Abstract
The extracellular matrix (ECM) is a fundamental component of multicellular organisms that provides mechanical and chemical cues that orchestrate cellular and tissue organization and functions. Degradation, hyperproduction or alteration of the composition of the ECM cause or accompany numerous pathologies. Thus, a better characterization of ECM composition, metabolism, and biology can lead to the identification of novel prognostic and diagnostic markers and therapeutic opportunities. The development over the last few years of high-throughput (“omics”) approaches has considerably accelerated the pace of discovery in life sciences. In this review, we describe new bioinformatic tools and experimental strategies for ECM research, and illustrate how these tools and approaches can be exploited to provide novel insights in our understanding of ECM biology. We also introduce a web platform “the matrisome project” and the database MatrisomeDB that compiles in silico and in vivo data on the matrisome, defined as the ensemble of genes encoding ECM and ECM-associated proteins. Finally, we present a first draft of an ECM atlas built by compiling proteomics data on the ECM composition of 14 different tissues and tumor types.
Keywords: Extracellular matrix, Matrisome, MatrisomeDB, ECM atlas, Bioinformatics, Proteomics
INTRODUCTION
The extracellular matrix (ECM), a complex meshwork of proteins, is a fundamental component of multicellular organisms. The ECM is the three-dimensional architectural scaffold that defines tissue boundaries and biomechanical properties [1] and cell polarity. It also serves as an adhesive substrate for cell migration and, by binding morphogens and growth factors [2], can create concentration gradients for haptotactic migration [3, 4] or pattern formation [5]. Extracellular matrix proteins provide biochemical cues interpreted by cell surface receptors, such as the integrins [6] and initiate signaling cascades controlling cell survival, cell proliferation, differentiation and stem cell state [7, 8].
Advances in genome sequencing have allowed one to trace the evolution of the ECM and have revealed that, although some domains or modules characteristic of ECM proteins existed in unicellular organisms, the elaboration of ECM proteins and ECMs appeared largely in metazoa [9, 10]. The multiplication and diversification of ECM proteins has accompanied major evolutionary innovations, in particular in the vertebrate phylum, including the appearance of a closed vascular system and of structures such as the neural crest, teeth, cartilage and bones [9, 10]. During development, ECM plays vital roles in stem cell niches and in guiding migration and polarity of cells and axonal projections and in morphogenesis and coherence of tissues. Remodeling of ECM is essential during angiogenesis and branching morphogenesis of glands and in wound healing, as it is in cancer invasion [5].
Mutations in ECM genes are causal of musculo-skeletal, cardio-vascular, renal, ocular and skin, diseases [11]. In addition, excessive deposition or, conversely, destruction of the ECM can also lead to pathologies such as fibrosis or osteoarthritis [12]. ECM deposition (or desmoplasia) is a hallmark of tumor progression and has been used by pathologists as a marker of tumors with poor prognosis even long before the composition and the complexity of the ECM were uncovered. Recent studies from us and others have revealed that the ECM plays a functional role in tumor progression and dissemination [13, 14].
We previously proposed that characterizing the global composition of the extracellular matrix proteome, or “matrisome” of normal and diseased tissues would lead to important discoveries. This proposition raised two important questions: how do we define the extracellular matrix and how can one study the composition of a compartment largely made of insoluble proteins? Here, we review the bioinformatic tools and experimental approaches that have been developed over the last few years to characterize the composition of extracellular matrices. We also introduce a web interface “the matrisome project” that hosts a novel database on ECM and ECM-associated genes and proteins, MatrisomeDB that compiles in silico and experimental resources and we illustrate their utility to analyze various “omics” datasets. Finally, we present here a first draft of an ECM atlas built by compiling proteomics data on the extracellular matrix of 14 different tissues and tumor types.
1. IN SILICO DEFINITION OF THE MATRISOME
Bioinformatic approach to define the matrisome
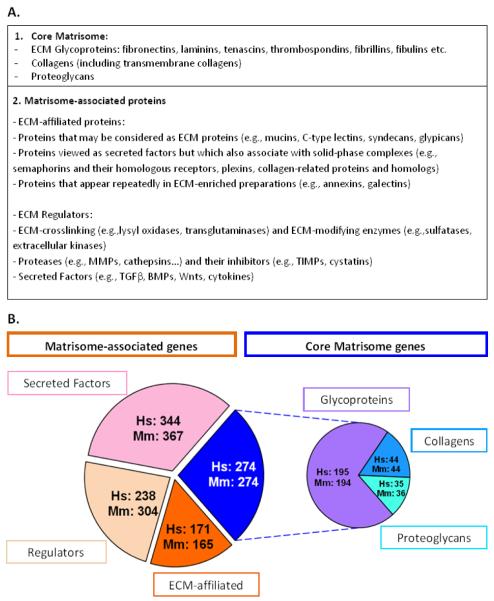
In 1984, Martin and colleagues coined the term “matrisome” in the context of basement membranes to define “supramolecular complexes of matrix components which are the functional units of the forming extracellular matrix” [15]. In 2012, we proposed to extend the definition of this term to include not only all the genes encoding structural ECM components but also genes encoding proteins that can (or may) interact with or remodel the ECM [16–18] (Figure 1A). To define the matrisome, we devised a bioinformatic approach to screen the human and mouse proteomes (or any other genomes of interest) using defining features of ECM proteins such as presence of signal peptide and of protein domains characteristic of ECM proteins [16, 19]. We further curated manually the lists obtained computationally and, based on structural or functional features, distinguished core matrisome proteins from matrisome-associated proteins. The core matrisome comprises ECM glycoproteins, collagens and proteoglycans (Figure 1A) (for reviews on each of these categories of proteins see [20–22]). ECM-associated proteins include ECM-affiliated proteins, ECM regulators and secreted factors that may interact with core ECM proteins (Figure 1A) [16].
Figure 1. Defining the matrisome.
A. Definition of matrisome categories. The core matrisome comprises ECM glycoproteins, collagens and proteoglycans. Matrisome-associated proteins include ECM-affiliated proteins, ECM regulators and secreted factors [16, 17].
B. Pie charts represent the number of human genes encoding core matrisome and matrisome-associated proteins. “Hs” indicates genes in the human genome and “Mm”, genes in the murine genome.
We note here that the criteria for assigning certain proteins to the matrisome categories can be debated. For example, the designation of the various secreted factors was deliberately inclusive since, although many of them are not known to associate with ECM, some clearly do and arguments can be adduced supporting the concept that many others do as well [2]. Equally, decisions such as where, or whether, to include some proteins containing short collagen triple helical domains or transmembrane segments are to some extent arbitrary. We view the current categorizations to be a working structure, subject to periodic revision in light of future discoveries.
Matrisome 2.0
The in silico definition of the matrisome relied on the interrogation of the protein database UniProt [23] using lists of domains from the InterPro [24] and SMART [25, 26] databases. The protein-centric lists originally defined were then turned into gene-centric lists using NCBI Entrez Gene as reference gene database [27]. Databases are, by nature, very dynamic and constantly updated. The release of a major update by UniProt in July 2014 with improved identification of isoforms, new knowledge (such as the identification of the first extracellular kinases [28]) and discussions with colleagues have prompted us to update the original matrisome lists. This updated version of the matrisome (v2.0) comprises 1027 genes for the human genome and 1110 genes for the mouse genome (Figure 1B). These numbers are slightly smaller than the ones we reported in 2012; this is mainly due to better annotations of pseudogenes (now removed from the matrisome lists) and removal of duplicate entries in the newer UniProt database. It is worth noting that the relatively large difference in the number of genes encoding ECM regulators in human and mouse is mostly due to gene duplication of proteases such as ADAMs (a disintegrin and metalloprotease domain proteins) and serpins (serine protease inhibitors) in the mouse genome. In addition to updating the gene-centric lists, we have also updated the list of UniProt accession numbers associated with each matrisome gene. These updated matrisome lists as well as updated domain lists are available for download from the web interface we recently developed http://matrisomeproject.mit.edu [29]. This website hosts a new database, matrisomeDB, that provides, for all human and murine matrisome genes, live cross-referencing to gene (HUGO Gene Nomenclature Committee [30] and Mouse Genome Informatics [31]) and protein (UniProt, InterPro) databases, and information on gene orthology. In addition to providing links to in silico resources on matrisome genes and proteins, matrisomeDB also incorporates experimental proteomics data generated in the Hynes lab. Database users can now easily determine whether or not a given ECM protein has been detected by proteomics in the tissues and tumors profiled (see below).
2. MATRISOME LISTS AS ANNOTATION TOOLS FOR BIG DATA
Identifying ECM signatures in gene and protein datasets
We previously noted that the matrisome lists and categories we defined are significantly more comprehensive than Gene Ontology’s “Cellular Component” categories for data mining and for posing questions relevant to ECM biology, since ECM proteins are currently scattered in multiple GO categories and comingled with non-ECM proteins [17]. In order to further facilitate the use of categorical matrisome lists for annotating genomic, transcriptomic or proteomic outputs and identifying ECM signatures within datasets, we derived a collection of ten matrisome gene sets (Table 1) and implemented them into the Molecular Signatures Database maintained by the Broad Institute. In addition to allowing rapid annotation of large datasets and facilitating the identification of ECM signatures, these gene sets can now readily be used to conduct Gene Set Enrichment Analysis (GSEA) studies [32, 33]. GSEA is a computational method that determines whether an a priori defined set of genes (for example, the matrisome) shows statistically significant, concordant differences between two biological states. Hussenet, Orend and collaborators used such an approach to identify a matrisome signature enriched in the transcriptome of angiogenic vs non-angiogenic oncogenic pancreatic islets, which they termed the “angiomatrix” signature [34]. Once identified, ECM signatures (of a given tissue or disease state) can be further analyzed with pathway or interaction analysis tools such as MatrixDB [35, 36].
Table 1. Matrisome gene sets available in the Molecular Signature Database (MSigDB v5.0) and integrated in the collection C2: Canonical Pathways.
This table provides a description of the 10 matrisome gene sets and links to their pages in MSigDB where lists of genes can be downloaded.
| Name | # Genes | Description |
|---|---|---|
| NABA_MATRISOME | 1027 | Ensemble of genes encoding extracellular matrix and extracellular matrix-associated proteins |
| NABA_CORE_MATRISOME | 274 | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| NABA_ECM_GLYCOPROTEINS | 195 | Genes encoding structural ECM glycoproteins |
| NABA_COLLAGENS | 44 | Genes encoding collagen proteins |
| NABA_PROTEOGLYCANS | 35 | Genes encoding proteoglycans |
| NABA_BASEMENT_MEMBRANES | 40 | Genes encoding structural components of basement membranes |
| NABA_MATRISOME_ASSOCIATED | 753 | Ensemble of genes encoding ECM-associated proteins including ECM-affiliated proteins, ECM regulators and secreted factors |
| NABA_ECM_AFFILIATED | 171 | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| NABA_ECM_REGULATORS | 238 | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| NABA_SECRETED_FACTORS | 344 | Genes encoding secreted soluble factors |
ECM signatures can also be used to interrogate other publicly available datasets. For example, we compared the ECM signatures of primary colorectal tumors and their liver metastases defined by proteomic analysis of small numbers of patient samples, with gene expression data from four clinical studies representing over 200 patients and demonstrated the association of a subset of the ECM proteins defined by proteomics with tumor progression [37].
Interrogating databases with matrisome lists
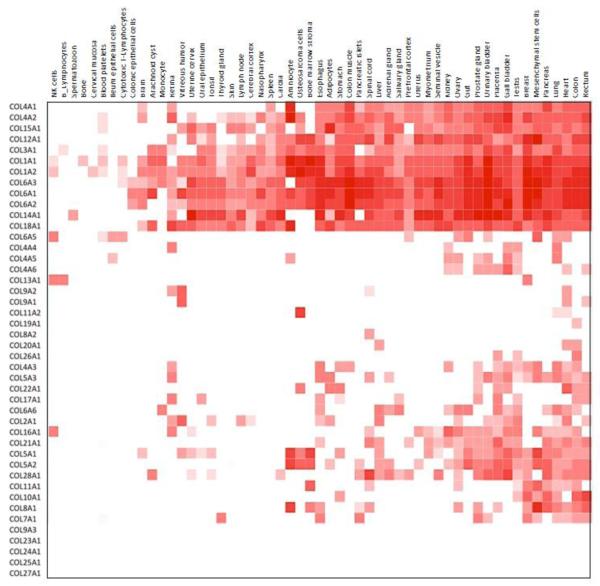
The two examples above illustrate the value of using limited experimental data on ECM genes or proteins to access and leverage the burgeoning pool of publicly available data from diverse modes of analysis. The development of high-throughput transcriptomics studies (microarray or RNAseq approaches) has been accompanied by the creation of databases to make the large body of data publicly available [38, 39]. Recent papers have also reported the first drafts of the human proteome [40–42] which has led to the development of a website http://proteomicsdb.org. One can therefore use the matrisome list or sublists and ask which ECM genes or proteins are detected in which tissues, at which developmental stage(s), etc. As an example, we interrogated proteomicsDB with the list of 44 human collagen genes (Figure 2). Although the most abundant and ubiquitous collagens (type I, III, IV, VI) were detected in most tissue and cell proteomes, less abundant or rarer collagens were seen only in a few tissues or in none so far. This may reflect reality but could also result from the fact that most studies included in proteomicsDB were not designed to look specifically for ECM proteins and may well have missed them. The very nature of ECM proteins, which are often very large, highly glycosylated, cross-linked, and difficult to solubilize, has made biochemical analyses of the composition of extracellular matrices challenging.
Figure 2. Expression of collagen chains across 55 tissue and cellular proteomes.
ProteomicsDB (April 2015 release) was interrogated with the 44 human collagen genes. The expression of 39 out of the 44 human collagen chains was reported in at least one of the 55 tissues or cell types for which global proteomics data were available. The five collagen chains not detected are: COL9A3, COL23A1, COL24A1, COL25A1, COL27A1.
3. EXPERIMENTAL CHARACTERIZATION OF IN VIVO MATRISOMES BY PROTEOMICS
Technical challenges and proposed solutions
ECM protein enrichment
The profiling of the protein composition of extracellular matrices by mass spectrometry is dependent on the availability of methods that selectively enrich for ECM proteins. Taking advantage of the insolubility of ECM proteins, we and others have reported the development of decellularization and ECM-enrichment methods that rely on the extraction of - somewhat more soluble - intracellular proteins [16, 43–46]. The insolubility of ECM proteins is, however, a challenge for subsequent proteomic analyses; indeed mass-spectrometry pipelines require proteins to be solubilized and then digested into peptides.
ECM protein solubilization
Complete ECM protein solubilization requires reagents in concentrations that are incompatible with mass spectrometry. In addition, a fraction of the extracellular matrix is resistant to solubilizing agents (such as SDS or urea) and would be lost if only solubilized material were carried forward. Therefore, we proposed that crude ECM-enriched samples be denatured, reduced and alkylated, deglycosylated, and digested by a combination of proteases (LysC and trypsin), all without intervening centrifugation, in order to minimize losses of otherwise insoluble materials. Using this approach, we could show that solubilization occurs as a concomitant of protease treatment [16, 43].
Identification of MS spectra and peptides
ECM proteins, in particular collagens, are known to contain extensive sites of unique posttranslational modifications such as hydroxylated lysines or prolines [47, 48]. It is thus important, when conducting database searches for identification of spectra, to allow for such posttranslational modifications.
Using pipelines combining ECM-enrichment procedures and mass spectrometry, we and others have reported the characterization of the composition of ECMs of several normal and diseased tissues (Table 2) [16, 13, 37, 46, 49–63]. As expected, these studies revealed the presence of largely ubiquitously expressed ECM proteins but also identified tissue-specific proteins. For example the Önnerfjord laboratory showed that different cartilages have different ECM compositions [58]. Moreover, using an approach combining quantitative and targeted mass spectrometry, they showed that the composition of articular cartilage presents spatial variation and demonstrated that asporin, tenascin-C, thrombospondin-4 and perlecan were the most abundant in the superficial cartilage layer, whereas mimecan and thrombospondin-1 were most abundant in the intermediate layer and aggrecan, osteomodulin, chondroadherin were enriched in deeper layers of articular cartilage. The Mayr laboratory reported the characterization of the cardiac ECM and aortic basement membrane and further identified ECM signatures of ischemia/reperfusion and abdominal aortic aneurysm [52, 55, 56]. Using human melanoma and mammary carcinoma xenografts in mouse, and the ability of mass spectrometry to distinguish human (tumor-derived) protein sequences from their murine (stroma or host-derived) counterparts, we have demonstrated that both the tumor cells and the stromal cells contribute to the production of the tumor ECM [13, 16]. We have also shown that several ECM proteins differentially expressed between poorly and highly metastatic tumors (including Latent TGFβ Binding Protein 3, LTBP3 and the protein Sushi, Nidogen, and EGF-like Domains 1, SNED1) were causal of a more metastatic phenotype [13]. Furthermore, the comparison of the matrisomes of normal and paired tumor samples identified consistent differences in the ECMs of (i) primary tumors as compared with normal surrounding tissues, (ii) metastases and the normal tissue in which they develop, and (iii) primary tumors as compared with metastases derived from them [37]. Finally, using both xenografts and human-patient-derived samples, we could show that some ECM proteins identified could serve as potential biomarkers for tumor progression, metastasis and survival [13, 37].
Table 2.
List of normal and diseased tissues, for which ECMs have been profiled by mass spectrometry
| Tissue_Species | Species | References |
|---|---|---|
| Normal tissues | ||
| Aorta | Human | Didangelos et al., 2010 [52] |
| Bone | Rat | Schrieweis et al., 2007 [50] |
| Cartilage; Growth plate cartilage | Mouse | Belluoccio et al., 2006 [49]; Wilson et al., 2010 [54] |
| Articular and tracheal cartilages, meniscus, intervertebral disc, ribs |
Human | Önnerfjord et al., 2012[58]; Müller et al., 2014 [61] |
| Colon | Mouse | Naba et al., 2012 [16] # |
| Colon | Human | Naba et al., 2014b [37] # |
| Lung | Human | Booth et al., 2012 [57] |
| Lung | Rat | Hill et al., 2015 [63] |
| Liver | Human | Naba et al., 2014b [37] # |
| Mammary gland | Rat | Hansen et al., 2009 [46]; O'Brien et al., 2012 [53] |
| Glomerular basement membrane | Human | Lennon et al., 2014 [60] # |
| Retinal vascular basement membrane, Lens capsule, Inner limiting membrane |
Human | Uechi et al., 2014 [62] # |
| Retinal vascular basement membrane | Chick embryo | Balasubramani et al., 2010 [51] |
| Diseased tissues | ||
| Poorly and highly metastatic melanoma xenografts |
Human/Mouse | Naba et al., 2012 [16] # |
| Poorly and highly metastatic mammary carcinoma xenografts |
Human/Mouse | Naba et al., 2014a [13] # |
| Metastatic primary colon carcinoma and derived liver metastases |
Human | Naba et al., 2014b [37] # |
| Fibrotic lung | Mouse | Decaris et al., 2014 [59] |
| Abdominal aortic aneurysm | Human | Didangelos et al., 2011 [55] |
| Cardiac ECM remodeling during ischemia/reperfusion |
Pig | Barallobre-Barreiro et al., 2012 [56] |
indicates studies for which raw mass spectrometry were made publicly available and used to build the draft of the ECM atlas (see Tables 3 & 4 and Supplementary Table 1).
Building an ECM atlas
The large amount of data generated in “omics” studies has prompted researchers to share their data with the scientific community through dedicated repositories such as Gene Expression Omnibus or GEO for transcriptomics data and ProteomeXchange for proteomics data. In fact more and more journals now strongly encourage or even require that the raw data accompanying publications be deposited in public databases.
We sought to build a first draft of an ECM atlas by integrating publicly available mass spectrometry data from studies designed specifically to characterize the global compositions of ECMs (Table 2). We gathered raw mass spectrometric data from our own group [13, 16, 37], data on the glomerular basement membrane from the Humphries lab [60] and data on three different ocular basement membranes (retinal vascular basement membrane, lens capsule, inner limiting membrane) from the Balasubramani group [62]. To provide consistent data analysis, all proteomics datasets were reanalyzed using the search pipeline we previously developed and peptides identified with a false discovery rate < 1.6% were assembled into proteins (using a UniProt database) and annotated as being ECM-derived or not (Supplementary Table 1) [16]. As the data were acquired on different mass spectrometers, we cannot readily derive quantitative information regarding the relative abundance of each protein across tissues. Nonetheless, this compilation provides a first draft of an ECM atlas.
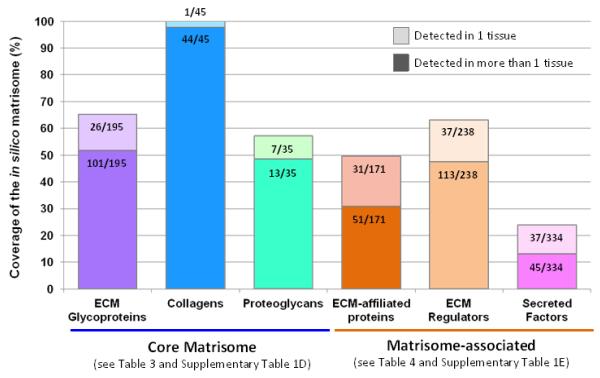
Compared with the “in silico” matrisome, this study shows that more than 60% of the predicted ECM glycoproteins were detected in one or more tissues (Figure 3). Interestingly, several ECM proteins, in particular members of the insulin-like-growth-factor-binding protein family (IGFBP3, IGFBP4, IGFBP5), members of the CCN family (CTGF and CYR61), thrombospondin-2 (Thbs2), tenascin-N (Tnn) and VWA9 were only detected, so far, in ECM-enriched preparations from tumor samples (colorectal or mammary carcinomas, melanomas) but not in those from normal tissues (Table 3, Supplementary Table 1C, 1D). All the collagen chains were detected in one or more tissues, even though some are often thought to be cartilage-specific (type II and type IX collagens) (Figure 3 and Table 3). Close to 60% of the predicted proteoglycans were detected. We hypothesize that the matrisome proteins not yet detected will be found in other tissues or at different developmental stages. For example, many ECM proteins are known to be exclusively expressed in teeth (such as ameloblastin, amelogenins, dentin sialophosphoprotein) [64], bones (such as bone gamma-carboxyglutamate (Gla) protein or osteocalcin, integrin-binding sialoprotein), the inner ear (such as the tectorins) or the nervous system [65, 66].
Figure 3. Experimental coverage of the in silico of the matrisome.
Bar chart represents, for each matrisome category, the percentage representation and number of ECM genes detected in one (lighter bars) or more than one (darker bars) tissue.
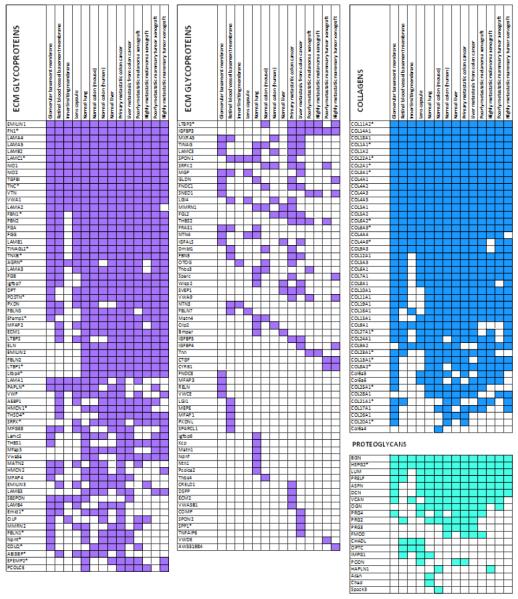
Table 3. ECM atlas part I: Core matrisome proteins detected by mass spectrometry.
List of ECM glycoproteins, collagens and proteoglycans detected with at least 2 peptides in one of the 14 tissues compiled in the ECM atlas: human glomerular basement membrane, human retinal vascular basement membrane, human inner limiting membrane (eye), human lens capsule, murine lung, murine colon, human colon, human liver, poorly and highly metastatic melanoma and mammary carcinoma xenografts.
indicates proteins for which isoform-specific peptides were detected (see Supplementary Table 1C for details).
ECM proteins exist in many different isoforms, and the presence or absence of certain spliced domains can change the function of a protein or be indicative of a diseased state. For example, fibronectin can include two fibronectin type III domains, each encoded by one exon (EIIIB and EIIIA), and the expression of the isoforms of fibronectin including one or both exons have been shown to be restricted to stages of the development, remodelling tumor vasculature and sites of injuries [67]. Similar results are true for spliced variants of tenascins [68] and other ECM proteins. The protein database UniProt we used to generate this first draft of the ECM atlas includes comprehensive nomenclature and data on protein isoforms. We were thus able to provide isoform information for several of the core matrisome proteins constituting the ECM atlas (see Table 3 and Supplementary Table 1C and 1D).
The experimental coverage of the predicted matrisome-associated proteins is not as high: 50% of the ECM-affiliated proteins, over 60% of the ECM regulators and only 25% of the secreted factors have been detected in at least one tissue and a large proportion of these proteins were detected in only one tissue (see lighter bars Figure 3 and Table 4 and Supplementary Table 1C and 1E). Whether this lower coverage and apparent tissue specificity is true or a consequence of the state of current analyses will need to be determined. Indeed, matrisome-associated proteins and, in particular secreted factors and ECM remodeling enzymes, are typically present in lower stoichiometry and their low abundance may compromise their identification by mass spectrometry. Some proteins apparently specific to a given tissue type may simply be present but in too low abundance in other tissues to be detected. This may be overcome by implementing “targeted” mass spectrometry approaches, which allow focus on the measurement of a subset of proteins suspected or known to be present in a given sample [69, 70]. Matrisome-associated proteins are also likely to be more soluble than core matrisome proteins and could be lost during decellularization. One approach to retrieve information on more soluble proteins would be to profile not only the composition of the insoluble ECM fraction generated by tissue decellularization but also the composition of the intermediate fractions generated during the decellularization process [43, 63] and the soluble components of tissue interstitial fluid. Of course, these intermediate fractions will also be enriched for intracellular components, thus robust data annotations using matrisome lists could assist with delineating which proteins are soluble matrisome proteins and which are contaminants. Finally, when we initially defined the list of matrisome-associated proteins, we wanted to be inclusive and have included entire families of proteins some of which may not be found in close association with core ECM proteins. This may have resulted in an over-prediction and, based on future experiments, we may revise our definition and distinguish more precisely those proteins belonging to the matrisome from those belonging to the secretome (the human protein atlas [71] predicts that over 3000 human genes have a secreted product).
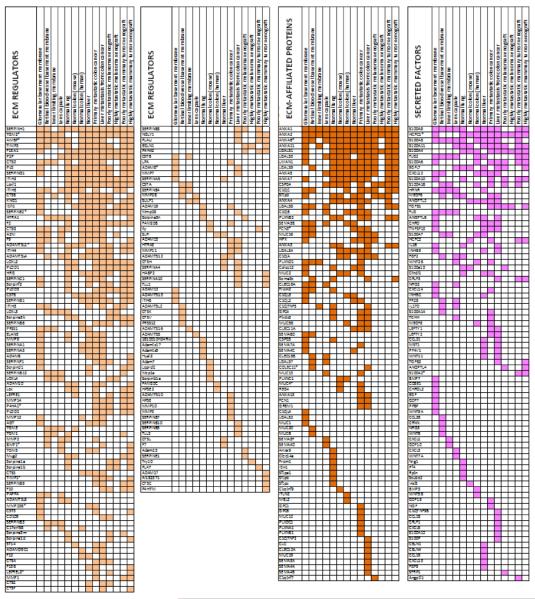
Table 4. ECM atlas part II: Matrisome-associated proteins detected by mass spectrometry.
List of ECM-affiliated proteins, ECM regulators and secreted factors detected with at least 2 peptides in one of the 14 tissues compiled in the ECM atlas: human glomerular basement membrane, human retinal vascular basement membrane, human inner limiting membrane (eye), human lens capsule, murine lung, murine colon, human colon, human liver, poorly and highly metastatic melanoma and mammary carcinoma xenografts.
|
indicates proteins for which isoform-specific peptides were detected (see Supplementary Table 1C for details).
4. FUTURE DIRECTIONS IN ECM RESEARCH
With the tools and methods now in place, we can postulate and certainly hope that the completion of a human ECM atlas will be achievable within the next few years. We would like to incorporate additional datasets (including the ones listed in Table 2) in the atlas if/when they become publicly accessible and we would like to encourage the deposition of all future research data in the public domain.
Time- and spatially-resolved matrisomes
The analysis of the composition of the ECM of tissues has already revealed novel or unsuspected components of the ECM characteristics of a diseased state or causal of a diseased state. Efforts should now focus on obtaining temporally- and spatially-resolved matrisomes. For those interested in the role of the ECM in diseases, we propose that quantitative proteomics will become a method of choice to profile the dynamic changes in the composition of the extracellular matrix that occur during disease progression or during the course of treatment [72]. We hypothesize that this type of study will allow the identification of novel prognostic or diagnostic ECM markers that could assist clinical decisions (see below).
Development of high-throughput approaches to map post-translational modifications of ECM proteins
ECM proteins undergo extensive post-translational modifications including hydroxylation, phosphorylation, sulfation, glycosylation, and crosslinking, that can have significant physiological and/or pathological implications [73]. We propose that beyond the profiling of the ECM proteome, the community should aim for broad implementation of novel “omics” approaches such as glycomics [74–76] to profile the post-translational modifications of ECM proteins.
ECM proteins also undergo proteolytic cleavage and release fragments as part of their physiological (or pathological) turnover. Increased ECM degradation is a hallmark of pathologies such as osteoarthritis, fibrosis, cancers [12]. ECM protein fragments displaying biological activities are termed matricryptins or matrikines [77]. These fragments are characterized by novel amino- and/or carboxy-terminal extremities and can thus be identified by mass spectrometry. The emergence of degradomics or terminomics approaches [78] offers interesting opportunities for ECM research. In addition, it has been proposed that the identification of cleaved fragments can also serve as proxy for identifying proteases (e.g. MMPs, cathepsins) active in a given tissue [79].
Translational applications
ECM proteins as biomarkers
Alterations of the extracellular matrix are responsible for, or accompany, the development of pathologies, such as skeletal and articular diseases, cardiovascular diseases, skin diseases, fibrosis and cancers. We have previously shown that comparison of the matrisomes of normal and cancerous tissues can lead to the identification of novel candidate biomarkers [37]. We thus propose that pursuing the effort of characterizing the composition of extracellular matrices could lead to the identification of novel biomarkers for other diseases in addition to cancers. It is worth noting that ECM proteins are particularly favorable candidate biomarkers for immunohistochemically based assays since they are readily accessible, abundant, and laid down in characteristic patterns. Once identified, disease-specific ECM proteins, or protein isoforms could also serve as anchors for imaging molecules (e.g. fluorescent molecules, radiotracers) or therapeutics (e.g. drugs, cytokines, radioisotopes) that could be coupled, for example, to anti-disease-specific ECM protein antibodies, on the model of systems developed in the Neri lab [80, 81]. In addition, proteolytic fragments of ECM proteins can be released in body fluids and could be used as readouts for disease progression or treatment efficiency [82–84].
ECM proteins as therapeutic targets
There are successful precedents for inhibiting integrins to treat thrombosis, auto-immune and inflammatory diseases, etc [85, 86]. However, there are also many unsuccessful examples, including the use of matrix metalloproteinase inhibitors, in clinical trials for cancer patients [87]. In a 2009 review, Järveläinen and colleagues noted that ECM proteins are often ignored in drug discovery efforts [88]. We postulate that this will change and that high-throughput approaches will permit the identification of novel disease-specific ECM proteins and protein isoforms and that the characterization of the molecular mechanisms downstream of these proteins will offer novel therapeutic opportunities. Therapeutic strategies could include (i) targeting ECM protein synthesis or post-translational modifications, (ii) targeting ECM remodeling (degradation or crosslinking) and (iii) targeting ECM/ECM receptor interactions [88] as well as the antibody-mediated targeting discussed above.
ECM and regenerative medicine
The ECM provides biophysical and biochemical signals that orchestrate organ formation and function and thus should play a central part in tissue engineering strategies [89]. In addition, the ECM is a fundamental component of stem cell niches [90, 91]. In fact, many stem cell markers are ECM receptors (e.g. CD49a-f are integrins α1-6, CD29 is the integrin β1 and Lgr5 is an R-spondin receptor) and laminins have been demonstrated to affect pluripotency and differentiation of stem cells [90]. Tissue engineers have exploited, to varying degrees, aspects of ECM biology, in particular its biomechanical properties (not discussed here), to design novel scaffolds to support tissue regeneration [92]. Allogeneic or xenogeneic ECMs are now being used routinely to aid the reconstruction of a variety of tissues (e.g. urinary bladder, skin) [93]. Whole organ engineering, if successful, should solve the problem of shortage of organs available to patients awaiting transplant. One approach to engineer a whole functional organ (heart, lung) consists in using decellularized organ scaffolds comprising ECM and some (generally unknown) associated materials and repopulating the scaffold with stem cells. However, so far this approach not yet succeeded in regenerating fully functional organs. This may be in part due to the fact that decellularization results, as shown by mass spectrometry, in the loss of some ECM components and associated growth factors [63, 94, 95]. At the opposite extreme, minimalistic approaches are parsimoniously incorporating features of ECM proteins such as RGD (integrin-binding) peptides or mimics in artificial scaffolds. However, proteomic studies have revealed that the ECMs of tissues are made of 150+ proteins and although reconstructing this complexity may be difficult (and perhaps unnecessary [96]), we propose that the results of proteomics studies aimed at characterizing in vivo ECMs should be exploited to guide the design of the next generation of bio-inspired scaffolds to support organ regeneration.
CONCLUSIONS
The extracellular matrix has long been considered as a structural component of tissue organization. Recognition of the roles of specific ECM receptors and the binding and presentation of secreted growth factors introduced new concepts of how ECM proteins affect cell behavior and the developing understanding of mechanochemical transduction of ECM-derived signals have all combined to implicate ECM in a wide range of biologically and medically important areas [97]. With the recent development of experimental techniques for thorough characterization of ECM composition and of bioinformatic means to exploit that information, we are at an exciting point in ECM research where we can deploy the power of broad-scale genomic, proteomic and other “omics” approaches to provide new insights into development, disease, therapy and regenerative medicine in addition to further fundamental understanding of the enduring fascinating mysteries of ECM biology.
Supplementary Material
HIGHLIGHTS.
The matrisome is defined as the ensemble of 1000+ genes encoding ECM and ECM-associated proteins.
Bioinformatic and experimental approaches to study the ECM/matrisome are discussed.
We introduce a novel website and database MatrisomeDB to centralize resources on the matrisome.
We present a draft of an ECM atlas compiling proteomics data on the ECM of 14 different tissues and tumors.
“Omics” data provide novel insights into ECM functions in development, homeostasis and disease.
ACKNOWLEDGMENTS
The authors would like to thank Drs. Jill Mesirov and Arthur Liberzon from the Broad Institute’s Computational Biology and Bioinformatics Organization for implementing the matrisome gene sets into the Molecular Signature Database.
The authors would also like to thank the laboratories of Drs. Martin Humphries (University of Manchester) and Manimalha Balasubramani (University of Pittsburgh) for making their raw mass spectrometry data publicly available.
This work was supported by grants from the National Cancer Institute – Tumor Microenvironment Network (U54 CA126515/CA163109), the Broad Institute of MIT and Harvard, the Howard Hughes Medical Institute, of which ROH is an Investigator and in part by Support Grant to the Koch Institute (P30-CA14051 from the National Cancer Institute). AN was supported by postdoctoral fellowships from the Howard Hughes Medical Institute and the Ludwig Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Humphrey JD, Dufresne ER, Schwartz MA. Mechanotransduction and extracellular matrix homeostasis. Nat Rev Mol Cell Biol. 2014;15:802–812. doi: 10.1038/nrm3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326:1216–9. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huttenlocher A, Horwitz AR. Integrins in cell migration. Cold Spring Harb Perspect Biol. 2011;3:a005074. doi: 10.1101/cshperspect.a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charras G, Sahai E. Physical influences of the extracellular environment on cell migration. Nat Rev Mol Cell Biol. 2014 doi: 10.1038/nrm3897. [DOI] [PubMed] [Google Scholar]
- 5.Ingber DE. Extracellular matrix: A solid-state regulator of cell form, function, and tissue Development. Comprehensive Physiology. 2011:541–556. [Google Scholar]
- 6.Campbell ID, Humphries MJ. Integrin structure, activation, and interactions. Cold Spring Harb Perspect Biol. 2011 doi: 10.1101/cshperspect.a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rozario T, DeSimone DW. The extracellular matrix in development and morphogenesis: a dynamic view. Dev Biol. 2010;341:126–140. doi: 10.1016/j.ydbio.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wickström SA, Radovanac K, Fässler R. Genetic analyses of integrin signaling. Cold Spring Harb Perspect Biol. 2011 doi: 10.1101/cshperspect.a005116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hynes RO. The evolution of metazoan extracellular matrix. J Cell Biol. 2012;196:671–679. doi: 10.1083/jcb.201109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keeley FW, Mecham RP, editors. Evolution of Extracellular Matrix. Springer; Berlin, Heidelberg: 2013. [Google Scholar]
- 11.Bateman JF, Boot-Handford RP, Lamandé SR. Genetic diseases of connective tissues: cellular and extracellular effects of ECM mutations. Nat Rev Genet. 2009;10:173–183. doi: 10.1038/nrg2520. [DOI] [PubMed] [Google Scholar]
- 12.Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naba A, Clauser KR, Lamar JM, et al. Extracellular matrix signatures of human mammary carcinoma identify novel metastasis promoters. eLife. 2014 doi: 10.7554/eLife.01308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014;15:1243–1253. doi: 10.15252/embr.201439246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin GR, Kleinman HK, Terranova VP, et al. The regulation of basement membrane formation and cell-matrix interactions by defined supramolecular complexes. Ciba Found Symp. 1984;108:197–212. doi: 10.1002/9780470720899.ch13. [DOI] [PubMed] [Google Scholar]
- 16.Naba A, Clauser KR, Hoersch S, et al. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014647. M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naba A, Hoersch S, Hynes RO. Towards definition of an ECM parts list: an advance on GO categories. Matrix Biol. 2012;31:371–372. doi: 10.1016/j.matbio.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hynes RO, Naba A. Overview of the matrisome--an inventory of extracellular matrix constituents and functions. Cold Spring Harb Perspect Biol. 2012 doi: 10.1101/cshperspect.a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hohenester E, Engel J. Domain structure and organisation in extracellular matrix proteins. Matrix Biol. 2002;21:115–28. doi: 10.1016/S0945-053X(01)00191-3. [DOI] [PubMed] [Google Scholar]
- 20.Hynes RO, Yamada KM, editors. Cold Spring Harbor Perspectives in Biology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2012. Extracellular Matrix Biology. [Google Scholar]
- 21.Iozzo RV, Schaefer L. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015;42:11–55. doi: 10.1016/j.matbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mecham R, editor. The Extracellular Matrix: an Overview. Springer Science & Business Media; 2011. [Google Scholar]
- 23.Consortium TU. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell A, Chang H-Y, Daugherty L, et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc Natl Acad Sci. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tagliabracci VS, Engel JL, Wen J, et al. Secreted kinase phosphorylates extracellular proteins that regulate biomineralization. Science. 2012;336:1150–1153. doi: 10.1126/science.1217817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naba A, Ding H, Whittaker CA, Hynes RO. 2015 http://matrisomeproject.mit.edu.
- 30.Gray KA, Daugherty LC, Gordon SM, et al. Genenames.org: the HGNC resources in 2013. Nucleic Acids Res. 2013;41:D545–552. doi: 10.1093/nar/gks1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson Laboratories Mouse Genome Informatics website. http://www.informatics.jax.org/
- 32.Liberzon A. A description of the Molecular Signatures Database (MSigDB) web site. Methods Mol Biol, Clifton, NJ. 2014;1150:153–160. doi: 10.1007/978-1-4939-0512-6_9. [DOI] [PubMed] [Google Scholar]
- 33.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langlois B, Saupe F, Rupp T, et al. AngioMatrix, a signature of the tumor angiogenic switch-specific matrisome, correlates with poor prognosis for glioma and colorectal cancer patients. Oncotarget. 2014;5:10529–10545. doi: 10.18632/oncotarget.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chautard E, Ballut L, Thierry-Mieg N, Ricard-Blum S. MatrixDB, a database focused on extracellular protein–protein and protein–carbohydrate interactions. Bioinformatics. 2009;25:690–691. doi: 10.1093/bioinformatics/btp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Launay G, Salza R, Multedo D, et al. MatrixDB, the extracellular matrix interaction database: updated content, a new navigator and expanded functionalities. Nucleic Acids Res. 2015;43:D321–D327. doi: 10.1093/nar/gku1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naba A, Clauser KR, Whittaker CA, et al. Extracellular matrix signatures of human primary metastatic colon cancers and their metastases to liver. BMC Cancer. 2014;14:518. doi: 10.1186/1471-2407-14-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapushesky M, Adamusiak T, Burdett T, et al. Gene Expression Atlas update—a value-added database of microarray and sequencing-based functional genomics experiments. Nucleic Acids Res. 2012;40:D1077–D1081. doi: 10.1093/nar/gkr913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petryszak R, Burdett T, Fiorelli B, et al. Expression Atlas update—a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 2014;42:D926–D932. doi: 10.1093/nar/gkt1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim M-S, Pinto SM, Getnet D, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilhelm M, Schlegl J, Hahne H, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 42.Uhlén M, Fagerberg L, Hallström BM, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 43.Naba A, Clauser KR, Hynes RO. Enrichment of extracellular matrix proteins from tissues and digestion into peptides for mass spectrometry analysis. J Vis Exp. 2015 doi: 10.3791/53057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barallobre-Barreiro J, Didangelos A, Yin X, et al. A Sequential Extraction Methodology for Cardiac Extracellular Matrix Prior to Proteomics Analysis. In: Vivanco F, editor. Heart Proteomics. Humana Press; Totowa, NJ: 2013. pp. 215–223. [DOI] [PubMed] [Google Scholar]
- 45.Wilson R, Belluoccio D, Bateman JF. Proteomic analysis of cartilage proteins. Methods. 2008;45:22–31. doi: 10.1016/j.ymeth.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 46.Hansen KC, Kiemele L, Maller O, et al. An in-solution ultrasonication-assisted digestion method for improved extracellular matrix proteome coverage. Mol Cell Proteomics. 2009;8:1648–57. doi: 10.1074/mcp.M900039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myllyharju J. Intracellular Post-Translational Modifications of Collagens. In: Brinckmann J, Notbohm H, Müller PK, editors. Collagen. Springer Berlin Heidelberg; 2005. pp. 115–147. [Google Scholar]
- 48.Yamauchi M, Sricholpech M. Lysine post-translational modifications of collagen. Essays Biochem. 2012;52:113–133. doi: 10.1042/bse0520113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Belluoccio D, Wilson R, Thornton DJ, et al. Proteomic analysis of mouse growth plate cartilage. Proteomics. 2006;6:6549–6553. doi: 10.1002/pmic.200600191. [DOI] [PubMed] [Google Scholar]
- 50.Schreiweis MA, Butler JP, Kulkarni NH, et al. A proteomic analysis of adult rat bone reveals the presence of cartilage/chondrocyte markers. J Cell Biochem. 2007;101:466–476. doi: 10.1002/jcb.21196. [DOI] [PubMed] [Google Scholar]
- 51.Balasubramani M, Schreiber EM, Candiello J, et al. Molecular interactions in the retinal basement membrane system: a proteomic approach. Matrix Biol. 2010;29:471–483. doi: 10.1016/j.matbio.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 52.Didangelos A, Yin X, Mandal K, et al. Proteomics characterization of extracellular space components in the human aorta. Mol Cell Proteomics. 2010;9:2048–2062. doi: 10.1074/mcp.M110.001693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Brien J, Fornetti J, Schedin P. Isolation of mammary-specific extracellular matrix to assess acute cell-ECM interactions in 3D culture. J Mammary Gland Biol Neoplasia. 2010;15:353–364. doi: 10.1007/s10911-010-9185-x. [DOI] [PubMed] [Google Scholar]
- 54.Wilson R, Diseberg AF, Gordon L, et al. Comprehensive profiling of cartilage extracellular matrix formation and maturation using sequential extraction and label-free quantitative proteomics. Mol Cell Proteomics. 2010;9:1296–1313. doi: 10.1074/mcp.M000014-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Didangelos A, Yin X, Mandal K, et al. Extracellular matrix composition and remodeling in human abdominal aortic aneurysms: a proteomics approach. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.008128. M111.008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barallobre-Barreiro J, Didangelos A, Schoendube FA, et al. Proteomics analysis of cardiac extracellular matrix remodeling in a porcine model of ischemia/reperfusion injury. Circulation. 2012;125:789–802. doi: 10.1161/CIRCULATIONAHA.111.056952. [DOI] [PubMed] [Google Scholar]
- 57.Booth AJ, Hadley R, Cornett AM, et al. Acellular normal and fibrotic human lung matrices as a culture system for in vitro investigation. Am J Respir Crit Care Med. 2012;186:866–876. doi: 10.1164/rccm.201204-0754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Önnerfjord P, Khabut A, Reinholt FP, et al. Quantitative proteomic analysis of eight cartilaginous tissues reveals characteristic differences as well as similarities between subgroups. J Biol Chem. 2012;287:18913–18924. doi: 10.1074/jbc.M111.298968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Decaris ML, Gatmaitan M, FlorCruz S, et al. Proteomic analysis of altered extracellular matrix turnover in bleomycin-induced pulmonary fibrosis. Mol Cell Proteomics. 2014;13:1741–1752. doi: 10.1074/mcp.M113.037267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lennon R, Byron A, Humphries JD, et al. Global analysis reveals the complexity of the human glomerular extracellular matrix. J Am Soc Nephrol. 2014;25:939–951. doi: 10.1681/ASN.2013030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Müller C, Khabut A, Dudhia J, et al. Quantitative proteomics at different depths in human articular cartilage reveals unique patterns of protein distribution. Matrix Biol. 2014;40:34–45. doi: 10.1016/j.matbio.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 62.Uechi G, Sun Z, Schreiber EM, et al. Proteomic view of basement membranes from human retinal blood vessels, inner limiting membranes, and lens capsules. J Proteome Res. 2014 doi: 10.1021/pr5002065. [DOI] [PubMed] [Google Scholar]
- 63.Hill RC, Calle EA, Dzieciatkowska M, et al. Quantification of extracellular matrix proteins from a rat lung scaffold to provide a molecular readout for tissue engineering. Mol Cell Proteomics MCP. 2015;14:961–973. doi: 10.1074/mcp.M114.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu JC-C, Yamakoshi Y, Yamakoshi F, et al. Proteomics and genetics of dental enamel. Cells Tissues Organs. 2005;181:219–231. doi: 10.1159/000091383. [DOI] [PubMed] [Google Scholar]
- 65.Zimmermann DR, Dours-Zimmermann MT. Extracellular matrix of the central nervous system: from neglect to challenge. Histochem Cell Biol. 2008;130:635–653. doi: 10.1007/s00418-008-0485-9. [DOI] [PubMed] [Google Scholar]
- 66.Barros CS, Franco SJ, Müller U. Extracellular matrix: functions in the nervous system. Cold Spring Harb Perspect Biol. 2011 doi: 10.1101/cshperspect.a005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwarzbauer JE, DeSimone DW. Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb Perspect Biol. 2011 doi: 10.1101/cshperspect.a005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chiquet-Ehrismann R, Tucker RP. Tenascins and the importance of adhesion modulation. Cold Spring Harb Perspect Biol. 2011 doi: 10.1101/cshperspect.a004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boja ES, Rodriguez H. Mass spectrometry-based targeted quantitative proteomics: achieving sensitive and reproducible detection of proteins. Proteomics. 2012;12:1093–1110. doi: 10.1002/pmic.201100387. [DOI] [PubMed] [Google Scholar]
- 70.Carr SA, Abbatiello SE, Ackermann BL, et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based asay development using a fit-for-purpose approach. Mol Cell Proteomics. 2014;13:907–917. doi: 10.1074/mcp.M113.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pontén F, Jirström K, Uhlen M. The Human Protein Atlas--a tool for pathology. J Pathol. 2008;216:387–393. doi: 10.1002/path.2440. [DOI] [PubMed] [Google Scholar]
- 72.Gillette MA, Carr SA. Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat Methods. 2013;10:28–34. doi: 10.1038/nmeth.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yalak G, Olsen BR. Proteomic database mining opens up avenues utilizing extracellular protein phosphorylation for novel therapeutic applications. J Transl Med. 2015;13:125. doi: 10.1186/s12967-015-0482-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sasisekharan R, Raman R, Prabhakar V. Glycomics approach to structure-function relationships of glycosaminoglycans. Annu Rev Biomed Eng. 2006;8:181–231. doi: 10.1146/annurev.bioeng.8.061505.095745. [DOI] [PubMed] [Google Scholar]
- 75.Lech M, Capila I, Kaundinya GV. Mass spectrometric methods for the analysis of heparin and heparan sulfate. Methods Mol Biol Clifton NJ. 2015;1229:119–128. doi: 10.1007/978-1-4939-1714-3_12. [DOI] [PubMed] [Google Scholar]
- 76.Chiu Y, Huang R, Orlando R, Sharp JS. GAG-ID: Heparan Sulfate and Heparin Glycosaminoglycan High-Throughput Identification Software. Mol Cell Proteomics MCP. 2015 doi: 10.1074/mcp.M114.045856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ricard-Blum S, Salza R. Matricryptins and matrikines: biologically active fragments of the extracellular matrix. Exp Dermatol. 2014;23:457–463. doi: 10.1111/exd.12435. [DOI] [PubMed] [Google Scholar]
- 78.Rogers LD, Overall CM. Proteolytic post-translational modification of proteins: proteomic tools and methodology. Mol Cell Proteomics. 2013;12:3532–3542. doi: 10.1074/mcp.M113.031310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schlage P, Auf dem Keller U. Proteomic approaches to uncover MMP function. Matrix Biol. 2015 doi: 10.1016/j.matbio.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 80.Santimaria M, Moscatelli G, Viale GL, et al. Immunoscintigraphic detection of the ED-B domain of fibronectin, a marker of angiogenesis, in patients with cancer. Clin Cancer Res. 2003;9:571–579. [PubMed] [Google Scholar]
- 81.Pasche N, Neri D. Immunocytokines: a novel class of potent armed antibodies. Drug Discov Today. 2012;17:583–590. doi: 10.1016/j.drudis.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 82.Yao L, Lao W, Zhang Y, et al. Identification of EFEMP2 as a serum biomarker for the early detection of colorectal cancer with lectin affinity capture assisted secretome analysis of cultured fresh tissues. J Proteome Res. 2012;11:3281–94. doi: 10.1021/pr300020p. [DOI] [PubMed] [Google Scholar]
- 83.Schierwagen R, Leeming DJ, Klein S, et al. Serum markers of the extracellular matrix remodeling reflect antifibrotic therapy in bile-duct ligated rats. Front Physiol. 2013;4:195. doi: 10.3389/fphys.2013.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bunatova K, Pesta M, Kulda V, et al. Plasma TIMP1 level is a prognostic factor in patients with liver metastases. Anticancer Res. 2012;32:4601–4606. [PubMed] [Google Scholar]
- 85.Goodman SL, Picard M. Integrins as therapeutic targets. Trends Pharmacol Sci. 2012;33:405–412. doi: 10.1016/j.tips.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 86.Estevez B, Shen B, Du X. Targeting integrin and integrin signaling in treating thrombosis. Arterioscler Thromb Vasc Biol. 2015;35:24–29. doi: 10.1161/ATVBAHA.114.303411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vandenbroucke RE, Libert C. Is there new hope for therapeutic matrix metalloproteinase inhibition? Nat Rev Drug Discov. 2014;13:904–927. doi: 10.1038/nrd4390. [DOI] [PubMed] [Google Scholar]
- 88.Järveläinen H, Sainio A, Koulu M, et al. Extracellular matrix molecules: potential targets in pharmacotherapy. Pharmacol Rev. 2009;61:198–223. doi: 10.1124/pr.109.001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dvir T, Timko BP, Kohane DS, Langer R. Nanotechnological strategies for engineering complex tissues. Nat Nanotechnol. 2011;6:13–22. doi: 10.1038/nnano.2010.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Watt FM, Huck WTS. Role of the extracellular matrix in regulating stem cell fate. Nat Rev Mol Cell Biol. 2013;14:467–473. doi: 10.1038/nrm3620. [DOI] [PubMed] [Google Scholar]
- 91.Gattazzo F, Urciuolo A, Bonaldo P. Extracellular matrix: A dynamic microenvironment for stem cell niche. Biochim Biophys Acta. 2014;1840:2506–2519. doi: 10.1016/j.bbagen.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Badylak SF, Weiss DJ, Caplan A, Macchiarini P. Engineered whole organs and complex tissues. Lancet. 2012;379:943–952. doi: 10.1016/S0140-6736(12)60073-7. [DOI] [PubMed] [Google Scholar]
- 93.Faulk DM, Johnson SA, Zhang L, Badylak SF. Role of the extracellular matrix in whole organ engineering. J Cell Physiol. 2014;229:984–989. doi: 10.1002/jcp.24532. [DOI] [PubMed] [Google Scholar]
- 94.Bonvillain RW, Danchuk S, Sullivan DE, et al. A nonhuman primate model of lung regeneration: detergent-mediated decellularization and initial In vitro recellularization with mesenchymal stem cells. Tissue Eng Part A. 2012;18:2437–2452. doi: 10.1089/ten.tea.2011.0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Petersen TH, Calle EA, Zhao L, et al. Tissue-engineered lungs for in vivo implantation. Science. 2010;329:538–541. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kyburz KA, Anseth KS. Synthetic mimics of the extracellular matrix: how simple is complex enough? Ann Biomed Eng. 2015:1–12. doi: 10.1007/s10439-015-1297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hynes RO. Stretching the boundaries of extracellular matrix research. Nat Rev Mol Cell Biol. 2014;15:761–763. doi: 10.1038/nrm3908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.