Abstract
Oxygen tension has emerged as a potent regulator of multiple erythrocyte properties, including glucose metabolism, cell volume, ATP release, and cytoskeletal organization. Because hemoglobin (Hb)1 binds to the cytoplasmic domain of band 3 (cdb3) in an oxygen dependent manner, with deoxyHb exhibiting significantly greater affinity for cdb3 than oxyHb, the deoxyHb-cdb3 interaction has been hypothesized to constitute the molecular switch for all O2-controlled erythrocyte processes. In this study, we describe a rapid and accurate method for quantitating the interaction of deoxyHb binding to cdb3. For this purpose, enhanced green fluorescent protein (eGFP) is fused to the COOH-terminus of cdb3, and the binding of Hb to the NH2-terminus of cdb3-eGFP is quantitated by Hb-mediated quenching of cdb3-eGFP fluorescence. As expected, the intensity of cdb3-eGFP fluorescence decreases only slightly following addition of oxyHb. However, upon deoxygenation of the same Hb-cdb3 solution, the fluorescence decreases dramatically (i.e. confirming that deoxyHb exhibits much greater affinity for cdb3 than oxyHb). Using this fluorescence quenching method, we not only confirm previously established characteristics of the Hb-cdb3 interaction, but also establish an assay that can be exploited to screen for inhibitors of the sickle Hb-cdb3 interaction that accelerates sickle Hb polymerization.
Keywords: red blood cells, protein structure, hemoglobin, anion exchanger 1, FRET analysis
Introduction
Molecular oxygen is well known to modulate the biological properties of mature human erythrocytes. First, the activities of several ion transport pathways, including Na/K/2Cl cotransport, Na/H antiport, and KCl cotransport are regulated by O2 pressure, the latter changing 20-fold between oxygenated and deoxygenated red blood cells (RBCs)1–4. Not surprisingly, this rapidly reversible O2 modulation leads to O2-dependent changes in cell hydration and volume5,6, raising the question regarding how a diatomic molecule like O2 can simultaneously control so many transporters? Second, erythrocyte deoxygenation stimulates ATP release from RBCs7, leading to activation of purinergic receptors on endothelial cells, the consequent production of NO, and an ensuing vasodilation8,9. Deoxygenation also reversibly shifts glucose consumption from the pentose phosphate pathway (PPP) to glycolysis10–12, acting as a switch to assure abundant reducing power (i.e. NADPH and glutathione) when the erythrocyte is saturated with O2 and abundant ATP when it is not. Finally, RBC deoxygenation reversibly dissociates the major bridge connecting the membrane to its cytoskeleton by severing the linkage between ankyrin and band 3 (also known as AE1, the anion transporter, the most abundant protein in the RBC membrane), the major anchor of the cytoskeleton to the bilayer13–16. (Fig. 1).
Fig. 1. Oxygen regulation of erythrocyte membrane protein interactions.
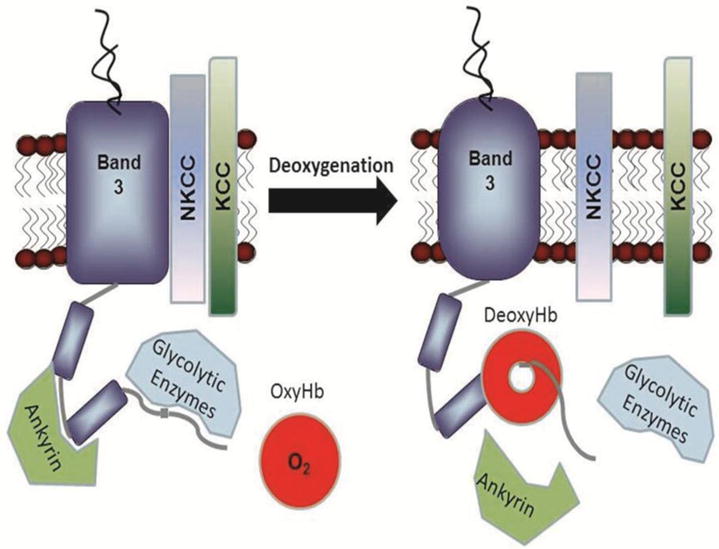
Glucose metabolism11, cation transport1, ATP release7 into circulation and ankyrin binding34 have all been reported to be oxygen-dependent processes in erythrocytes. Because the cytoplasmic domain of band 3 (cdb3) constitutes the only known Hb binding site on the erythrocyte membrane and since the Hb-cdb3 interaction is strongly O2-dependent, many if not all oxygen-regulated pathways in erythrocytes have been postulated to depend on the reversible association of deoxyHb with cdb317. This sketch depicts the binding of deoxyHb to cdb3 and the consequent displacement of several cdb3-associated proteins. Published crystal structure data reveal that deoxyHb binds cdb3 like a “donut on a string”, with the NH2-terminus of cdb3 extending 1.8 nm into the central cavity of deoxyHb51.
Consideration of possible mechanisms to explain the above O2-regulated processes prompted us to hypothesize that the reversible association of deoxyhemoglobin (deoxyHb) with band 3 might constitute a molecular switch that could trigger each of the above physiological changes. Indeed, there is considerable in vitro support for this hypothesis. First, deoxy- but not oxyHb has been found to bind avidly to the cytoplasmic domain of human band 3 (cdb3), specifically to residues 12–23 of the polypeptide17,18. Since no other O2 binding protein has been identified in RBCs, and because the only established deoxyHb binding site in the membrane exists on band 3, the reversible association of deoxyHb with band 3 seemed like a plausible molecular switch. Second, a crystal structure of a complex between deoxyHb and the NH2-terminus of cdb3 reveals that the NH2-terminus of band 3 extends 18Ǻ into a central cavity of deoxyHb18; i.e. the same cavity that closes upon Hb oxygenation. Thus, a mechanism immediately exists to explain why the deoxyHb-band 3 interaction is O2 dependent. Multiple lines of evidence also suggest that this reversible association of deoxyHb with band 3 changes the global conformation of the anion transporter19–21, displacing several signaling enzymes from cdb322–24 and thereby enabling communication of the oxygenation state of the cell to other membrane proteins. Moreover, band 3 has been shown to directly bind proteins thought to be involved in O2-regulated processes, including syk and lyn tyrosine kinases24–28, casein kinase I29, tyrosine phosphatases25,30, a glycolytic enzyme complex31–33, ankyrin34,35, protein 4.136,37, protein 4.238–40, adducin41, two glycophorins42,43, and several transporters16,44. In brief, the raw material for band 3 to mediate O2 regulation of RBC functions is present in the human erythrocyte. However, to test whether band 3 was intrinsically involved, we required an assay that would allow us to identify mutations in band 3 that might eliminate band 3’s affinity for deoxyHb and thereby the O2 regulation of RBC function. The focus of this paper was therefore to design and optimize an assay for measuring the affinity of deoxyHb for band 3 in order to study the interaction under different conditions.
Materials and Methods
Materials
Dialysis was performed using dialysis bags from Spectrum. When desired, proteins were concentrated by ultrafiltration in Vivaspin tubes obtained from GE Healthcare Life Sciences, and final protein concentrations were assayed using a MicroBCA kit from Thermo Scientific according to the manufacturer’s instructions. This MicroBCA assay is based on reduction of Cu2+ to Cu1+ by proteins in an alkaline solution. Protease inhibitors were purchased from Research Products International. All other materials and reagents were purchased from Sigma-Aldrich.
Design of cdb3 – eGFP fusion proteins
In order to assay the interaction of Hb with cdb3, we required fusion proteins comprised of wild type and mutant cdb3s linked at their COOH-termini to enhanced green fluorescent protein (eGFP). Moreover, to facilitate purification of the expressed fusion proteins, a histidine (His8) tag was attached to the COOH-terminus of each eGFP. The required cDNAs for murine cytoplasmic domain of band3 (corresponding to amino acids 1 to 398), murine kidney cdb3 (amino acids 80 to 398), and human cdb3 (amino acids 1 to 379) were PCR-amplified from the corresponding full length band 3 cDNA clones using forward primers containing an NdeI cleavage site followed by the start codon and reverse primers containing a XhoI cleavage site. These primers were:
for wild type mouse cdb3, forward: 5′-CATATGGGGGACATGCGGGACCAC-3′; reverse: 5′-CTCGAGAAAGATCCGGCCTGTGCG-3′
for mouse kidney cdb3, forward: 5′-GCGCATATGGACCAGAGGAACCAG-3′; reverse: 5′-CTCGAGAAAGATCCGGCCTGTGCG-3′
for human cdb3, forward: 5′-CGCCATATGGAGGAGCTGCAGGATGAT-3′; reverse: 5′-CTCGAGG AAGAGCTGGCCTGTCTG-3′.
Because the original eGFP gene contained an additional NdeI cleavage site within its coding sequence, we mutated this site to an alternate codon encoding the same amino acid. The amplified cDNA product was ligated into a pGEM-T easy vector (Promega), and after amplification, removed and inserted upstream of the eGFP sequence in a eGFP-fusion vector. Finally, all PCR-amplified cDNA fragments were sequenced to ensure fidelity of amplification.
Expression and purification of cdb3- eGFP proteins
Fusion protein expression was explored in various E. coli BL21(DE3) strains (Invitrogen) under the conditions described in the Results section. Optimal induction of fusion protein was obtained in BL21(DE3)pLysS bacteria grown in 2XYT media (16g tryptone, 10g yeast extract, and 5g NaCl per litter) at 28°C for 2 to 3 hours. The bacterial pellet was stored overnight at −80°C, thawed in lysis buffer (20mM NaH2PO4, 500mM NaCl, 15mM imidazole, 1 uM AEBSF, pH 7.5), and lysed on ice in a French press (SLM-Aminco). The bacterial lysate was centrifugated at 17,000×g in a Sorval SS-35 rotor for 30 minutes, and the supernatant was filtered and loaded onto a nickel-affinity column (GE Healthcare Life Sciences) equilibrated in lysis buffer. After loading, the column was washed with lysis buffer containing 40mM imidazole and then eluted in a gradient of the same buffer increasing to 250mM imidazole.
Hemoglobin preparation and handling
All animal procedures were approved by Purdue Animal Care and Use Committee in accordance with the guidelines from the National Institute of Health. Human blood was obtained with the understanding and written consent of each subject. The study methodologies were approved by the Purdue Institutional Review Board and conformed to the standards set by the Declaration of Helsinki. Human and murine erythrocyte hemoglobin were obtained from fresh whole blood as described previously11. Briefly, plasma and the buffy-coat were removed by centrifugation, and RBCs were washed in PBS (137mM NaCl, 2.7mM KCl, 8.1mM NaH2PO4, 1.5mM KH2PO4, pH 7.4). The packed RBCs were stored at 4°C for one week to begin depletion of 2,3-DPG, after which the cells were lysed in 5 volumes of cold water and spun at 20,000×g at 4°C. To remove any residual 2,3-DPG, the supernatant was first dialyzed for 12h in 1M NaCl, then 0.1M NaCl, and finally in 10 mM BisTris acetate, pH 6.5. Hemoglobin concentration was determined by measuring absorbance at 540nm using an extinction coefficient of 14.17 liters mmol−1 cm−1 45. Deoxygenation was performed by flushing the Hb solutions in a sealed cuvette with humidified argon until the change in absorption spectrum confirmed maximal deoxygenation of the sample (Fig. 2 insert, full absorption spectrum is not shown)45. Purification of hemoglobin to homogeneity was not routinely done, since initial studies demonstrated that deoxyHb binding to cdb3 is not affected by further purification steps.
Fig. 2. Selection of a fluorescence resonance energy transfer donor for FRET quenching by Hb using spectral analysis of oxy- and deoxyHb.
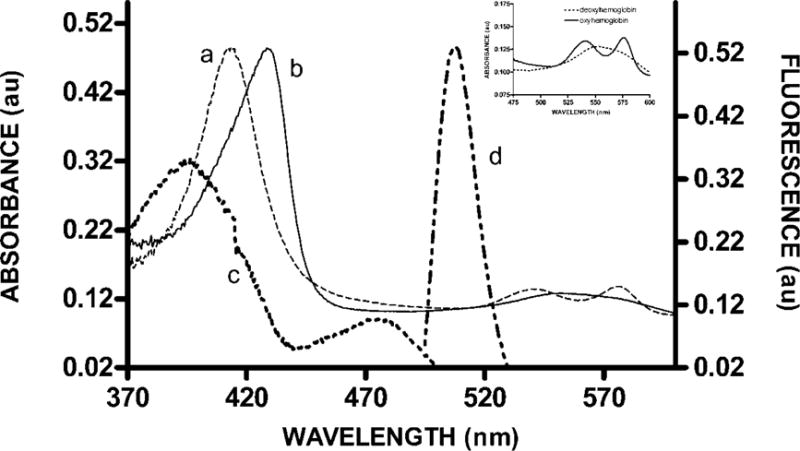
The absorption spectra of oxyHb (a) and deoxyHb (b) are identical at 380nm and 510nm. To prevent differential absorption of either donor excitation or emission as a function of oxygen tension, a FRET donor was required that could be excited at 380nm and would then emit at 510nm. As demonstrated by the excitation (c) and emission (d) spectra of eGFP, this protein complies with these spectral requirements and was therefore selected as the FRET donor. To show complete deoxygenation under our conditions the absorbance spectra of hemoglobin between 475–600nm before and after deoxygenation are shown as an inset in this figure.
Fluorescence resonance energy transfer (FRET) analysis of Hb affinity for band 3
All fluorescence measurements were obtained on an AMINCO-Bowman luminescence spectrometer, using an excitation wavelength of 380 nm and an emission spectrum scanned from 475 to 550 nm. The emission spectrum of the fully oxygenated solution of cdb3-eGFP plus hemoglobin was recorded first, and then the same solution was deoxygenated and the emission spectrum recorded again. The control sample with free eGFP (not fused to cdb3) was used to assess the magnitude of inner filter effects, as discussed under the Results section.
Calculation of the Forster distance between Hb and eGFP
R0, the Forster distance, is defined as the distance at which energy transfer efficiency from a fluorescence donor to a fluorescence acceptor pair is 50%. This R0 value depends both on the overlap integral of the emission spectrum of the donor with the absorption spectrum of the acceptor and their mutual molecular orientations. For the eGFP-Hb pair, R0 was computed to be 5 nm, according to the formula
where k is the orientation factor between donor and acceptor (in general, it is assumed that k2 = 2/3), n is the refractive index (1.33 for water, 1.4 for proteins), QYD is the fluorescence quantum yield of the donor in the absence of the acceptor (0.79 for eGFP) and J is the overlap integral. J was calculated using the a|e - UV-Vis Spectral Software 1.0 (www.FluorTools.com). The values for k2, QYD and n were obtained from46.
Results
Design of a band 3 reporter molecule that changes fluorescence upon binding of deoxyHb
In order to determine the impact of band 3 mutations on the relative affinity of band 3 for oxy- versus deoxyHb, we needed an assay that would generate a strong signal upon hemoglobin binding. For this purpose, we fused enhanced green fluorescent protein (eGFP) to the COOH-terminus of the cytoplasmic domain of band 3 (cdb3) with the expectation that binding of Hb to the NH2-terminus of cdb3 would lead to quenching of eGFP fluorescence at the NH2-terminus. Our reason for anticipating a change in eGFP quantum yield upon Hb binding was that the deoxyHb binding site in the crystal structure of human cdb3 resides only 1.8nm from the site of eGFP fusion to cdb3. Because the R0 for the eGFP-Hb FRET couple is only 5 nm, one would expect eGFP quenching upon hemoglobin binding.
Although the fluorescence quencher had to be Hb, we selected eGFP as the fluorescence donor for two important reasons. First, the emission spectrum of eGFP overlaps significantly with the absorption spectrum of both oxyHb and deoxyHb (Fig. 2). Second, we needed a fluorescence donor with both an emission and excitation maximum at wavelengths where oxy- and deoxyHb exhibit identical absorbances (i.e. to avoid any differences in the absorption of the fluorescence excitation and emission of eGFP during changes in Hb oxygenation). Comparison of the absorption spectra of oxy and deoxyHb revealed that at most wavelengths their absorption spectra are different (Fig. 2). However, at 380 nm and 510 nm eGFP can be excited and emit fluorescence without any differential absorption by oxy- or deoxyHb, respectively. Thus, by setting the excitation and emission monochrometers to 380 and 510 nm, respectively, changes in Hb oxygenation will not affect the FRET analyses unless the changes in oxygenation alter Hb binding and thereby affect eGFP quenching.
Expression, purification and characterization of cdb3-eGFP fusion proteins
The expression vector for the cdb3-eGFP fusion protein was prepared from a commercial vector containing a T7 polymerase promoter upstream from a multi-cloning site linked to the eGFP gene. As explained in the Methods, the cdb3 gene was inserted into this multi-cloning site between NdeI and XhoI restriction sites, resulting in the desired fusion protein with eGFP attached to residue 379 or 398 of the COOH-terminus of human or mouse cdb3, respectively. To assure optimal protein expression, the vector was transformed into E. coli strain BL21(DE3) containing the T7 polymerase gene and an IPTG inducible promoter. Initial attempts to express cdb3-eGFP (37°C for 5 hours) were unsuccessful due to its rapid proteolysis. To remedy this problem, we elected to: i) prevent protein expression in the absence of IPTG by using an E. coli strain containing T7 lysozyme (pLysS strain), which is an inhibitor of T7 polymerase; ii) reduce fusion protein digestion during IPTG induction by stimulation with IPTG at a reduced temperature (28°C), iii) inhibit further cdb3-eGFP proteolysis by inclusion of 1mM AEBSF in the lysis buffer; and iv) shorten the length of the IPTG induction time from 5 to 3 hours. With these modifications, acceptable levels of both human and murine cdb3-eGFP fusion proteins could be expressed and purified (Fig. 3).
Fig. 3. SDS-PAGE analysis of various cdb3-eGFP constructs following expression and purification from bacteria.
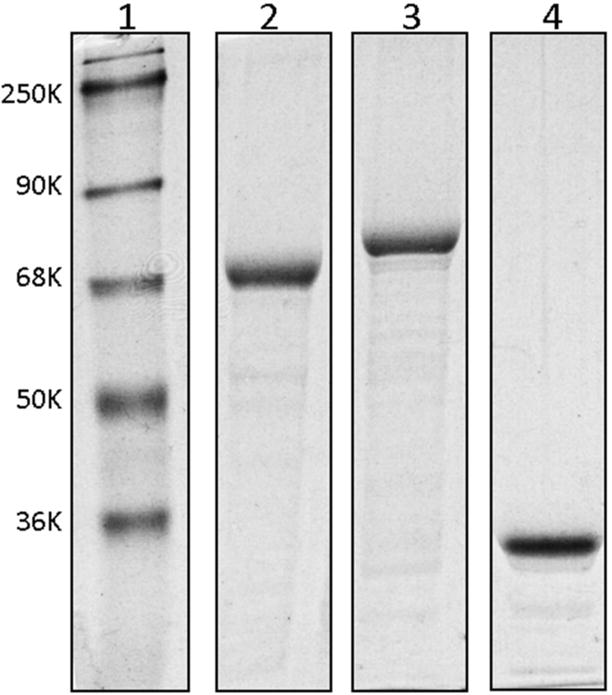
Cdb3-eGFP proteins were expressed in E. coli strain BL21(DE3) pLys S, purified using an affinity column, and analyzed by SDS-PAGE: lane 1, molecular markers; lane 2, human cdb3-eGFP fusion protein containing human cdb3 residues 1–379; lane 3, mouse cdb3-eGFP fusion protein containing mouse cdb3 residues 1–398; lane 4, eGFP protein which is not fused with cdb3.
Use of FRET quenching to evaluate hemoglobin binding to cdb3-eGFP
Because of the intense absorption of Hb, Hb binding to cdb3-eGFP had to be performed in very dilute solutions (Hb=0.07–8μM; [cdb3-eGFP]=0.2–0.5 μM). However, to assure that binding would still occur at these low protein concentrations, solution conditions were selected to maximize Hb-cdb3 affinity. Based on previously published observations47,48, these conditions required use of low ionic strength and slightly acidic pH. Thus, for all FRET analyses, both proteins were dialyzed overnight against 10 mM BisTris-acetate buffer, pH6.5.
As seen in Fig. 4, both eGFP alone and cdb3-eGFP yielded the maximum relative fluorescence in the absence of Hb. Addition of oxyHb to cdb3-eGFP resulted in significant (20%) eGFP quenching, probably due to a combination of some FRET-mediated quenching due to weak oxyHb binding and inner filter effects from free Hb. The latter effect commonly arises when molecules present throughout the solution absorbs either excitation or emission light49. Comparison of the absorption spectrum of Hb with the excitation and emission spectra of eGFP confirms that Hb will absorb both excitation and emission light (Fig. 2).
Fig.4. Quenching of cdb3-eGFP fluorescence by human oxyHb under various conditions.
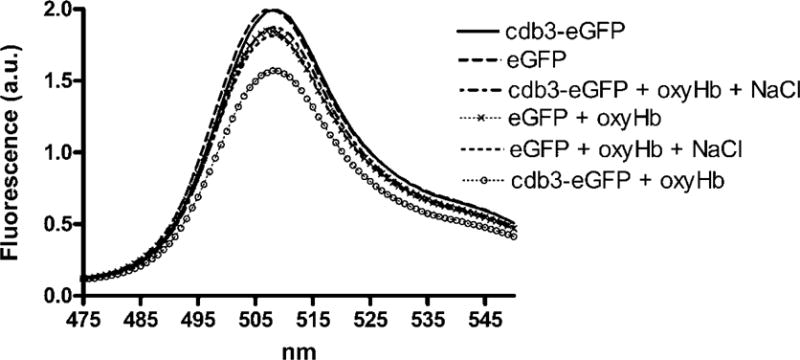
Either cdb3-eGFP or eGFP alone were incubated in 10 mM BisTris buffer (pH 6.5) under the following conditions: 1) in the absence of Hb; 2) in the presence of Hb; 3) in the presence of Hb and 50 mM NaCl. The emission spectrum was then recorded from 475nm to 550nm following excitation at 380nm. Each experiment was analyzed in triplicate with essentially identical results.
To determine the fraction of fluorescence quenching due to inner filter effects, NaCl was added to induce dissociation of the Hb-cdb3 interaction47,48, thereby removing any contribution of FRET-induced quenching from the total decrease in eGFP fluorescence. As seen in Fig. 4, addition of NaCl to the cdb3-eGFP and Hb mixture returned cdb3-eGFP fluorescence to nearly its nonquenched control value, suggesting that most of the oxyHb-induced quenching of cdb3-eGFP is due to direct binding of the two proteins. To confirm that only a small fraction of the Hb-induced quenching of cdb3-eGFP is due to inner filter effects of Hb, a similar set of measurements were obtained on free eGFP solutions in the absence of cdb3. As seen in Fig. 4, eGFP alone displayed the same fluorescence as cdb3-eGFP alone, but addition of Hb lowered the fluorescence of the eGFP solution only to the level of the Hb and cdb3-eGFP solution in the presence of NaCl. We therefore conclude that inner filter effects contribute only a small fraction of the total quenching seen upon addition of Hb. Nevertheless, to accurately correct for any inner filter effects in our calculations of FRET quenching due to Hb binding, the following formula was used:
where F is the fluorescence of cdb3-eGFP (Fprotein) or free eGFP (Fgfp) in the presence of oxy or deoxyHb.
Analysis of the binding affinity of Hb for cdb3-eGFP was performed by measuring the quenching of cdb3-eGFP as a function of Hb concentration and plotting the data using a standard noncooperative single binding site algorithm to determine Kd. Evaluation of the binding of oxyHb to cdb3-eGFP under the low ionic strength conditions described above (Fig. 5) reveal Kd values for human and mouse cdb3 of 1.8 × 10−7 M and 5.5 × 10−7M, respectively. These results are similar to those obtained under related low ionic strength conditions for binding of oxyHb to isolated erythrocyte membranes50.
Fig. 5. Measurement of Hb binding to cdb3-eGFP by analysis of FRET.
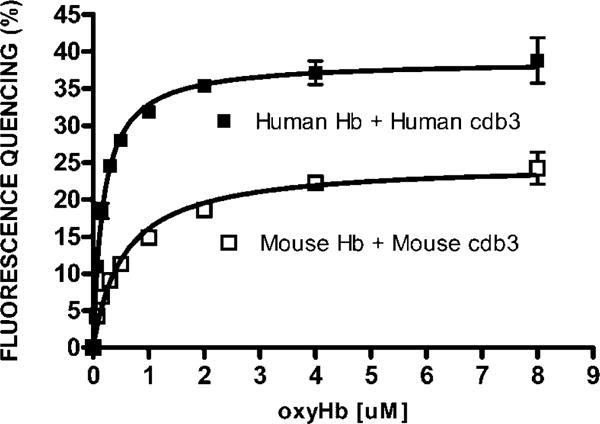
Increasing concentrations of Hb (0.07μM to 8μM) were incubated for 30 min at room temperature with cdb3-eGFP (0.2μM) in 10 mM BisTris buffer (pH6.5). Emission spectra were then recorded and the fluorescence intensity at 510nm was used to calculate the % fluorescence quenching using the equation described in the text. Prism software was employed to determine a Kd of human oxyhemoglobin for human cdb3 of 1.8 × 10−7 M; and the Kd of murine oxyhemoglobin for murine cdb3 of 5.5 × 10−7M. The data constitute the mean +/− S.D. of two experiments.
Analysis of the effect of oxygen on Hb binding to cdb3-eGFP was performed with the same cdb3-eGFP and Hb mixtures, only the solutions were subsequently deoxygenated by exposure to a stream of humidified argon for 45 min. In this protocol, the fluorescence intensity of the oxygenated sample was recorded first and then the same sample was deoxygenated and the fluorescence intensity recorded again. Deoxygenation was then confirmed by determining the absorption spectrum of the sample. As shown in Fig. 6, quenching of the cdb3-eGFP and deoxyHb sample was significantly greater than quenching of the corresponding oxyHb sample, suggesting that differential quenching can be used to map the binding site of deoxyHb on cdb3. To confirm this contention, cdb3-eGFP lacking residues 1–79 of murine cdb3 was examined and found to exhibit little deoxygenation-induced fluorescence quenching (fig. 6). Because deletion of residues 1–79 removes residues that contain the previously identified deoxyHb binding site on murine cdb3, the absence of deoxyHb binding to this deletion mutant confirms the validity of the analytical procedure.
Fig. 6. Measurement of the effect of oxygen on murine cdb3-eGFP binding to murine hemoglobin by FRET.
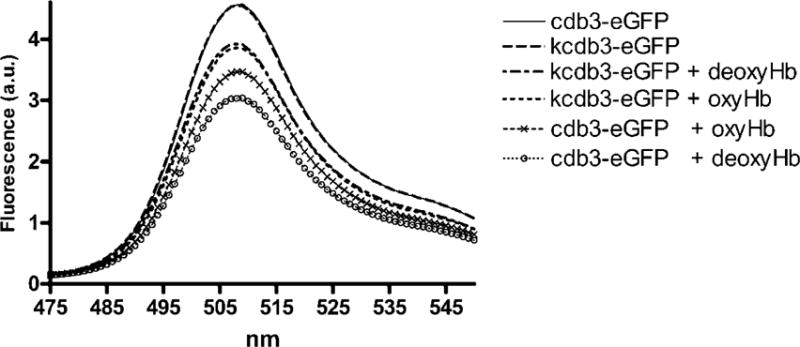
0.5μM of cdb3 was incubated with 1μM Hb under atmospheric conditions, and the emission spectrum of the solution was recorded. This same solution was then deoxygenated under a stream of humidified argon, and the emission spectrum of the solution was again recorded. Deoxygenation was confirmed by monitoring the absorbance of the solution. The kidney spliceoform of cdb3 (kcdb3) was used as a negative control, because it lacks the entire NH2-terminus of cdb3 (residues 1–79) required for deoxyHb binding.
Discussion
In this study, we have developed a FRET assay to quantitate the interaction of Hb with cdb3. For this purpose, eGFP was fused to the COOH-terminus of cdb3 at a distance of 1.75 nm from the docking site for Hb at the NH2-terminus, based on crystal structure calculations51. Since this distance between sites is <R0 (i.e. 5 nm), the interaction between Hb and cdb3 can be studied by measuring FRET-mediated quenching of eGFP. We believe this FRET assay may be preferred over other methods for quantitating the O2-dependence of Hb-cdb3 interactions47,48 for the following reasons: 1) in comparison to previous pull-down assays, our FRET assay is not encumbered by non-specific interactions between Hb and the substrate (e.g. beads) used to immobilize cdb3; 2) omission of all washing steps in our FRET assay avoids loss of weakly bound Hb which must be measured when investigating binding to low affinity cdb3 mutants; 3) very low concentrations of protein are required for this assay, rendering it especially suitable for valuable or difficult to obtain mutant proteins; 4) because the assay is performed in a sealed cuvette that can be repeatedly oxygenated and deoxygenated through inserted ports, the reversibility of the interaction can be accurately evaluated; and 5) using a multi-well plate and a plate reader with a gas-controlled detection chamber (e.g. infinite 200 PRO plate reader from Tecan), the assay may be adaptable to high throughput screening applications.
One possible use of the potential drug screening application of this assay arises from the observation that cdb3 accelerates the polymerization of deoxygenated sickle hemoglobin (deoxyHbS) in sickle cell disease52. This rate of HbS sickling is critical to manifestation of disease symptoms, since if sickling can be postponed until the erythrocyte reaches venous circulation, where the cells are not constrained by a narrow vasculature, the cells can reach the lungs where reoxygenation can induce HbS depolymerization. In order to identify drugs that might prevent HbS sickling and thereby treat sickle cell disease, an assay of small membrane-permeable molecules for their abilities to block deoxyHb-cdb3 interactions might prove useful.
Finally, because the NH2-terminus of cdb3 is known to be the anchor of numerous erythrocyte membrane proteins, a slightly modified version of our FRET assay could prove useful for characterization of a number of cdb3-peripheral protein interactions. For example, the association of glycolytic enzymes32, protein 4.136, protein 4.238, ankyrin34, and syk and lyn tyrosine kinases28 with cdb3 should all be measurable using this method as long as the aforementioned protein can be labeled with an appropriate FRET pair for cdb3-eGFP. By this method, many otherwise difficult to study cdb3 interactions can now be quantitatively evaluated.
Acknowledgments
This work was supported by National Institutes of Health grant GM24417-33.
Abbreviations
- PPP
pentose phosphate pathway
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barvitenko NN, Adragna NC, Weber RE. Erythrocyte signal transduction pathways, their oxygenation dependence and functional significance. Cell Physiol Biochem. 2015;15(1–4):1–18. doi: 10.1159/000083634. [DOI] [PubMed] [Google Scholar]
- 2.Gibson JS, Cossins AR, Ellory JC. Oxygen-sensitive membrane transporters in vertebrate red cells. J Exp Biol. 2000;203(Pt 9):1395–1407. doi: 10.1242/jeb.203.9.1395. [DOI] [PubMed] [Google Scholar]
- 3.Khan AI, Drew C, Bal SE, Ball V, Ellory JC, Gibson JS. Oxygen dependence of K(+)-Cl- cotransport in human red cell ghosts and sickle cells. Bioelectrochemistry. 2004;62(2):141–146. doi: 10.1016/j.bioelechem.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Joiner CH, Jiang M, Fathallah H, Giraud F, Franco RS. Deoxygenation of sickle red blood cells stimulates KCl cotransport without affecting Na+/H+ exchange. Am J Physiol. 1998;274(6 Pt 1):C1466–1475. doi: 10.1152/ajpcell.1998.274.6.C1466. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs M, Parpart AK. The influence of pH, Temperature, and oxygen Tension on Hemolysis by Hypotonic Solutions. The Biological Bulletin. 1931;60(2):95–119. [Google Scholar]
- 6.Rinehart J, Gulcicek EE, Joiner CH, Lifton RP, Gallagher PG. Determinants of erythrocyte hydration. Curr Opin Hematol. 2010;17(3):191–197. doi: 10.1097/MOH.0b013e32833800d0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergfeld GR, Forrester T. Release of ATP from human erythrocytes in response to a brief period of hypoxia and hypercapnia. Cardiovasc Res. 1992;26(1):40–47. doi: 10.1093/cvr/26.1.40. [DOI] [PubMed] [Google Scholar]
- 8.Ellsworth ML, Ellis CG, Goldman D, Stephenson AH, Dietrich HH, Sprague RS. Erythrocytes: oxygen sensors and modulators of vascular tone. Physiology (Bethesda) 2009;24:107–116. doi: 10.1152/physiol.00038.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sprague RS, Stephenson AH, Ellsworth ML. Red not dead: signaling in and from erythrocytes. Trends Endocrinol Metab. 2007;18(9):350–355. doi: 10.1016/j.tem.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Kinoshita A, Tsukad K, Soga T, Hishiki T, Ueno Y, Nakayama Y, Tomita M, Suematsu M. Roles of hemoglobin Allostery in hypoxia-induced metabolic alterations in erythrocytes: simulation and its verification by metabolome analysis. J Biol Chem. 2007;282(14):10731–10741. doi: 10.1074/jbc.M610717200. [DOI] [PubMed] [Google Scholar]
- 11.Messana I, Orlando M, Cassiano L, Pennacchietti L, Zuppi C, Castagnola M, Giardina B. Human erythrocyte metabolism is modulated by the O2-linked transition of hemoglobin. FEBS Lett. 1996;390(1):25–28. doi: 10.1016/0014-5793(96)00624-2. [DOI] [PubMed] [Google Scholar]
- 12.Lewis IA, Campanella ME, Markley JL, Low PS. Role of band 3 in regulating metabolic flux of red blood cells. Proc Natl Acad Sci U S A. 2009;106(44):18515–18520. doi: 10.1073/pnas.0905999106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu F, Satchwell TJ, Toye AM. Anion exchanger 1 in red blood cells and kidney: Band 3’s in a pod. Biochem Cell Biol. 2011;89(2):106–114. doi: 10.1139/o10-146. [DOI] [PubMed] [Google Scholar]
- 14.Vince JW, Reithmeier RA. Structure of the band 3 transmembrane domain. Cell Mol Biol. 1996;42(7):1041–1051. [PubMed] [Google Scholar]
- 15.Bruce L. Mutations in band 3 and cation leaky red cells. Blood Cells Mol Dis. 2006;36(3):331–336. doi: 10.1016/j.bcmd.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Bruce LJ, Beckmann R, Ribeiro ML, Peters LL, Chasis JA, Delaunay J, Mohandas N, Anstee DJ, Tanner MJ. A band 3-based macrocomplex of integral and peripheral proteins in the RBC membrane. Blood. 2003;101(10):4180–4188. doi: 10.1182/blood-2002-09-2824. [DOI] [PubMed] [Google Scholar]
- 17.Chu H, Breite A, Ciraolo P, Franco RS, Low PS. Characterization of the deoxyhemoglobin binding site on human erythrocyte band 3: implications for O2 regulation of erythrocyte properties. Blood. 2008;111(2):932–938. doi: 10.1182/blood-2007-07-100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walder JA, Chatterjee R, Steck TL, Low PS, Musso GF, Kaiser ET, Rogers PH, Arnone A. The interaction of hemoglobin with the cytoplasmic domain of band 3 of the human erythrocyte membrane. J Biol Chem. 1984;259(16):10238–10246. [PubMed] [Google Scholar]
- 19.Salhany JM, Cordes KA, Gaines ED. Light-scattering measurements of hemoglobin binding to the erythrocyte membrane. Evidence for transmembrane effects related to a disulfonic stilbene binding to band 3. Biochemistry. 1980;19(7):1447–1454. doi: 10.1021/bi00548a028. [DOI] [PubMed] [Google Scholar]
- 20.Salhany JM, Cassoly R. Kinetics of p-mercuribenzoate binding to sulfhydryl groups on the isolated cytoplasmic fragment of band 3 protein. Effect of hemoglobin binding on the conformation. J Biol Chem. 1989;264(3):1399–1404. [PubMed] [Google Scholar]
- 21.Tellone E, Ficarra S, Scatena R, Giardina B, Koty A, Russo A, Colucci D, Bellocco E, Lagana G, Galtieri A. Influence of gemfibrozil on sulfate transport in human erythrocytes during the oxygenation-deoxygenation cycle. Physiol Res. 2008;57(4):621–629. doi: 10.33549/physiolres.931251. [DOI] [PubMed] [Google Scholar]
- 22.Siciliano A, Turrini F, Bertoldi M, Matte A, Pantaleo A, Olivieri O, De Franceschi L. Deoxygenation affects tyrosine phosphoproteome of red cell membrane from patients with sickle cell disease. Blood Cells Mol Dis. 2010;44(4):233–242. doi: 10.1016/j.bcmd.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Merciris P, Claussen WJ, Joiner CH, Giraud F. Regulation of K-Cl cotransport by Syk and Src protein tyrosine kinases in deoxygenated sickle cells. Pflugers Arch. 2003;446(2):232–238. doi: 10.1007/s00424-003-1025-z. [DOI] [PubMed] [Google Scholar]
- 24.Merciris P, Hardy-Dessources MD, Giraud F. Deoxygenation of sickle cells stimulates Syk tyrosine kinase and inhibits a membrane tyrosine phosphatase. Blood. 2000;98(10):3121–3127. doi: 10.1182/blood.v98.10.3121. [DOI] [PubMed] [Google Scholar]
- 25.Bordin L, Brunati AM, Donella-Deana A, Baggio B, Toninello A, Clari G. Band 3 is an anchor protein and a target for SHP-2 tyrosine phosphatase in human erythrocytes. Blood. 2002;100(1):276–282. doi: 10.1182/blood.v100.1.276. [DOI] [PubMed] [Google Scholar]
- 26.Pantaleo A, Ferru E, Giribaldi G, Mannu F, Carta F, Matte A, de Franceschi L, Turrini F. Oxidized and poorly glycosylated band 3 is selectively phosphorylated by Syk kinase to form large membrane clusters in normal and G6PD-deficient red blood cells. Biochem J. 2009;418(2):359–367. doi: 10.1042/BJ20081557. [DOI] [PubMed] [Google Scholar]
- 27.Bordin l, Ion-Popa F, Brunati AM, Clari G, Low PS. Effector-induced Syk-mediated phosphorylation in human erythrocytes. Biochim. Biophys Acta. 2005;1745(1):20–28. doi: 10.1016/j.bbamcr.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 28.Brunati AM, Bordin L, Clari G, James P, Quadroni M, Baritono E, Pinna LA, Donella-Deana A. Sequential phosphorylation of protein band 3 by Syk and Lyn tyrosine kinases in intact human erythrocytes: identification of primary and secondary phosphorylation sites. Blood. 2000;96(4):1550–1557. [PubMed] [Google Scholar]
- 29.Wang CC, Tao M, Wei T, Low PS. Identification of the major casein kinase I phosphorylation sites on erythrocyte band 3. Blood. 1997;89(8):3019–3024. [PubMed] [Google Scholar]
- 30.Zipser Y, Kosower NS. Phosphotyrosine phosphatase associated with band 3 protein in the human erythrocyte membrane. Biochem J. 1996;314(Pt 3):881–887. doi: 10.1042/bj3140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campanella ME, Chu H, Low PS. Assembly and regulation of a glycolytic enzyme complex on the human erythrocyte membrane. Proc Natl Acad Sci U S A. 2005;102(7):2402–2407. doi: 10.1073/pnas.0409741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu H, Low PS. Mapping of glycolytic enzyme-binding sites on human erythrocyte band 3. Biochem J. 2006;400(1):143–151. doi: 10.1042/BJ20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campanella ME, Chu H, Wandersee NJ, Peters LL, Mohandas N, Gilligan DM, Low PS. Characterization of glycolytic enzyme interactions with murine erythrocyte membranes in wild-type and membrane protein knockout mice. Blood. 2008;112(9):3900–3906. doi: 10.1182/blood-2008-03-146159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett V, Stenbuck PJ. Association between ankyrin and the cytoplasmic domain of band 3 isolated from the human erythrocyte membrane. J Biol Chem. 1980;255(13):6424–6432. [PubMed] [Google Scholar]
- 35.Chang SH, Low PS. Identification of a critical ankyrin-binding loop on the cytoplasmic domain of erythrocyte membrane band 3 by crystal structure analysis and site-directed mutagenesis. J Biol Chem. 2003;278(9):6879–6884. doi: 10.1074/jbc.M211137200. [DOI] [PubMed] [Google Scholar]
- 36.Pasternack GR, Anderson RA, Leto TL, Marchesi VT. Interactions between protein 4.1 and band 3. An alternative binding site for an element of the membrane skeleton. J Biol Chem. 1985;260(6):3676–3683. [PubMed] [Google Scholar]
- 37.An XL, Takakuwa Y, Nunomura W, Manno S, Mohandas N. Modulation of band 3-ankyrin interaction by protein 4.1. Functional implications in regulation of erythrocyte membrane mechanical properties. J Biol Chem. 1996;271(52):33187–33191. doi: 10.1074/jbc.271.52.33187. [DOI] [PubMed] [Google Scholar]
- 38.Cohen CM, Dotimas E, Korsgren C. Human erythrocyte membrane protein band 4.2 (pallidin) Semin Hematol. 1993;30(2):119–137. [PubMed] [Google Scholar]
- 39.Rybicki AC, Schwartz RS, Hustedt EJ, Cobb CE. Increased rotational mobility and extractability of band 3 from protein 4.2-deficient erythrocyte membranes: evidence of a role for protein 4.2 in strengthening the band 3-cytoskeleton linkage. Blood. 1996;88(7):2745–2753. [PubMed] [Google Scholar]
- 40.Peters LL, Jindel HK, Gwynn B, Korsgren C, John KM, Lux SE, Mohandas N, Cohen CM, Cho MR, Golan DE, Brugnara C. Mild spherocytosis and altered red cell ion transport in protein 4. 2-null mice. J Clin Invest. 1999;103(11):1527–1537. doi: 10.1172/JCI5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anong WA, Franco T, Chu H, Weis TL, Devlin EE, Bodine DM, An X, Mohandas N, Low PS. Adducin forms a bridge between the erythrocyte membrane and its cytoskeleton and regulates membrane cohesion. Blood. 2009;114(9):1904–1912. doi: 10.1182/blood-2009-02-203216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruce LJ, Ring SM, Anstee DJ, Reid ME, Wilkinson S, Tanner MJ. Changes in the blood group Wright antigens are associated with a mutation at amino acid 658 in human erythrocyte band 3: a site of interaction between band 3 and glycophorin A under certain conditions. Blood. 1995;85(2):541–547. [PubMed] [Google Scholar]
- 43.Mohandas N, Chasis JA. Red blood cell deformability, membrane material properties and shape: regulation by transmembrane, skeletal and cytosolic proteins and lipids. Semin Hematol. 1993;30(3):171–192. [PubMed] [Google Scholar]
- 44.Janoshazi A, Solomon AK. Interaction among anion, cation and glucose transport proteins in the human red cell. J Membr Biol. 1989;112(1):25–37. doi: 10.1007/BF01871161. [DOI] [PubMed] [Google Scholar]
- 45.van Kampen EJ, Zijlstra WG. Spectrophotometry of hemoglobin and hemoglobin derivatives. Adv Clin Chem. 1983;23:199–257. doi: 10.1016/s0065-2423(08)60401-1. [DOI] [PubMed] [Google Scholar]
- 46.Fessenden JD. Forster resonance energy transfer measurements of ryanodine receptor type 1 structure using a novel site-specific labeling method. PLoS One. 2009;4(10):e7338. doi: 10.1371/journal.pone.0007338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chetrite G, Cassoly R. Affinity of hemoglobin for the cytoplasmic fragment of human erythrocyte membrane band 3. Equilibrium measurements at physiological pH using matrix-bound proteins: the effects of ionic strength, deoxygenation and of 2,3-diphosphoglycerate. J Mol Biol. 1985;185(3):639–644. doi: 10.1016/0022-2836(85)90076-2. [DOI] [PubMed] [Google Scholar]
- 48.Shaklai N, Yguerabide J, Ranney HM. Interaction of hemoglobin with red blood cell membranes as shown by a fluorescent chromophore. Biochemistry. 1977;16(25):5585–5592. doi: 10.1021/bi00644a031. [DOI] [PubMed] [Google Scholar]
- 49.Puchalski MM, Morra MJ, von Wandruszka R. Assesment of inner filter effect corrections in fluorimetry. Fresenius’. J Anall Chem. 1991;340(6):341–344. [Google Scholar]
- 50.Shaklai N, Yguerabide J, Ranney HM. Classification and localization of hemoglobin binding sites on the red blood cell membrane. Biochemistry. 1977;16(25):5593–5597. doi: 10.1021/bi00644a032. [DOI] [PubMed] [Google Scholar]
- 51.Zhang D, Kiyatkin A, Bolin JT, Low PS. Crystallographic structure and functional interpretation of the cytoplasmic domain of erythrocyte membrane band 3. Blood. 2000;96(9):2925–2933. [PubMed] [Google Scholar]
- 52.Rotter MA, Chu H, Low PS, Ferrone FA. Band 3 catalyzes sickle hemoglobin polymerization. Biophys Chem. 2010;146(2–3):55–59. doi: 10.1016/j.bpc.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


