Abstract
High-levels of transcription through a gene stimulate spontaneous mutation rate, a phenomenon termed transcription-associated mutation (TAM). While transcriptional effects on specific mutation classes have been identified using forward mutation and frameshift-reversion assays, little is yet known about transcription-associated base substitutions in yeast. To address this issue, we developed a new base substitution reversion assay (the lys2-TAG allele). We report a 22-fold increase in overall reversion rate in the high- relative to the low-transcription strain (from 2.1- to 47- × 10−9). While all detectable base substitution types increased in the high-transcription strain, G→T and G→C transversions increased disproportionately by 58- and 52-fold, respectively. To assess a potential role of DNA damage in the TAM events, we measured mutation rates and spectra in individual strains defective in the repair of specific DNA lesions or null for the error-prone translesion DNA polymerase zeta (Pol zeta). Results exclude a role of 8-oxoGuanine, general oxidative damage, or apurinic/apyrimidinic sites in the generation of TAM G→T and G→C transversions. In contrast, the TAM transversions at GC base pairs depend on Pol zeta for occurrence implicating DNA damage, other than oxidative lesions or AP sites, in the TAM mechanism. Results further indicate that transcription-dependent G→T transversions in yeast differ mechanistically from equivalent events in E. coli reported by others. Given their occurrences in repair-proficient cells, transcription-associated G→T and G→C events represent a novel type of transcription-associated mutagenesis in normal cells with potentially important implications for evolution and genetic disease.
Keywords: transcription-associated mutation, base substitutions, yeast, Saccharomyces cerevisiae, REV3, spontaneous mutation rate
INTRODUCTION
Spontaneous mutations occur via a variety of mechanisms including replication errors, spontaneous DNA lesions (both those resulting from endogenous mutagens and those resulting from hydrolytic attack to DNA) that escape repair, and bypass of replication-blocking lesions by error-prone translesion synthesis DNA polymerases. Among the most common endogenous DNA lesions are oxidation products and apurinic/apyrimidinic (AP) sites [for a review, see Friedberg et al. 2006]. Beginning with the former, guanine is highly susceptible to oxidative attack resulting in 7,8-dihydro-8-oxoguanine (8-oxoG) lesions in DNA, which mispair with dATP during replication forming G→T transversions. In yeast, the OGG1 gene encodes 8-OxoGuanine DNA N-glycosylase, which excises 8-oxoG lesions from DNA creating AP sites that are targets for the base excision repair (BER) pathway [Thomas et al., 1997]. Moreover, organisms possess anti-oxidant enzymes such as thioredoxin peroxidases that reduce levels of reactive oxygen species and hence reduce mutation rates due to general oxidative damage. While yeast (Saccharomyces cerevisiae) possesses five thioredoxin peroxidases, the Tsa1 protein provides the greatest suppression of spontaneous mutagenesis due to reactive oxygen species [Huang et al., 2003; Wong et al., 2004].
In addition to removal of damaged bases by lesion-specific N-glycosylases, AP sites also result from base loss via hydrolytic attack to the glycosidic bond connecting each nitrogenous base to its respective deoxyribose sugar in DNA. The APN1 gene encodes the Apn1 protein that is responsible for greater than 97% of both AP endonuclease and 3′-repair diesterase activities in yeast [Popoff et al., 1990]. Moreover, Apn1 null strains accumulate unrepaired AP sites and exhibit a mutator phenotype [Ramotar et al., 1991]. While cells contain robust DNA repair pathways to reduce mutagenic burdens from endogenous and exogenous mutagens, not all DNA repair is faithful, especially regarding potentially lethal replication-blocking lesions. Such lesions trigger either an error-free or error-prone damage-tolerance pathway [for a review see Friedberg et al., 2006]. The REV3 gene in yeast encodes the catalytic subunit of the error-prone translesion DNA polymerase zeta (Polζ), which is responsible for a majority of spontaneous and induced mutagenesis [Lawrence 2002]. Our present understanding of spontaneous mutagenesis owes much to the previous identification of mutator/antimutator phenotypes and to the characterization of mutation spectra. Given the role of spontaneous mutagenesis in evolution and genetic disease, it is important to identify and characterize additional factors that influence mutation rate.
Transcription is one such factor that contributes to spontaneous mutagenesis in prokaryotes [Beletskii and Bhagwat, 1996; Wright et al., 1999] and yeast [Datta and Jinks-Robertson, 1995] in addition to induced mutagenesis in E. coli [Fix et al., 2008] and human cells [Hendriks et al., 2008]. Specifically, high-levels of transcription through a gene stimulate spontaneous mutation rate (transcription-associated mutation or TAM) and mitotic recombination (transcription-associated recombination or TAR). Thus far, several mechanisms have been postulated or shown to operate in TAM and/or TAR. Of particular relevance to the present study, high levels of transcription create genomic regions with greater susceptibility to DNA damage likely due either to changes in chromatin structure and/or formation of transient single-stranded DNA regions [Datta and Jinks-Robertson, 1995; Beletskii and Bhagwat, 1996; Garcia-Rubio et al., 2003]. Alternative sources of transcription-associated genomic instability include (i) interference or collisions between the replication and transcription machineries [Prado and Aguilera, 2005; Wellinger et al., 2006] and (ii) generation of mutagenic/recombinogenic intermediates via transcription-associated topological changes, specifically supercoiling that is the target of topoisomerase enzymes [Lippert et al., 2011; Takahashi et al., 2011; Garcia-Rubio and Aguilera, 2012] and the supercoiling-induced formation of R-loops (RNA:DNA hybrids) that form behind the transcription bubble [reviewed by Aguilera and Garcia-Muse, 2012]. Several recent reviews have been written on TAM and/or TAR [Aguilera and Gomez-Gonzalez, 2008; Gottipati and Helleday, 2009; Hendriks et al., 2010b; Kim and Jinks-Robertson, 2012].
Focusing on spontaneous mutagenesis, high levels of transcription have been shown to stimulate various mutation classes. In yeast, Datta and Jinks-Robertson first identified TAM using both a LYS2 forward mutation assay and a lys2 +1 frameshift (FS) allele that detects −1 and other events that restore the Lys+ phenotype [Datta and Jinks-Robertson, 1995]. Using multiple FS reversion assays, Jinks-Robertson et al. demonstrated that TAM FS rates increased in strains defective in nucleotide excision repair or homologous recombination. Moreover, a majority of TAM FSs depended on Polζ for occurrence supporting a role of spontaneous DNA damage in the generation of TAM FSs [Datta and Jinks-Robertson, 1995; Morey et al., 2000; Kim et al., 2007]. Using the LYS2 forward mutation assay, which detects a wide variety of mutation classes, we identified a greater than 71-fold specific increase in 2-4 bp deletion rate under high-transcription conditions, thus identifying 2-4 bp deletions as unique signature events of TAM [Lippert et al., 2004].
More recently, we and others recapitulated the TAM signature events at a second gene locus (CAN1) in yeast. Specifically, we demonstrated that TAM 2-5 bp deletion rate remained unaffected in homologous recombination-defective or Rev3 null backgrounds but absolutely required topoisomerase 1, thus suggesting topoisomerase cleavage complexes as sites of TAM deletion occurrence [Lippert et al., 2011; Takahashi et al., 2011]. In addition to the striking increase in TAM short deletions, other mutation classes also increased, albeit to a lesser extent, in the high-transcription strain. In particular, base substitutions (BSs) increased modestly (1.8-2.8 fold) in the high- relative to the low-transcription strain [Lippert et al., 2004, 2011]; however, because short deletions dominated the high-transcription forward mutation spectrum, only small numbers of BSs were recovered. Hence, the exploration of TAM BSs using a forward mutation assay would be labor intensive. TAM BSs, consequently, have been understudied in yeast. In the current study we extend previous work on TAM that used FS reversion and forward mutation assays by developing a BS reversion assay. Using the new lys2-TAG allele, we identify a strong transcription-dependent BS mutator-phenotype and further identify G→T and G→C transversions as predominant events. To further assess a potential role of DNA damage in TAM BSs, we tested three hypotheses: (i) transcription-associated BSs would occur at sites of oxidative lesions (ii) AP sites would factor in the mechanism underlying TAM BSs, and (iii) Polζ would play a role in the generation of TAM BSs.
MATERIALS AND METHODS
Media and Growth Conditions
Yeast (Saccharomyces cerevisiae) strains were grown in nonselective YEPA medium (1% yeast extract, 2% peptone, and 0.3 g/l adenine sulfate) supplemented with 2% glycerol and 2% ethanol (YEPAGE) or 2% dextrose (YEPAD). Lys+ revertants were selected on synthetic dextrose (SD) medium (1.7 g/l yeast nitrogen base without amino acids, 0.25 g/l adenine sulfate, 2% agar, and 2% dextrose) supplemented with 2.0 g/l drop-out mix synthetic minus lysine (SC-Lys, US Biological, Swampscott, MA). Canavanine-resistant (CanR) mutants were selected on canavanine medium [SD medium supplemented with 2.0 g/l drop-out mix synthetic minus arginine (US Biological) and 60 μg/ml of filter-sterilized l-canavanine sulfate]. For strain constructions, transformants were selected on Geneticin medium (YEPAD medium supplemented with 2% agar and 200 mg Geneticin/l) or SC-Ura medium [SD medium supplemented with 2.0 g/l drop out mix synthetic minus uracil (US Biological)]. All incubations were at 30°C.
Yeast Strains and Strain Constructions
All yeast strains are listed in Table I. Previously the low-transcription wild type strain, MB49-102, was isolated from strain SJR282 (MATα ade2-101oc his3Δ200 suc2 ura3ΔNco gal80Δ::HIS3) [Datta and Jinks-Robertson, 1995] in a LYS2 forward mutation assay and the mutation was identified as a C→T transition at nucleotide position 1696 of the LYS2 gene [Lippert et al., 2004]. The high-transcription wild type strain, MJL7, was created by replacing 50 nucleotides upstream of LYS2 (nucleotides −50 to −1 relative to the LYS2 open reading frame) with the pGAL promoter as described previously [Lippert et al., 2004]. Briefly, a kanMX6-PGAL1 gene-modifying cassette possessing 60 nucleotides of LYS2 homology at each end was PCR-amplified using plasmid pFA6a-kanMX6-PGAL1 [Longtine et al., 1998] as template DNA with primers 5′-ATAAGTAACAAGCAGCCAATAGTATAAAAAAAAATCTGAGTTTATTACCTTTCCTGGAATGAATTCGAGCTCGTTTAAAC-3′ and 5′-ATGTGGTAACACTGAAAGAGTTGGATTATCCAACTTCTCTATCCAGACCTTTTCGTTAGTCATTTTGAGATCCGGGTTTT-3′. Transformations were performed using the lithium acetate method [Gietz et al., 1995] and transformants were selected on Geneticin medium. Correct replacement of the pLYS with the pGAL promoter was confirmed by PCR.
TABLE I.
Yeast Strains
| Strain | Genotype | Ref. or Source |
|---|---|---|
| MB49-102 | MATα ade2-101oc his3Δ200 suc2 ura3Δ Nco gal80Δ::HIS3 lys2-TAG | Lippert et al., 2004 |
| MJL7 | MB49-102, kanMX6-pGAL-lys2-TAG | Lippert et al., 2004 |
| MJL40 | MB49-102, ogg1Δ::loxP-URA3Kl-loxP | This study |
| MJL41 | MJL7, ogg1Δ::loxP-URA3Kl-loxP | This study |
| MJL42 | MB49-102, rev3Δ::loxP-URA3Kl-loxP | This study |
| MJL43 | MJL7, rev3Δ::loxP-URA3Kl-loxP | This study |
| MJL44 | MB49-102, tsa1Δ::loxP-URA3Kl-loxP | This study |
| MJL45 | MJL7, tsa1Δ::loxP-URA3Kl-loxP | This study |
| MJL90 | MB49-102, apn1Δ::loxP-URA3Kl-loxP | This study |
| MJL91 | MJL7, apn1Δ::loxP-URA3Kl-loxP | This study |
The ogg1Δ, tsa1Δ, apn1Δ, and rev3Δ single deletion strains were constructed by transforming the low- (MB49-102) and high- (MJL7) transcription strains with PCR-generated cassettes containing the URA3 gene from Kluyveromyces lactis (URA3Kl) flanked by 60 nucleotides of homology at each end to the appropriate target gene. Specifically, cassettes were amplified using plasmid pUG72 [Gueldener et al., 2002] as template DNA and primer pairs 5′-CAGCGGAAGAAGGCATTTGAAGCGTCCTGATTCATAATTGCGATTTTATTTATCAACCAGCAGCTGAAGCTTCGTACGC-3′ and 5′-ATGTATCGCCTTTTCGGTCGCGTGCTTTTATCGTGGTATTTACTATGACTTTTTAAGCGCATAGGCCACTAGTGGATCTG-3′ for the ogg1Δ cassette, 5′-GGGCCTTCCCCTCGTTCAATTGCTCACAACCAACCACAACTACATACACATACATACACACAGCTGAAGCTTCGTACGC-3′ and 5′-TATAAACGTAAAGAGTGAATTTTAAATAAGTAGTCATTTAGACAACTCTGCAAGCGTCGCATAGGCCACTAGTGGATCTG-3′ for the tsa1Δ cassette, 5′-TCCGAATAAGAAACACAAAACGCAACATTAATAAGCTTTTGGCATATCGGAACCATCGTACAGCTGAAGCTTCGTACGC-3′ and 5′-ACGTACGTTGAGATAATCTACAAAAATTGATTACGTATTTAAAATTCTTCTCGCTTCTCAGCATAGGCCACTAGTGGATCTG-3′ for the apn1Δ cassette, and 5′-AAGAGAAAGTATTTGAGTCAATACAAAACTACAAGTTGTGGCGAAATAAAATGTTTGGAACAGCTGAAGCTTCGTACGC-3′ and 5′-CGTTATACATAGAAACAAATAACTACTCATCATTTTGCGAGACATATCTGTGTCTAGAGCATAGGCCACTAGTGGATCTG-3′ for the rev3Δ cassette. After selection on SC-Ura medium, correct transformants were confirmed by PCR. For each new strain, two independent isolates were constructed. Rates and spectra were measured using both strain isolates, which provided parallel results in every case.
Mutation Rate Measurements
Mutation rates were determined as described by Spell and Jinks-Robertson [2004]. Briefly, 2-day old colonies on YEPAD plates were inoculated into 5 ml YEPAGE medium and grown for 3 days in a roller bottle apparatus (New Brunswick Scientific, Edison, NJ). Cells were washed once with sterile dH2O and resuspended with 1 ml sterile dH2O. Cultures were plated in 0.1 ml volumes onto SC-Lys plates to determine the number of mutants per culture. Dilutions were plated at low density onto YEPAD plates to determine the total cell population. Colonies were counted after 2–3 days. For determination of canavanine-resistant (CanR) mutation rate, cells were grown as described above for Lys+ experiments except that cultures were plated at high density onto canavanine medium rather than SC-Lys medium. Lys+ reversion rates and 95% confidence intervals were determined by the maximum likelihood method using the SALVADOR computer program [Zheng, 2005]. CanR mutation rates and 95% confidence intervals were determined by the method of the median [Lea and Coulson, 1949] using an Excel spreadsheet as described by Spell and Jinks-Robertson [2004]. Mutation rates were based on a minimum of 14 cultures. Statistical significance at the 0.05 level was identified by nonoverlap between the 95% confidence intervals surrounding two mutation rates [Spell and Jinks-Robertson, 2004].
DNA Sequence Analysis
A single Lys+ revertant was isolated per culture by streaking for purification onto SC-Lys agar. Genomic DNA was isolated using the Puregene Yeast genomic DNA isolation kit (Gentra Systems). A 871 bp portion of the LYS2 gene corresponding to nucleotides 1218–2089 was PCR-amplified using primers 5′-AGGTGTTGTAGTTGGACCAGATT-3′ and 5′-TACCGCAACATTCACAGTCA-3′. PCR samples and sequencing primer, 5′-TTCCAACCCAACCCTATCTT-3′, were sent to the High Throughput Genomics Unit at the University of Seattle for DNA sequence analysis.
RESULTS
The New lys2-TAG Allele
In a previous LYS2 forward mutation study, we isolated a Lys- mutant, MB49-102, and identified the mutation as a C→T transition at LYS2 nucleotide position 1696 [Lippert et al., 2004]. As the mutation created a TAG nonsense codon in place of the wild type glutamine (CAG) codon, we named it the lys2-TAG allele. In spontaneous mutation rate experiments, Lys+ revertants occurred at a rate of 2.1 × 10−9 (Table II, see wild type low-transcription strain). Figure 1 illustrates nine possible BSs at the TAG stop codon relative to the non-transcribed strand. Of these nine, G→A transitions would result in an alternate (TAA) stop codon, precluding the recovery of G:C→A:T events by this assay. To determine all BS types detected by the lys2-TAG assay, we sequenced 53 independent Lys+ revertants (Table II). When focusing on events relative to the non-transcribed strand, all six potential transversions and T→C, but not A→G, transitions were recovered. Simple BSs at the TAG stop codon comprised 94% (50 of 53 revertants) of events. We grouped the three additional events, a multiple BS (see Table II footnote) and two instances where no mutation existed within the TAG target, as “other” events in Table II and Figure 2. Of the seven simple BS types recovered, G→T transversions comprised the largest proportion (24% or 13 of 53 events), followed by T→C transitions and A→T transversions (17% each), T→G transversions (13%), and T→A, A→C, and G→C transversions (4 of 53 or 7.5% each) (Table II).
TABLE II.
lys2-TAG Reversion Rates and Spectra in Low- and High-Transcription Strains
| WT |
ogg1Δ
|
tsa1Δ
|
apn1Δ
|
rev3Δ
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Trans-cription level | Mutations | No. | Ratea × 10−9 | No. | Rate × 10−9 | No. | Rate × 10−9 | No. | Rate × 10−9 | No. | Rate × 10−9 |
| Low | T→C | 9 | 0.36 [1×] | 8 | 0.82 | 4 | 0.91 | 8 | 0.77 | 19 | 0.32 [0.89×] |
| T→G | 7 | 0.28 [1×] | 4 | 0.41 | 8 | 1.8 [6.5×] | 2 | 0.19 | 4 | 0.067 | |
| T→A | 4 | 0.16 [1×] | 1 | 0.10 | 1 | 0.23 | 5 | 0.48 [3×] | 0 | <0.017 | |
| A→C | 4 | 0.16 [1×] | 8 | 0.82 [5.1×] | 10 | 2.3 [14×] | 15 | 1.45 [9×] | 3 | 0.050 | |
| A→T | 9 | 0.36 [1×] | 2 | 0.20 | 7 | 1.6 | 5 | 0.48 | 1 | 0.017 | |
| G→T | 13 | 0.52 [1×] | 29 | 3.0 [5.7×] | 12 | 2.7 | 4 | 0.39 | 12 | 0.20 | |
| G→C | 4 | 0.16 [1×] | 1 | 0.10 | 9 | 2.0 [13×] | 4 | 0.39 | 5 | 0.084 [0.52×] | |
| Otherb | 3 | 0.12 [1×] | 0 | <0.10 | 0 | <0.23 | 4 | 0.39 [3.2×] | 0 | <0.017 | |
| Total (95% CI)c | 53 | 2.1 [1×] (1.4–3.1) | 53 | 5.4 [2.6×] (3.9–7.1) | 51 | 11.6 [5.5×] (9.6–14) | 47 | 4.5 [2.1×] (3.4–5.8) | 44 | 0.74 [0.35×] (0.50–1.0) | |
| High | T→C | 5 | 3.2 | 0 | <1.1 | 5 | 4.7 | 0 | <4.4 | 30 | 1.3 [3.6×] |
| T→G | 1 | 0.64 | 1 | 1.1 | 6 | 5.7 | 0 | <4.4 | 1 | 0.042 | |
| T→A | 3 | 1.9 | 0 | <1.1 | 3 | 2.8 | 0 | <4.4 | 3 | 0.13 | |
| A→C | 0 | <0.64 | 2 | 2.2 | 3 | 2.8 | 38 | 168 [1050×] | 1 | 0.042 | |
| A→T | 4 | 2.6 | 10 | 10.8 | 9 | 8.5 | 4 | 18 | 10 | 0.42 | |
| G→T | 47 | 30 [58×] | 30 | 32 [61×] | 32 | 30 [58×] | 3 | 13 | 19 | 0.80 | |
| G→C | 13 | 8.3 [52×] | 9 | 9.7 [61×] | 9 | 8.5 [53×] | 0 | <4.4 | 2 | 0.085 | |
| Other | 0 | <0.64 | 1 | 1.1 | 1 | 0.95 | 2 | 9 | 1 | 0.042 | |
| Total (95% CI) | 73 | 47 [22×] (39–54) | 53 | 57 [27×] (47–67) | 68 | 64 [30×] (56–72) | 47 | 208 [99×] (179–236) | 67 | 2.8 [1.3×] (2.2–3.6) | |
Rates for individual BS types were calculated by multiplying the total rate by the proportion of each mutation type recovered. If zero events were recovered, a rate was calculated for a single event and expressed as an upper limit. For all overall mutation rates and some BS type-specific rates, bracketed values represent the fold-increase relative to the low-transcription wild type overall and BS subtype-specific rates, respectively. In order to highlight those BS types that increased disproportionately, BS type-specific fold-increases are reported only if they exceed the total fold-increase for each strain.
“Other” mutations indicate Lys+ revertants that showed no change at the lys2-TAG allele with two exceptions: one multiple BS was recovered from the WT low-transcription and the tsa1Δ high-transcription strains, a TAG (stop) → ATC (Ile) event and a TAG (stop) →TTA (Leu) event, respectively.
CI, confidence interval.
Fig. 1.
All base substitution (BS) types potentially detected by the lys2-TAG assay in yeast (Saccharomyces cerevisiae). The TAG nonsense codon located at LYS2 nucleotide positions 1696–1698 is shown above potential BS types (followed by resulting codons and amino acid residues).
Fig. 2.
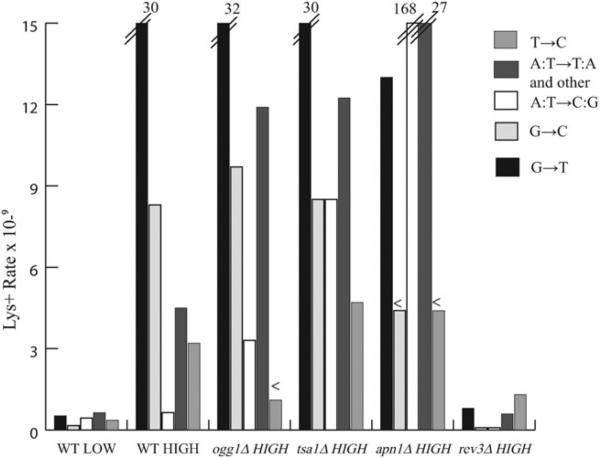
Spontaneous reversion rate of the lys2-TAG allele reported by mutation class. Luria-Delbruck fluctuation analysis was performed and overall reversion rates were determined by the maximum likelihood method. Rates for individual base substitution types were calculated by multiplying the total Lys+ rate for each strain by the proportion of each mutation category recovered. Proportions are based on sample sizes ranging from 44 to 73 independent Lys+ revertants (see Table II). Rates that exceed the Y-axis range are indicated with slash marks (//) and text corresponding to the correct value. When zero events were recovered, an upper limit is reported for that category and a “less than” symbol (<) is placed above the bar.
High Levels of Transcription Stimulate G→T and G→C Transversions
In strain MB49-102, the normal low-level pLYS promoter regulates the chromosomal lys2-TAG allele. To generate the high-transcription strain, MJL7, we replaced pLYS with the highly inducible pGAL promoter. The MB49-102 strain background lacks the GAL80 gene (Table I; see Methods), which encodes a negative regulator of pGAL transcription [Johnston, 1987]; hence, high levels of transcription through the pGAL-lys2-TAG allele occur in the presence or absence of galactose when grown in YEPAGE medium. Hereafter, we refer to strains MB49-102 and MJL7 as the wild type low- and high-transcription strains, respectively.
In the high-transcription strain, reversion rate increased 22-fold (from 2.1- to 47- × 10−9) and G→T transversions predominated (64% or 47 of 73 events). When the transcription-associated increase in mutation rate is considered, G→ T transversions increased 58-fold in the high- (30 × 10−9) relative to the low- (0.52 × 10−9) transcription strain. Another BS type, G→C transversions, increased disproportionately 52-fold (from 0.16- to 8.3- × 10−9) (Table II, Fig. 2). While transversions at GC bp comprised the most striking transcription-associated increases, it should be noted that rates of four other BS types also increased in the high- relative to the low-transcription strain: T→A, T→C, A→T, and T→G BSs increased 12-, 8.9-, 7.1-, and 2.3-fold, respectively (Table II). Given the strong increase in transversions at GC bp, hereafter we focus primarily on those events and refer to the transcription-associated G→T and G→C transversions as TAM-G events or TAM-G transversions. Next we explored potential roles of various DNA lesions in occurrences of TAM-G events.
If TAM occurs via increased susceptibility of highly transcribed regions to endogenous DNA damage, one would expect a synergistic effect of elevated DNA damage under high-transcription conditions relative to the individual effects of: (i) elevated DNA damage alone and (ii) a high-transcription level alone. We therefore created gene deletion strains that experience elevated levels of specific DNA lesions and measured mutation rates and spectra in low- and high-transcription strains.
Role of Oxidative DNA Damage and Abasic Sites in TAM-G Transversions
The major oxidative lesion, 8-oxoG, accumulates in repair-defective strains that lack 8-OxoGuanine DNA N-glycosylase (ogg1Δ strains) resulting in a specific increase in G→T transversions [Thomas et al., 1997]. To investigate a potential role of 8-OxoG lesions in TAM, we measured mutation rates and spectra in ogg1Δ strains (Table II and Fig. 2). Under low-transcription conditions, overall mutation rate increased 2.6-fold (from 2.1- to 5.4- × 10−9) and G→T transversions, specifically, increased 5.7-fold (from 0.52- to 3.0- × 10−9) in the ogg1Δ relative to the wild type strains. In the high-transcription strain, the overall ogg1Δ mutation rate (57 × 10−9) did not differ significantly from wild type strains (47 × 10−9) based on overlapping 95% confidence intervals. Moreover as illustrated in Figure 2, TAM-G transversions occurred at nearly identical rates in ogg1Δ and wild type high-transcription strains: 32- versus 30- × 10−9, respectively, for G→T events and 9.7- versus 8.3- × 10−9, respectively, for G→C events.
To determine if oxidized lesions other than 8-OxoG generated the TAM-G events, we examined mutagenesis in strain backgrounds null for the major thioredoxin peroxidase antioxidant enzyme, Tsa1. In a Tsa1 deficient backgrounΔ, Lys+ mutation rate increased 5.5-fold (from 2.1- to 11.6- × 10−9) in the low-transcription strain (Table II). The high-transcription tsa1Δ strain exhibited a slight increase in overall mutation rate relative to the wild type strain, 64- versus 47- × 10−9, respectively; however, specific rates of G→T and G→C transversions were indistinguishable between high-transcription wild type (30- and 8.3- × 10−9, respectively) and high-transcription tsa1Δ (30- and 8.5- × 10−9, respectively) strains (Table II and Fig. 2). We next explored the potential role of a second common spontaneous lesion, the AP site, in TAM BSs.
It is well established that Apn1-deficient (apn1Δ) strains experience elevated levels of AP sites resulting in a mutator phenotype characterized largely by elevated levels of A:T→C:G transversions and, to a lesser extent, A:T→T:A and other BS types [Kunz et al., 1994]. Because guanine is the base most susceptible to hydrolytic attack resulting in base loss [Lindahl and Nyberg, 1972], we reasoned that guanine-derived AP sites might play a role in TAM. We therefore measured mutation rates and spectra in Apn1-deficient strains (Table II and Fig. 2). The low-transcription apn1Δ strain exhibited a 2.1-fold increase in mutation rate relative to the wild type strain, 4.5- and 2.1- × 10−9, respectively. In contrast to this modest mutator effect under low-transcription conditions, the apn1Δ high-transcription strain exhibited a synergistic 99-fold increase (208 × 10−9) relative to the wild type low-transcription strain (2.1 × 10−9). Moreover, A→C transversions dominated the apn1Δ high-transcription spectrum, comprising 38 of 47 or 81% of events, whereas zero T→G events (A→C transversions in the transcribed strand) occurred in the sample of 47 Lys+ revertants. In contrast to the striking increase in A→C transversions, only three G→T and zero G→C transversions occurred among the 47 revertants. The recovery of so few events provides no clarity regarding whether TAM-G rates decreased modestly or remained unaffected in the apn1Δ relative to the wild type high-transcription strains (Table II and Fig. 2).
Transcription-Dependent BSs Exhibit a Nearly Complete Dependence on Polζ
REV3 encodes the catalytic subunit of DNA polymerase-zeta (Polζ), an error-prone translesion polymerase involved in the bypass of replication-blocking damage. The majority of spontaneous mutagenesis in yeast depends on Polζ for occurrence, consequently, Rev3 null strains exhibit an antimutator phenotype [for a review, see Lawrence, 2002]. Table II and Figure 2 illustrate mutation rates and spectra recovered from rev3Δ strains. In the low-transcription strain, overall mutation rate decreased 65% in the rev3Δ relative to the wild type strain, 0.74- and 2.1 × 10−9, respectively. In the rev3Δ high-transcription strain, Lys+ rate decreased 94% relative to the wild type high-transcription strain, 2.8- and 47- × 10−9, respectively, to a value similar to the wild type low-transcription strain (2.1 × 10−9). An analysis of 67 independent reversions revealed reductions in all BS types in the rev3Δ relative to the wild type high-transcription strains. Moreover with the exception of T→C transitions, rates of all BS types decreased to levels seen in the wild type low-transcription strain. For example, G→T rate decreased 97% relative to the wild type high-transcription strain (from 30- to 0.8- × 10−9) corresponding to the rate exhibited by the wild type low-transcription strain (0.52 × 10−9). Whereas the rate of T→C transitions also decreased relative to the wild type high-transcription strain (from 3.2- to 1.3- × 10−9, a 59% reduction), T→C events remained 3.6-fold elevated relative to the wild type low-transcription strain, 1.3- versus 0.36- × 10−9, respectively (Table II and Fig. 2).
Mutation Rate at a Second Gene That Is Regulated by Its Normal Promoter
To determine whether the TAM mutator phenotype is specific to the highly transcribed lys2-TAG allele as opposed to a generalized effect, we measured forward mutation rates at the CAN1 gene, which is regulated by its normal promoter in these strains. CAN1 mutation rates did not differ significantly between the wild type “low”- and “high”-transcription strains, 14.8- and 17.6- × 10−8, respectively (Table III). Regarding the low-transcription deletion strains, CanR rate increased 3.5-fold in the ogg1Δ strain background (from 14.8- to 52- × 10−8), increased 2.6-fold in the tsa1Δ background (from 14.8- to 38- × 10−8), and decreased approximately 60% in the rev3Δ background (from 14.8- to 6.0- × 10−8). Similar to the wild type strains, CanR rates in the ogg1Δ (52- and 63- × 10−8), tsa1Δ (38- and 42- × 10−8), and rev3Δ (6.0- and 7.0- × 10−8) backgrounds did not differ between the low- (pLYS-lys2-TAG), and high- (pGAL-lys2-TAG) transcription strains, respectively.
TABLE III.
CAN1 Forward Mutation Rates in Strains Containing a pLYS- or pGAL-Regulated lys2-TAG Allele
| lys2-TAG Transcription level | CanR Ratea × 10−8 |
|||
|---|---|---|---|---|
| WT (95% CIb) | ogg1Δ (95% CI) | tsa1Δ (95% CI) | rev3Δ (95% CI) | |
| Low | 14.8 [1×] (11–18) |
52 [3.5×] (50–64) |
38 [2.6×] (26–73) |
6.0 [0.41×] (2.6–9.0) |
| High | 17.6 [1.2×] (14–31) |
63 [4.3×] (50–108) |
42 [2.8×] (32–72) |
7.0 [0.47×] (5.2–9.8) |
Bracketed values represent fold-increases relative to the low-transcription wild type rate.
CI, confidence interval.
DISCUSSION
We previously used LYS2 and CAN1 forward mutation assays to characterize TAM in yeast [Lippert et al., 2004; Lippert et al., 2011]. This approach was highly successful in identifying 2-5 bp deletions as the predominant transcription-associated mutations. Other mutation classes such as base substitutions (BSs) increased 1.8- to 2.8-fold in the high- relative to the low-transcription strains; however, BSs comprised small proportions of the high-transcription spectra making it difficult to analyze trends in specific BS types. While forward mutation assays detect a wide range of mutation classes, reversion assays are highly sensitive to a specific mutation class. Moreover, the smaller mutation target allows for the determination of spectra with relative ease. In this study we developed a BS assay that detects events that revert a LYS2 TAG nonsense codon, the lys2-TAG allele. The lys2-TAG assay detects all BS types except G:C→A:T transitions. Because zero A→G transitions (at position two of the TAG target) were recovered in a total of 556 Lys+ revertants analyzed to date, tryptophan at this amino acid position (see Fig. 1) likely results in a nonfunctional Lys2 protein. In addition to the simple BSs recovered using the lys2-TAG allele, a small proportion (10 of 556 or 2%) of Lys+ revertants possessed no mutation either within the TAG mutation target or within approximately 800 adjacent bps (Table II). These rare events likely represent extragenic suppressor mutations. Furthermore as discussed below, the lys2-TAG assay exhibits expected mutator/antimutator effects and characteristic spectra in low-transcription single knock-out ogg1Δ, tsa1Δ, apn1Δ, and rev3Δ strain backgrounds [Kunz et al., 1994; Roche et al., 1994; Thomas et al., 1997; Huang and Kolodner 2005]. The broad range of BS types recovereΔ, low occurrences of suppressor mutations, and characteristic responses to known mutator/antimutator strain backgrounds validate the new lys2-TAG assay for this and future studies focused on BS mutagenesis.
In the low-transcription strain, mutations occurred in roughly equal proportions (38, 24, and 32%) among the first (T), second (A), and third (G) positions, respectively, of the TAG mutation target (Table II). In response to high levels of transcription through the lys2-TAG allele, mutation rate increased 22-fold, and the mutation spectrum shifted dramatically. While mutations at GC bp comprised only 32% of the low-transcription spectrum, G→T and G→C events accounted for 82% of the high-transcription spectrum, and G→T transversions alone comprised the majority (64%) of events. BS rates originating at AT bp also increased in the high-transcription strain, albeit to a lesser extent. While unlikely, a trivial explanation for these results would be a generalized mutator phenotype associated with lys2-TAG overexpression. We excluded this possibility by demonstrating that high levels of transcription through the lys2-TAG allele had no effect on CanR rate (Table III). Altogether these results demonstrate an unambiguous effect of transcription level on BS mutagenesis in yeast and identify G→T and G→C transversions as major transcription-associated events.
The strong bias for TAM transversions at GC bp identified in the present study suggests that guanine and/or cytosine bases may experience increased damage during high levels of transcription. We therefore tested whether oxidative damage or AP sites played a role in the origin of transcription-associated BSs. Specifically, we looked for a synergistic increase in the rate of G→T or G→C transversions in high-transcription genetic deletion strains with elevated levels of 8-oxoG lesions in DNA (ogg1Δ background), increased general oxidative damage (tsa1Δ background), or elevated levels of AP sites (apn1Δ background). Overall mutation rate as well as specific rates of G→T (ogg1Δ strain only) and G→C (tsa1Δ strain only) events increased significantly in the low-transcription strains (Table II) as expected of single mutant ogg1Δ and tsa1Δ strain backgrounds [Thomas et al., 1997; Huang and Kolodner, 2005]. However, no synergistic increase in TAM-G transversions resulted in either high-transcription strain background; we therefore rejected the hypotheses that 8-oxoG lesions or general oxidative damage generated the TAM-G transversions in wild type cells.
In contrast to the ogg1Δ and tsa1Δ results, the apn1Δ high-transcription strain exhibited a synergistic 99-fold increase in total mutation rate relative to the low-transcription wild type strain when compared to the individual effects of the apn1Δ background alone (3.2-fold increase) and a high level of transcription alone (22-fold increase) (Table II). However, G→T and G→C rates either decreased moderately or remained unchanged in the high-transcription apn1Δ relative to the high-transcription wild type strain indicating that AP sites play little if any role in the generation of TAM-G events. Rather, the synergistic increase in overall reversion rate resulted from a specific increase in A→C transversions (Table II; Fig. 2), which we discuss below.
Finally, we explored a role of Polζ in the TAM BS occurrences. The 94% reduction in overall Lys+ reversion rate under high-transcription conditions indicates that the vast majority of TAM BSs require Polζ for occurrence. In the rev3Δ high-transcription strain, only T→C transitions remained partially elevated (1.3 × 10−8) relative to the wild type high-transcription strain (3.2 × 10−8) when compared with the wild type low-transcription strain (0.36 × 10−8), thus corresponding to a 59% reduction and a 3.6-fold elevation relative to the wild type high- and low-transcription strains, respectively. Therefore, 41% of TAM T→C BSs occur via a Polζ independent mechanism. All BS types, except T→C transitions, decreased in the rev3Δ high-transcription strain to levels exhibited by the wild type low-transcription strain (Table II and Fig. 2), thus indicating a complete dependence on Polζ for their occurrences and implicating DNA damage in the underlying mechanism. The Polζ dependence of TAM BSs parallels results reported previously for TAM simple and complex frameshifts [Datta and Jinks-Robertson, 1995; Kim et al., 2007], suggesting that the different TAM events might share a similar mechanism or mutagenic intermediate. Consistent with this possibility, TAM simple −1 FSs also exhibit a strong bias for GC bp [Morey et al., 2000].
Whereas the majority of previous work on TAM in yeast used forward mutation and frameshift reversion assays, Kim and Jinks-Robertson [2010] recently developed a BS assay based on a LYS2 TAA nonsense allele that is regulated by the highly inducible pTET promoter (i.e., the pTET-lys2-TAA allele). In the authors’ study, the lys2-TAA allele exhibited a similar mutation rate under low-transcription conditions (2.8 × 10−9) as the lys2-TAG allele (2.1 × 10−9; Table II) described in this study. Under high-transcription conditions, lys2-TAA reversion rate increased ~10-fold [Kim and Jinks-Robertson, 2010] compared to the 22-fold induction reported here for the lys2-TAG allele. This between-assay difference in TAM induction might result from different strain backgrounds, promoters, sequence contexts, and/or mutation-target locations within LYS2. However, we favor the possibility that the different mutation targets (TAA versus TAG) account for some of the 2-fold difference in rate because the lys2-TAA assay detects events at AT bp only, whereas G→T and G→C transversions dominated the lys2-TAG high-transcription spectrum.
Previous studies on TAM in prokaryotes identified that transcriptional induction stimulated a variety of BS types [Klapacz and Bhagwat, 2002; Hudson et al., 2003]. In E. coli, Bhagwat and colleagues identified transcription-dependent C→T transitions and G→T transversions specific to the nontranscribed strand resulting from cytosine deamination and 8-oxoG lesions, respectively [Beletskii and Bhagwat, 1996; Klapacz and Bhagwat, 2005]. Given the far greater susceptibility of single DNA strands to chemical mutagens relative to double-stranded DNA [Lindahl and Nyberg, 1972; Frederico et al., 1990; Lindahl, 1993], the authors proposed that the nontranscribed strand experiences greater damage when transiently single-stranded at the transcription bubble relative to the transcribed strand, which the transcription machinery likely protects. Whereas both E. coli and yeast exhibit TAM G→T transversions, the role of 8-oxoG lesions in the E. coli events [Klapacz and Bhagwat, 2005] contrasts with results reported here in yeast and indicates that, at least regarding G→T events, different TAM mechanisms operate in the two organisms. However, the data presented here provide no mechanistic insight into this surprising result.
While highly sensitive to a variety of BS types, the lys2-TAG allele cannot detect C:G→T:A transitions. In addition to the transcription-associated C→T mutator phenotype identified in E. coil [Beletskii and Bhagwat, 1996], a second group reported a transcription-dependent UV-induced C→T mutator phenotype in nucleotide excision repair-defective mammalian cells. Moreover, the predominant CpC→TpT and C→T transitions that occurred under high-transcription conditions resulted from cytosine deamination in UV-induced photolesions located specifically in the transcribed strand [Hendriks et al., 2010a]. In yeast, identification of potential roles of cytosine deamination and strand specificity in TAM BSs await future studies. We are currently developing a reversion assay capable of detecting C:G→T:A transitions.
While unrelated to TAM-G transversions, we observed a striking increase in transcription-dependent A→C transversions in the apn1Δ strain background. Moreover, these A:T→C:G transversions at the second position of the TAG stop codon almost certainly represent strand-specific events because zero T→G transversions (A→C transversions in the transcribed strand) occurred at the first position of the TAG target in a sample of 47 revertants (Table II). These data confirm the published results of Kim and Jinks-Robertson [2010] using the pTET-lys2-TAA BS assay (described above). The authors reported a 201-fold increase in A→C transversions in apn1Δ strain backgrounds relative to wild type cells under high-transcription conditions [Kim and Jinks-Robertson, 2010], a result which corresponds well to the greater than 262-fold increase (from < 0.64- to 168- × 10−9) reported in the present study (Table II). The authors further identified the underlying mechanism: under high-transcription conditions, dUTP incorporates into DNA in place of dTTP, and uracil DNA glycosylase creates an AP site that is subject to mutagenic bypass by Polζ [Kim and Jinks-Robertson, 2009, 2010]. In contrast to the TAM A→C events, which occur at elevated levels only in Apn1-deficient backgrounds (Table II) [Kim and Jinks-Robertson 2009; Kim and Jinks-Robertson 2010], the TAM-G transversions reported in the present study occur in repair-proficient cells.
In summary, we report the development and use of a new assay to focus on transcription-associated BSs. The lys2-TAG allele is highly sensitive to TAM, detecting a 22-fold overall increase in mutation rate and a dramatic shift in mutation spectra. Specifically a high level of transcription through the lys2-TAG allele resulted in approximately 55-fold increases in transversions at GC bp. Using a genetic approach, we excluded a role of either oxidative damage or AP sites in the TAM-G mutator phenotype and identified a near complete dependence on Polζ for all TAM BS types (except T→C transitions, which exhibited a partial dependence) implicating DNA damage in the underlying mechanism of action. The new lys2-TAG assay provides a useful tool for the rapid identification of further genetic requirements. Importantly, TAM-G transversions occur in DNA repair proficient cells. Thus, transcription-associated G→T and G→C events represent a novel type of transcription-associated mutagenesis in normal cells with potentially important implications for evolution and genetic disease.
ACKNOWLEDGMENTS
The authors thank Sue Jinks-Robertson for providing helpful feedback on the manuscript and Mark Longtine for his gift of plasmid pFA6a-kanMX6-pGAL1.
Grant sponsor: National Institutes of Health Grant; Grant Number: R15 GM079778, Grant sponsors: Supported summer research, Saint Michael's College Summer Research Award, Undergraduate Research Fellowship from the American Society for Microbiology.
Footnotes
AUTHOR CONTRIBUTIONS
M.J.L. designed the study. M.P.A., K.J.B., W.C.C., M.P.H., and M.J.L. collected and analyzed data. M.J.L. prepared the manuscript draft along with figures and tables, with contributions from M.P.A. and W.C.C on an early draft. All authors had complete access to the study data and approved the final manuscript.
REFERENCES
- Abdulovic AL, Minesinger BK, Jinks-Robertson S. The effect of sequence context on spontaneous Polzeta-dependent mutagenesis in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:2082–2093. doi: 10.1093/nar/gkn054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Aguilera A, Gomez-Gonzalez B. Genome instability: A mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- Beletskii A, Bhagwat AS. Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:13919–13924. doi: 10.1073/pnas.93.24.13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Jinks-Robertson S. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]
- Fix D, Canugovi C, Bhagwat AS. Transcription increases methyl-methane sulfonate-induced mutations in alkB strains of Escherichia coli. DNA repair. 2008;7:1289–1297. doi: 10.1016/j.dnarep.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: Determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd ed. ASM Press; Washington, D.C.: 2006. p. 1118. [Google Scholar]
- Garcia-Rubio M, Huertas P, Gonzalez-Barrera S, Aguilera A. Recombinogenic effects of DNA-damaging agents are synergistically increased by transcription in Saccharomyces cerevisiae. New insights into transcription-associated recombination. Genetics. 2003;165:457–466. doi: 10.1093/genetics/165.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rubio ML, Aguilera A. Topological constraints impair RNA polymerase II transcription and causes instability of plasmid-borne convergent genes. Nucleic Acids Res. 2012;40:1050–1064. doi: 10.1093/nar/gkr840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Gottipati P, Helleday T. Transcription-associated recombination in eukaryotes: Link between transcription, replication and recombination. Mutagenesis. 2009;24:203–210. doi: 10.1093/mutage/gen072. [DOI] [PubMed] [Google Scholar]
- Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks G, Calleja F, Besaratinia A, Vrieling H, Pfeifer GP, Mullenders LH, Jansen JG, de Wind N. Transcription-dependent cyto-sine deamination is a novel mechanism in ultraviolet light-induced mutagenesis. Curr Biol. 2010a;20:170–175. doi: 10.1016/j.cub.2009.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks G, Calleja F, Vrieling H, Mullenders LH, Jansen JG, de Wind N. Gene transcription increases DNA damage-induced mutagenesis in mammalian stem cells. DNA repair. 2008;7:1330–1339. doi: 10.1016/j.dnarep.2008.04.015. [DOI] [PubMed] [Google Scholar]
- Hendriks G, Jansen JG, Mullenders LH, de Wind N. Transcription and replication: Far relatives make uneasy bedfellows. Cell cycle. 2010b;9:2300–2304. doi: 10.4161/cc.9.12.11987. [DOI] [PubMed] [Google Scholar]
- Huang ME, Kolodner RD. A biological network in Saccharomyces cerevisiae prevents the deleterious effects of endogenous oxidative DNA damage. Mol Cell. 2005;17:709–720. doi: 10.1016/j.molcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Huang ME, Rio AG, Nicolas A, Kolodner RD. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc Natl Acad Sci USA. 2003;100:11529–11534. doi: 10.1073/pnas.2035018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RE, Bergthorsson U, Ochman H. Transcription increases multiple spontaneous point mutations in Salmonella enterica. Nucleic Acids Res. 2003;31:4517–4522. doi: 10.1093/nar/gkg651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Abdulovic AL, Gealy R, Lippert MJ, Jinks-Robertson S. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair. 2007;6:1285–1296. doi: 10.1016/j.dnarep.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S. dUTP incorporation into genomic DNA is linked to transcription in yeast. Nature. 2009;459:1150–1153. doi: 10.1038/nature08033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S. Abasic sites in the transcribed strand of yeast DNA are removed by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2010;30:3206–3215. doi: 10.1128/MCB.00308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S. Transcription as a source of genome instability. Nat Rev Genet. 2012;13:204–214. doi: 10.1038/nrg3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapacz J, Bhagwat AS. Transcription-dependent increase in multiple classes of base substitution mutations in Escherichia coli. J Bacteriol. 2002;184:6866–6872. doi: 10.1128/JB.184.24.6866-6872.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapacz J, Bhagwat AS. Transcription promotes guanine to thy-mine mutations in the non-transcribed strand of an Escherichia coli gene. DNA repair. 2005;4:806–813. doi: 10.1016/j.dnarep.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Kunz BA, Henson ES, Roche H, Ramotar D, Nunoshiba T, Demple B. Specificity of the mutator caused by deletion of the yeast structural gene (APN1) for the major apurinic endonuclease. Proc Natl Acad Sci USA. 1994;91:8165–8169. doi: 10.1073/pnas.91.17.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CW. Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair. 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- Lippert MJ, Freedman JA, Barber MA, Jinks-Robertson S. Identification of a distinctive mutation spectrum associated with high levels of transcription in yeast. Mol Cell Biol. 2004;24:4801–4809. doi: 10.1128/MCB.24.11.4801-4809.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert MJ, Kim N, Cho JE, Larson RP, Schoenly NE, O'Shea SH, Jinks-Robertson S. Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proc Natl Acad Sci USA. 2011;108:698–703. doi: 10.1073/pnas.1012363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Bra-chat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Morey NJ, Greene CN, Jinks-Robertson S. Genetic analysis of transcription-associated mutation in Saccharomyces cerevisiae. Genetics. 2000;154:109–120. doi: 10.1093/genetics/154.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoff SC, Spira AI, Johnson AW, Demple B. Yeast structural gene (APN1) for the major apurinic endonuclease: Homology to Escherichia coli endonuclease IV. Proc Natl Acad Sci USA. 1990;87:4193–4197. doi: 10.1073/pnas.87.11.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Aguilera A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 2005;24:1267–1276. doi: 10.1038/sj.emboj.7600602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramotar D, Popoff SC, Gralla EB, Demple B. Cellular role of yeast Apn1 apurinic endonuclease/3′-diesterase: repair of oxidative and alkylation DNA damage and control of spontaneous mutation. Mol Cell Biol. 1991;11:4537–4544. doi: 10.1128/mcb.11.9.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche H, Gietz RD, Kunz BA. Specificity of the yeast rev3Δ anti-mutator and REV3 dependency of the mutator resulting from a defect (rad1Δ) in nucleotide excision repair. Genetics. 1994;137:637–646. doi: 10.1093/genetics/137.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spell RM, Jinks-Robertson S. Determination of mitotic recombination rates by fluctuation analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2004;262:3–12. doi: 10.1385/1-59259-761-0:003. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Burguiere-Slezak G, Van der Kemp PA, Boiteux S. Topoisomerase 1 provokes the formation of short deletions in repeated sequences upon high transcription in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2011;108:692–697. doi: 10.1073/pnas.1012582108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Scot AD, Barbey R, Padula M, Boiteux S. Inactivation of OGG1 increases the incidence of G.C→T.A transversions in Saccharomyces cerevisiae: Evidence for endogenous oxidative damage to DNA in eukaryotic cells. Mol Gen Genet. 1997;254:171–178. doi: 10.1007/s004380050405. [DOI] [PubMed] [Google Scholar]
- Wellinger RE, Prado F, Aguilera A. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol Cell Biol. 2006;26:3327–3334. doi: 10.1128/MCB.26.8.3327-3334.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CM, Siu KL, Jin DY. Peroxiredoxin-null yeast cells are hypersensitive to oxidative stress and are genomically unstable. J Biol Chem. 2004;279:23207–23213. doi: 10.1074/jbc.M402095200. [DOI] [PubMed] [Google Scholar]
- Wright BE, Longacre A, Reimers JM. Hypermutation in dere-pressed operons of Escherichia coli K12. Proc Natl Acad Sci USA. 1999;96:5089–5094. doi: 10.1073/pnas.96.9.5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q. New algorithms for Luria-Delbruck fluctuation analysis. Math Biosci. 2005;196:198–214. doi: 10.1016/j.mbs.2005.03.011. [DOI] [PubMed] [Google Scholar]