 9032–9041 [PubMed] [Google Scholar]
- 16. Diamandis E. P., and Yousef G. M. (2002) Human tissue kallikreins: a family of new cancer biomarkers. Clin. Chem. 48, 1198–1205 [PubMed] [Google Scholar]
- 17. Shaw J. L., and Diamandis E. P. (2007) Distribution of 15 human kallikreins in tissues and biological fluids. Clin. Chem. 53, 1423–1432 [DOI] [PubMed] [Google Scholar]
- 18. Blankley R. T., Fisher C., Westwood M., North R., Baker P. N., Walker M. J., Williamson A., Whetton A. D., Lin W., McCowan L., Roberts C. T., Cooper G. J., Unwin R. D., and Myers J. E. (2013) A label-free selected reaction monitoring workflow identifies a subset of pregnancy specific glycoproteins as potential predictive markers of early-onset pre-eclampsia. Mol. Cell. Proteomics 12, 3148–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shi T., Gao Y., Quek S. I., Fillmore T. L., Nicora C. D., Su D., Zhao R., Kagan J., Srivastava S., Rodland K. D., Liu T., Smith R. D., Chan D. W., Camp D. G. 2nd, Liu A. Y., and Qian W. J. (2014) A highly sensitive targeted mass spectrometric assay for quantification of AGR2 protein in human urine and serum. J. Proteome Res. 13, 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tighe P. J., Ryder R. R., Todd I., and Fairclough L. C. (2015) ELISA in the multiplex era: potentials and pitfalls. Proteomics Clin. Appl. 9, 406–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Percy A. J., Chambers A. G., Yang J., Hardie D. B., and Borchers C. H. (2014) Advances in multiplexed MRM-based protein biomarker quantitation toward clinical utility. Biochim. Biophys. Acta 1844, 917–926 [DOI] [PubMed] [Google Scholar]
- 22. Drabovich A. P., Pavlou M. P., Dimitromanolakis A., and Diamandis E. P. (2012) Quantitative analysis of energy metabolic pathways in MCF-7 breast cancer cells by selected reaction monitoring assay. Mol. Cell. Proteomics 11, 422–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burgess M. W., Keshishian H., Mani D. R., Gillette M. A., and Carr S. A. (2014) Simplified and efficient quantification of low-abundance proteins at very high multiplex via targeted mass spectrometry. Mol. Cell. Proteomics 13, 1137–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huttenhain R., Soste M., Selevsek N., Rost H., Sethi A., Carapito C., Farrah T., Deutsch E. W., Kusebauch U., Moritz R. L., Nimeus-Malmstrom E., Rinner O., and Aebersold R. (2012) Reproducible quantification of cancer-associated proteins in body fluids using targeted proteomics. Sci. Transl. Med. 4, 142ra194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Obiezu C. V., Shan S. J., Soosaipillai A., Luo L. Y., Grass L., Sotiropoulou G., Petraki C. D., Papanastasiou P. A., Levesque M. A., and Diamandis E. P. (2005) Human kallikrein 4: quantitative study in tissues and evidence for its secretion into biological fluids. Clin. Chem. 51, 1432–1442 [DOI] [PubMed] [Google Scholar]
- 26. Drabovich A. P., Pavlou M. P., Schiza C., and Diamandis E. P. (2016) Dynamics of protein expression reveals primary targets and secondary messengers of estrogen receptor alpha signaling in MCF-7 breast cancer cells. Mol. Cell. Proteomics 15, 2093–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kishi T., Grass L., Soosaipillai A., Shimizu-Okabe C., and Diamandis E. P. (2003) Human kallikrein 8: immunoassay development and identification in tissue extracts and biological fluids. Clin. Chem. 49, 87–96 [DOI] [PubMed] [Google Scholar]
- 28. Kishi T., Soosaipillai A., Grass L., Little S. P., Johnstone E. M., and Diamandis E. P. (2004) Development of an immunofluorometric assay and quantification of human kallikrein 7 in tissue extracts and biological fluids. Clin. Chem. 50, 709–716 [DOI] [PubMed] [Google Scholar]
- 29. Shaw J. L., Grass L., Sotiropoulou G., and Diamandis E. P. (2007) Development of an immunofluorometric assay for human kallikrein 15 (KLK15) and identification of KLK15 in tissues and biological fluids. Clin. Biochem. 40, 104–110 [DOI] [PubMed] [Google Scholar]
- 30. Diamandis E. P., Yousef G. M., Soosaipillai A. R., Grass L., Porter A., Little S., and Sotiropoulou G. (2000) Immunofluorometric assay of human kallikrein 6 (zyme/protease M/neurosin) and preliminary clinical applications. Clin. Biochem. 33, 369–375 [DOI] [PubMed] [Google Scholar]
- 31. Kapadia C., Chang A., Sotiropoulou G., Yousef G. M., Grass L., Soosaipillai A., Xing X., Howarth D. H., and Diamandis E. P. (2003) Human kallikrein 13: production and purification of recombinant protein and monoclonal and polyclonal antibodies, and development of a sensitive and specific immunofluorometric assay. Clin. Chem. 49, 77–86 [DOI] [PubMed] [Google Scholar]
- 32. Drabovich A. P., Jarvi K., and Diamandis E. P. (2011) Verification of male infertility biomarkers in seminal plasma by multiplex selected reaction monitoring assay. Mol. Cell. Proteomics 10, M110 004127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu C., Shi T., Brown J. N., He J., Gao Y., Fillmore T. L., Shukla A. K., Moore R. J., Camp D. G. 2nd, Rodland K. D., Qian W. J., Liu T., and Smith R. D. (2014) Expediting SRM assay development for large-scale targeted proteomics experiments. J. Proteome Res. 13, 4479–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xi Z., K. T., Korkmaz K., Kurys P., Elbi C., Risberg B., Danielsen H., Loda M., Saatcioglu F. (2004) Kallikrein 4 is a predominantly nuclear protein and is overexpressed in prostate cancer. Cancer Res. 64, 2365–2370 [DOI] [PubMed] [Google Scholar]
- 35. Dong Y., Bui L. T., Odorico D. M., Tan O. L., Myers S. A., Samaratunga H., Gardiner R. A., and Clements J. A. (2005) Compartmentalized expression of kallikrein 4 (KLK4/hK4) isoforms in prostate cancer: nuclear, cytoplasmic and secreted forms. Endocr. Relat. Cancer 12, 875–889 [DOI] [PubMed] [Google Scholar]
- 36. Lequin R. M. (2005) Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA). Clin. Chem. 51, 2415–2418 [DOI] [PubMed] [Google Scholar]
- 37. Rauh M. (2012) LC-MS/MS for protein and peptide quantification in clinical chemistry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 883- 884, 59–67 [DOI] [PubMed] [Google Scholar]
- 38. Grant R. P., and Hoofnagle A. N. (2014) From lost in translation to paradise found: enabling protein biomarker method transfer by mass spectrometry. Clin. Chem. 60, 941–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Anderson N. L., Anderson N. G., Pearson T. W., Borchers C. H., Paulovich A. G., Patterson S. D., Gillette M., Aebersold R., and Carr S. A. (2009) A human proteome detection and quantitation project. Mol. Cell. Proteomics 8, 883–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Korbakis D., Brinc D., Schiza C., Soosaipillai A., Jarvi K., Drabovich A. P., and Diamandis E. P. (2015) Immunocapture-selected reaction monitoring screening facilitates the development of ELISA for the measurement of native TEX101 in biological fluids. Mol. Cell. Proteomics 14, 1517–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stahl-Zeng J., Lange V., Ossola R., Eckhardt K., Krek W., Aebersold R., and Domon B. (2007) High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol. Cell. Proteomics 6, 1809–1817 [DOI] [PubMed] [Google Scholar]
- 42. Drabovich A. P., Martinez-Morillo E., and Diamandis E. P. (2015) Toward an integrated pipeline for protein biomarker development. Biochim. Biophys. Acta 1854, 677–686 [DOI] [PubMed] [Google Scholar]
- 43. Drabovich A. P., and Diamandis E. P. (2010) Combinatorial peptide libraries facilitate development of multiple reaction monitoring assays for low-abundance proteins. J. Proteome Res. 9, 1236–1245 [DOI] [PubMed] [Google Scholar]
- 44. Whiteaker J. R., Zhao L., Anderson L., and Paulovich A. G. (2010) An automated and multiplexed method for high throughput peptide immunoaffinity enrichment and multiple reaction monitoring mass spectrometry-based quantification of protein biomarkers. Mol. Cell. Proteomics 9, 184–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huggins G. R., and Preti G. (1981) Vaginal odors and secretions. Clin. Obstet. Gynecol. 24, 355–377 [DOI] [PubMed] [Google Scholar]
- 46. Shaw J. L., Smith C. R., and Diamandis E. P. (2007) Proteomic analysis of human cervico-vaginal fluid. J. Proteome Res. 6, 2859–2865 [DOI] [PubMed] [Google Scholar]
- 47. Komatsu N., Saijoh K., Toyama T., Ohka R., Otsuki N., Hussack G., Takehara K., and Diamandis E. P. (2005) Multiple tissue kallikrein mRNA and protein expression in normal skin and skin diseases. Br. J. Dermatol. 153, 274–281 [DOI] [PubMed] [Google Scholar]
- 48. Komatsu N., Tsai B., Sidiropoulos M., Saijoh K., Levesque M. A., Takehara K., and Diamandis E. P. (2006) Quantification of eight tissue kallikreins in the stratum corneum and sweat. J. Invest. Dermatol. 126, 925–929 [DOI] [PubMed] [Google Scholar]
- 49. Burian M., Velic A., Matic K., Gunther S., Kraft B., Gonser L., Forchhammer S., Tiffert Y., Naumer C., Krohn M., Berneburg M., Yazdi A. S., Macek B., and Schittek B. (2015) Quantitative proteomics of the human skin secretome reveal a reduction in immune defense mediators in ectodermal dysplasia patients. J. Invest. Dermatol. 135, 759–767 [DOI] [PubMed] [Google Scholar]
- 50. Csosz E., Emri G., Kallo G., Tsaprailis G., and Tozser J. (2015) Highly abundant defense proteins in human sweat as revealed by targeted proteomics and label-free quantification mass spectrometry. J. Eur. Acad. Dermatol. Venereol. 29, 2024–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Raiszadeh M. M., Ross M. M., Russo P. S., Schaepper M. A., Zhou W., Deng J., Ng D., Dickson A., Dickson C., Strom M., Osorio C., Soeprono T., Wulfkuhle J. D., Petricoin E. F., Liotta L. A., and Kirsch W. M. (2012) Proteomic analysis of eccrine sweat: implications for the discovery of schizophrenia biomarker proteins. J. Proteome Res. 11, 2127–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Brattsand M., Stefansson K., Lundh C., Haasum Y., and Egelrud T. (2005) A proteolytic cascade of kallikreins in the stratum corneum. J. Invest. Dermatol. 124, 198–203 [DOI] [PubMed] [Google Scholar]
- 53. Drabovich A. P., Saraon P., Jarvi K., and Diamandis E. P. (2014) Seminal plasma as a diagnostic fluid for male reproductive system disorders. Nat. Rev. Urol. 11, 278–288 [DOI] [PubMed] [Google Scholar]
- 54. Drabovich A. P., Dimitromanolakis A., Saraon P., Soosaipillai A., Batruch I., Mullen B., Jarvi K., and Diamandis E. P. (2013) Differential diagnosis of azoospermia with proteomic biomarkers ECM1 and TEX101 quantified in seminal plasma. Sci. Transl. Med. 5, 212ra160. [DOI] [PubMed] [Google Scholar]
- 55. Heijnsdijk E. A., Wever E. M., Auvinen A., Hugosson J., Ciatto S., Nelen V., Kwiatkowski M., Villers A., Paez A., Moss S. M., Zappa M., Tammela T. L., Makinen T., Carlsson S., Korfage I. J., Essink-Bot M. L., Otto S. J., Draisma G., Bangma C. H., Roobol M. J., Schroder F. H., and de Koning H. J. (2012) Quality-of-life effects of prostate-specific antigen screening. N. Engl. J. Med. 367, 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vegvari A., Sjodin K., Rezeli M., and Marko-Varga G. (2013) Quantification of human kallikrein-2 in clinical samples by selected reaction monitoring. J. Proteome Res. 12, 4612–4616 [DOI] [PubMed] [Google Scholar]
- 57. Vickers A. J., Cronin A. M., Roobol M. J., Savage C. J., Peltola M., Pettersson K., Scardino P. T., Schroder F. H., and Lilja H. (2010) A four-kallikrein panel predicts prostate cancer in men with recent screening: data from the European Randomized Study of Screening for Prostate Cancer, Rotterdam. Clin. Cancer Res. 16, 3232–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Korkmaz K. S., Korkmaz C. G., Pretlow T. G., and Saatcioglu F. (2001) Distinctly different gene structure of KLK4/KLK-L1/prostase/ARM1 compared with other members of the kallikrein family: intracellular localization, alternative cDNA forms, and Regulation by multiple hormones. DNA Cell Biol. 20, 435–445 [DOI] [PubMed] [Google Scholar]
- 59. Matsumura M., Bhatt A. S., Andress D., Clegg N., Takayama T. K., Craik C. S., and Nelson P. S. (2005) Substrates of the prostate-specific serine protease prostase/KLK4 defined by positional-scanning peptide libraries. Prostate 62, 1–13 [DOI] [PubMed] [Google Scholar]
- 60. Takayama T. K., McMullen B. A., Nelson P. S., Matsumura M., and Fujikawa K. (2001) Characterization of hK4 (prostase), a prostate-specific serine protease: activation of the precursor of prostate specific antigen (pro-PSA) and single-chain urokinase-type plasminogen activator and degradation of prostatic acid phosphatase. Biochemistry 40, 15341–15348 [DOI] [PubMed] [Google Scholar]
- 61. Klokk T. I., Kilander A., Xi Z., Waehre H., Risberg B., Danielsen H. E., and Saatcioglu F. (2007) Kallikrein 4 is a proliferative factor that is overexpressed in prostate cancer. Cancer Res. 67, 5221–5230 [DOI] [PubMed] [Google Scholar]
- 62. Stephenson S. A., Verity K., Ashworth L. K., and Clements J. A. (1999) Localization of a new prostate-specific antigen-related serine protease gene, KLK4, is evidence for an expanded human kallikrein gene family cluster on chromosome 19q13.3–13.4. J. Biol. Chem. 274, 23210–23214 [DOI] [PubMed] [Google Scholar]
- 63. Day C. H., Fanger G. R., Retter M. W., Hylander B. L., Penetrante R. B., Houghton R. L., Zhang X., McNeill P. D., Filho A. M., Nolasco M., Badaro R., Cheever M. A., Reed S. G., Dillon D. C., and Watanabe Y. (2002) Characterization of KLK4 expression and detection of KLK4-specific antibody in prostate cancer patient sera. Oncogene 21, 7114–7120 [DOI] [PubMed] [Google Scholar]
- 64. Obiezu C. V., Soosaipillai A., Jung K., Stephan C., Scorilas A., Howarth D. H., and Diamandis E. P. (2002) Detection of human kallikrein 4 in healthy and cancerous prostatic tissues by immunofluorometry and immunohistochemistry. Clin. Chem. 48, 1232–1240 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.