Abstract
Only few molecular studies have addressed the age of bacterial pathogens that infected humans before the beginnings of medical bacteriology, but these have provided dramatic insights. The global genetic diversity of Helicobacter pylori, which infects human stomachs, parallels that of its human host. The time to the most recent common ancestor (tMRCA) of these bacteria approximates that of anatomically modern humans, i.e. at least 100 000 years, after calibrating the evolutionary divergence within H. pylori against major ancient human migrations. Similarly, genomic reconstructions of Mycobacterium tuberculosis, the cause of tuberculosis, from ancient skeletons in South America and mummies in Hungary support estimates of less than 6000 years for the tMRCA of M. tuberculosis. Finally, modern global patterns of genetic diversity and ancient DNA studies indicate that during the last 5000 years plague caused by Yersinia pestis has spread globally on multiple occasions from China and Central Asia. Such tMRCA estimates provide only lower bounds on the ages of bacterial pathogens, and additional studies are needed for realistic upper bounds on how long humans and animals have suffered from bacterial diseases.
Keywords: history of disease, plague, tuberculosis, gastritis, ancient DNA, comparative genomics
1. Background
The oldest bacterial pathogens in microbiological strain collections date from the 1890s, soon after medical bacteriology was introduced. Older, historical descriptions of clinical disease are rarely sufficiently precise to provide definitive attributions to individual bacterial pathogens, and only a few bacterial diseases leave characteristic lesions on skeletons. In recent years, comparative genomics has reconstructed the short-term evolutionary history of multiple bacterial clades whose diversity converges on a recent common ancestor (MRCA) that existed decades ago. One example is a clone of antibiotic-resistant Staphylococcus aureus that has been a common cause of hospital disease in Europe and other continents [1] (electronic supplementary material, table S1). Another example is food-borne gastroenteritis caused by Salmonella enterica serovar Agona, whose MRCA dates to the 1930s, and which spread globally in the 1960s via contaminated fish meal from South America [2]. However, despite some claims to the contrary, we have not identified any special properties that are generally characteristic for clones that spread globally. Instead, with few exceptions [3,4], it has not been possible to definitively identify any genetic changes in bacteria that have resulted in increased virulence or transmissibility, and could therefore have accounted for recent expansions of the bacterial populations [5]. Even though antibiotic-resistant mutants are repeatedly selected by the use of antibiotics for medical and veterinary treatment of disease, many such mutants are soon replaced by their antibiotic-sensitive kin [5,6]. Similarly, antigenic variants that were selected for resistance to specific antibodies were soon eliminated once the bacteria had spread to immunologically naive populations, leaving only the parental variants expressing the original antibody targets [7]. We also lack an understanding of why many pathogenic clones are transient, flaring and spreading for decades, only to be replaced by others. One possible explanation for our lack of understanding of these phenomena is that our time frame of comparison is too short, and as a result focuses on random genetic drift, whereas important changes primarily occur over longer periods.
Any analysis of changes during long-term evolution needs to know when such changes happened, which, in turn, depends on dating genetic events. Until several years ago, microbiologists believed that bacteria accumulate mutations at a constant molecular clock rate of approximately 3.4 × 10−9 substitutions per nucleotide per year, which was based on an estimate that the bacterial species Escherichia coli and S. enterica separated about 160 million years ago [8]. This universal clock rate was used to calculate the ages of other bacterial taxa based on their genetic diversity, resulting in estimates of 70 million years for the age of Moraxella catarrhalis [9], 10 000–43 000 years for S. enterica serovar Typhi [6] and 7000 years for E. coli O157:H7 [10]. However, these estimated ages are almost certainly wrong, as are all other estimates published before 2009, because protein-coding genes mutate at different rates in different taxa [11], and the original calibrations that supported a constant clock rate are invalid [12]. Instead, comparative genomics of multiple bacterial pathogens have indicated that their clock rates vary from taxon to taxon, with extreme values that differ by orders of magnitude (electronic supplementary material, table S1).
Even our newest estimates of bacterial clock rates are too simplistic: they assume that clock rates are constant over time, and uniform within lineages. However, it has been argued that the clock rate slows down with time owing to purifying selection [13], which would result in an underestimate of the time to the most recent common ancestor (tMRCA). This concept has received little attention for bacteria, although it may apply to Helicobacter pylori. For H. pylori, the average clock rate was 1000-fold slower for bacterial lineages that separated more than 50 000 years ago (2.6 × 10−7) [12] than the short-term clock rate (3 × 10−4) measured immediately after initiating an infection [14], and chronic infections over years were associated with intermediate rates (1.4 × 10−5) [15]. Short-term rates would therefore be inappropriate for calculating the tMRCA of H. pylori. Calculations based on the laboratory mutation rate are even worse because they differ by at least sevenfold between different strains of H. pylori [12], and it is not possible to convert from the mutations per generation calculated in the laboratory into substitutions per year. Clock rates in nature are also not necessarily independent of demography. For example, clock rates seem to have accelerated temporarily within Y. pestis during the large bacterial population expansions that occurred during plague outbreaks [16].
Here, I concentrate on selected examples where the ages of pathogenic bacteria were investigated over longer time frames through combinations of studies on extant bacteria plus genomic data from ancient DNA (aDNA).
2. Helicobacter pylori and anatomically modern humans
Helicobacter pylori infects the stomachs of approximately 50% of all humans, but the frequency of infection varies regionally, ranging from very low infection rates in young North Americans and Europeans to more than 90% infection rates in large parts of Africa [17]. Infection almost always results in chronic gastritis; approximately 10–15% of infected individuals develop gastric or peptic ulcers and gastric or MALT lymphoma also arises in approximately 1%. The only known natural hosts for H. pylori are humans, but these bacteria can also infect some laboratory animals and are transmitted from humans to captive primates in zoos. Helicobacter acinonychis, the closest relative of H. pylori, seems to have arisen as a distinct species after a host jump of H. pylori from humans to large felines [18].
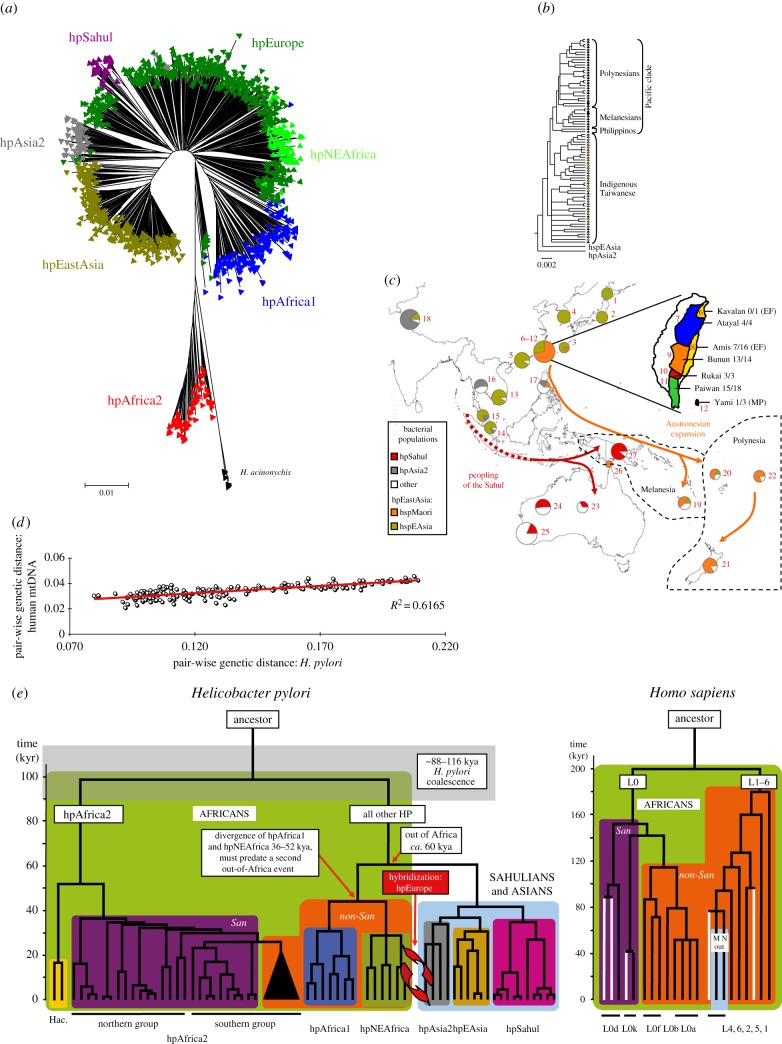
In the late 1990s, I became intrigued by why the genetic diversity of H. pylori seemed to reflect their geographical origins [19]. My co-workers and I collected bacteria from highly diverse geographical sources and distinct human populations, including former hunter–gatherers (San in South Africa: [20]; Baka pygmies in Cameroon [21]; aboriginals in Taiwan and Australia [22]) as well as highlanders in Papua New Guinea [22], Amerinds in North and South America [23] and Buddhists and Muslims in the Himalayas (Ladakh) [24]. Over 2000 strains from diverse geographical sources were genotyped for seven housekeeping gene fragments, confirming that H. pylori from distinct continents or subcontinents belonged to distinct bacterial populations. These populations were assigned mnemonic designations referring to their geographical or ethnic associations (figure 1a). Many of the geographical associations correspond to regions first colonized by anatomically modern humans in sequential migrations during the last 60 000 years (figure 1c). As a result of the bottlenecks associated with those migrations, and subsequent isolation by distance, the pairwise genetic distances between human populations increase with distance and their genetic diversity drops with distance from sub-Saharan Africa [26]. Quantitatively similar patterns were found for H. pylori [25], and the pairwise genetic distances between H. pylori correlated strongly with genetic distances between human mtDNA sequences from corresponding geographical areas (figure 1d). The obvious interpretation was that H. pylori have accompanied humans since their out of Africa migrations about 60 kya. In further support of this conclusion, a non-recombinant phylogenetic tree of the sequences of housekeeping gene fragments of H. pylori shares important branching patterns with that of mtDNA from humans (figure 1e). We therefore used six dates when modern humans first reached certain geographical areas to calibrate the corresponding nodes in the H. pylori phylogenetic tree, and estimated a tMRCA for H. pylori of about 100 000 years (figure 1e) [20].
Figure 1.
Population structure, dated phylogeny and ancient migrations of Helicobacter pylori based on sequences of seven housekeeping gene fragments. (a) Global population structure (based on electronic supplementary material, figure S3 [25] updated with additional data from [22]). (b,c) Migrations into the Pacific of hspMaori and hpSahul (modified from figure 1 of [22]). (b) A phylogenetic tree of hspMaori, a subpopulation of hpEastAsia, shows patterns of serial descent from bacteria from indigenous Taiwanese of six ethnic groupings through isolates from Filipinos, Melanesians and Polynesians. (c) Frequencies of hspMaori (orange) and hpSahul (red) in pie charts according to geographical source. (c inset; Taiwan subdivided by indigenous ethnic group; number of hspMaori/number of all H. pylori (Austronesian language family)). (d) Quantitative parallel patterns of pairwise genetic distances between H. pylori and human mtDNA samples from corresponding geographical areas. (source: figure 7 of [20]). (e) Comparison of phylogenetic tree of H. pylori housekeeping gene fragments (left) and human mtDNA (right) (source: figure 6 of [20]). African lineages are shown on a green background, whereas the background for lineages outside Africa is light blue. San clades are purple, non-San clades are orange and H. acinonychis is yellow. San mtDNA lineages that were identified in this study are shown as white lines.
A striking number of parallels were obtained between the genetic patterns and sources of native human populations and their associations with populations of H. pylori, including native Americans who migrated from Asia across the Bering Strait (hspAmerind, a subpopulation of hpEastAsia); Austronesians who migrated from Taiwan to the Pacific (hspMaori, a subpopulation of hpEastAsia; figure 1b,c); Bantu who migrated from West Africa to East and South Africa (hpAfrica1) and the original inhabitants of the highlands of Papua New Guinea and central Australia (hpSahul) [20,22,23,25]. It is also striking that the click-speaking San, former hunter–gatherers whose mtDNA defines a basal lineage for humans, were found to be the original host of hpAfrica2, which, together with the feline H. acinonychis, forms a basal lineage within H. pylori (figure 1e).
Not all geographical patterns are concordant between H. pylori and humans. For example, we had hoped to identify ancestral variants of H. pylori within Baka pygmies in Cameroon. However, the H. pylori that infected Baka pygmies belonged to the same hpAfrica1 and hpNEAfrica populations as those from their Bantu neighbours who had first migrated to that area within the last 3000–6000 years [21]. If Baka pygmies were previously infected by other H. pylori, those bacteria have likely been lost through the lineage extinction that would be associated with human population sizes that are too small to stably maintain infection [21]. A similar pattern was observed with Amerinds in the Americas, who were predominantly infected with hpEurope and hpAfrica1 that were imported after 1492 [23]. In South America, hspAmerind H. pylori were only isolated from Amerinds living in extremely remote areas of the Amazon basin, and who have had only limited contact with Europeans and Africans.
Possibly the most dramatic apparent discrepancy between human population structure and that of H. pylori is represented by the hpEurope bacterial population. Similar to Europeans, hpEurope is predominant from Western Asia throughout Europe. However, hpEurope is a hybrid population, representing the descendents of admixture between the ancestors of hpNEAfrica, now largely restricted to northeastern Africa, and hpAsia2, which is found throughout central and northern Asia. This admixture event seems to have occurred long after humans and H. pylori left Africa [20] (figure 1e). Indeed, analyses of ancient H. pylori DNA from the Iceman, a 5300-year-old copper age mummy from the Italian Alps, showed that its genomic structure predates that admixture event, and its ancestry was almost exclusively from hpAsia2 [27]. In contrast, the only modern hpAsia2 H. pylori that were isolated in Europe in modern times were from the Bangladeshi community. Finding pure hpAsia2 in the Italian Alps 5000 years ago suggests that the admixture that led to hpEurope was even more recent than 5000 years. However, the human migrations that are known to have occurred since the Bronze age [28] do not include a potential wave of migrants who might have introduced hpNEAfrica throughout Western Eurasia.
3. Over 5000 years of plague
Bubonic plague is an invasive disease of humans that often presents with buboes (inflamed lymph nodes in the inguinal, axillary and cervical regions) as clinical symptoms [29]. Prior to the introduction of antibiotic therapy in the mid-twentieth century, the mortality rate of bubonic plague was extremely high, and even higher for pneumonic plague, an infection of the lungs which is transmitted from human to human by droplets. In 1894, marine shipping from Hong Kong distributed Yersinia pestis from China to shipping ports around the globe, thus initiating the third plague pandemic (figure 2d). In some areas, these bacteria caused novel chains of endemic and epidemic infections of local rodents whose fleas were able to infect humans, and caused large outbreaks of bubonic plague. Rats were the primary rodent hosts in Hong Kong itself, as well as in India and Madagascar. However, in most other geographical areas, including the USA, human plague infections are usually acquired from other species of rodents. Pneumonic plague was less common than bubonic plague, with the notable exception of Manchuria, where the transmission of Y. pestis from infected marmots to fur trappers resulted in large outbreaks after 1910 [29].
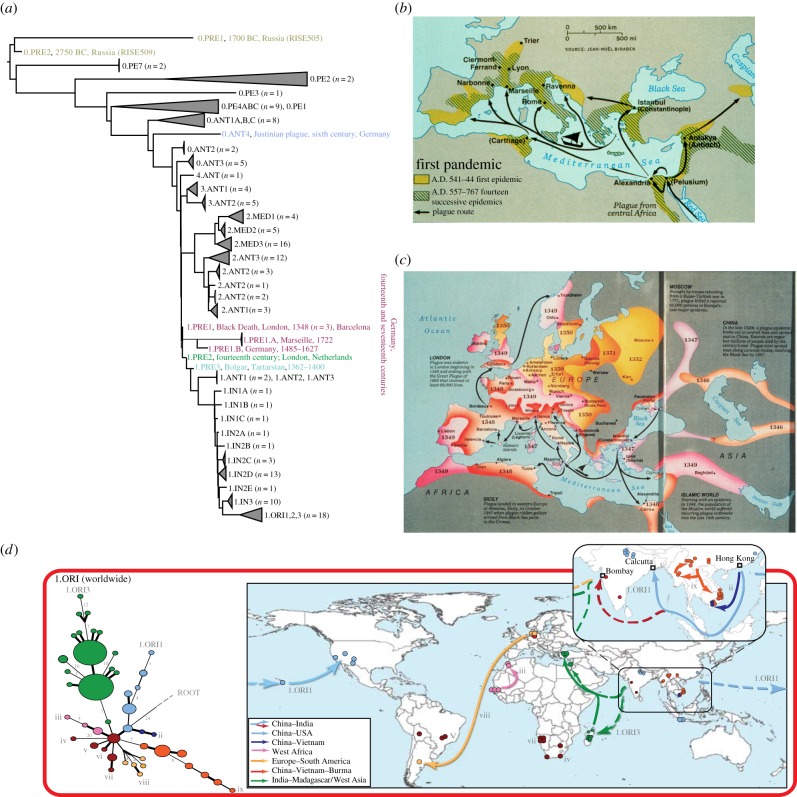
Figure 2.
Plague pandemics and phylogeny of Yersinia pestis. (a) Manual concatenation of the topologies of phylogenies described in [30], [16] and [31], including additional ancient genomes and genotypes (see below), plus proposed mnemonics for those populations. Sources of ancient genomes and genotypes: the Bronze Age (0.PRE1, 0.PRE2; grey) [31], the Justinianic Pandemic (0.ANT4; blue) [32]; the Black Death (1.PRE1; maroon; London, 1348 [33]; Barcelona 1300–1420 [34]; SNP-based genotypes from Germany (fourteenth and seventeenth centuries) [35]) and descendent branches of 1.PRE from Ellwangen (1.PRE1B; 1485–1627) [34] and Marseille (1.PRE1.A, 1722) [36]). One SNP further along branch 1 are the populations 1.PRE2 (London, 1362–1400 [36] and the related low-resolution genotype found in Berg-Op-Zoom, The Netherlands from the fourteenth century [37]; green). One further SNP down the branch is a short branch leading to a genome from Western Asia (1.PRE3; Bolgar City, Tatarstan, Russia; light blue) [34]. (b,c) Maps of the spread of the Justinianic (b) and second plague (c) pandemics. With permission from Elisabeth Carniel. (d) Reconstruction of waves of transmission of individual lineages within the third pandemic. Modified from supplementary figure 3 from [30] (http://research.ucc.ie/NG1/index.html).
Historical records from two prior plague pandemics had already described an association of buboes with lethal epidemic diseases. These pandemics are referred to as the Justinianic (First) Pandemic (multiple waves between 541 and 767; figure 2b) [38] and the Second Pandemic that recurred in multiple waves through to the early-nineteenth century after reaching Europe in 1348 (the Black Death; figure 2c) [39]. Within 1–2 years after these pandemics began, plague had already spread by marine routes to a large part of the Mediterranean basin and Western Europe (henceforth collectively referred to as Europe), and then continued to spread inland at great speed. Each of those two pandemics killed a significant proportion of the European population at that time. The rapid inland geographical transmission and the high proportion of lethality across European populations differs from the epidemiological patterns of modern plague, which spreads much more slowly, and does not kill more than a small fraction of the local human population during a major outbreak. As a consequence, epidemiologists, zoologists and historians long questioned whether Y. pestis was indeed responsible for the first two plague pandemics, and presented a variety of seemingly convincing arguments for alternative aetiological agents [40]. However, human skeletons dating between 300 and 5000 years ago have now yielded aDNA derived from Y. pestis genotypes, and the evolutionary branches defined by these genotypes are intertwined among those of extant Y. pestis. As a result, there is now a general consensus that Y. pestis has been a common cause of plague for millennia. There is a total lack of convincing evidence for the existence of any additional factors or other pathogens that may have been responsible for the special epidemiological patterns of the first two pandemics of historical plague. Indeed, the power of the ‘new plague paradigm’ is now stimulating historians to articulate new questions about the history of these deadly pandemics [41].
(a). Populations and phylogeny
When I first developed an interest in Y. pestis in the late 1990s, multiple technical issues hindered definitive population genetic analyses. First, Y. pestis is a category A potential agent for biocrimes or bioterrorism, whose transport is prohibited except for military organizations or WHO reference laboratories. As a result, no single laboratory has yet assembled a collection of Y. pestis bacteria or their DNA, that is broadly representative of its entire extant global diversity. Second, endemic plague is primarily a disease of wild rodents, but most microbiologists and historians focus on human plague even though it represents a rare consequence of occasional spillover from the primary, rodent hosts [42]. Third, unlike H. pylori, in which every third nucleotide is polymorphic, only a few thousand nucleotides are polymorphic over the entire genome of Y. pestis [16], a frequency of approximately 1/1000. Fourth, the greatest genetic diversity of Y. pestis is found in Central to Eastern Asia [16,30]. However, Central Asia is underrepresented in modern analyses because its microbiological surveillance of rodent infections largely disappeared after the collapse of the Soviet Union in 1990. Fortunately, extensive historical collections of bacteria that were isolated during the twentieth century still exist at the Institut Pasteur in Paris [43] and Madagascar [44], the Beijing Institute of Microbiology and Epidemiology [16] and the Bundeswehr Institute of Microbiology in Munich [45]. These facilitated access to DNA from bacterial strains isolated from a variety of global sources, including Africa, the USA, the Middle East, South East Asia, China and Mongolia as well as a few strains from Central Asia. Furthermore, sequencing and SNP typing have become much simpler since the early 2000s, and genome sequencing and genotyping of genetically monomorphic bacteria [46] such as Y. pestis is now quite straightforward. As a result, my extensive global collaborations with scientists in France, China, the USA, the UK and Madagascar have now resulted in hundreds of complete and draft genomes and SNP-based genotypes [16,30]. These are quite easy to interpret, because, unlike H. pylori, there are no traces of homologous recombination within Y. pestis. Instead, almost all of the polymorphic nucleotides (SNPs) in the genome of Y. pestis represent unique (non-homoplastic) mutations that have each only occurred once in its evolutionary history [16,30]. These SNPs define the same topology independent of the phylogenetic algorithm used, including that of a unique maximum-parsimony tree. An important consequence of this genetic simplicity is that results from SNP testing, and full genome sequences from both extant and ancient sequences can be readily combined in a single evolutionary tree (figure 2a).
The Y. pestis evolutionary tree is unambiguously rooted within Yersinia pseudotuberculosis, of which Y. pestis is a genetically monomorphic clade [47]. Branches and populations within that tree that are defined by extant diversity have already been assigned characteristic names [16,30,48], for example branch 0 for the ancestral branch leading from Y. pseudotuberculosis. I have now taken the liberty of arbitrarily assigning similar designations to ancient genotypes in figure 2a in order to facilitate the following summary.
Branch 0 begins with ancient genomes from the teeth of Bronze-age individuals who died up to 5000 years ago [31] (populations 0.PRE1, 0.PRE2). These are followed by a variety of extant populations found today in eastern and central Asia (0.PE7, 0.PE1-4, 0.ANT1) [16,30] and then by a genome from the Justinianic pandemic in Germany (0.ANT4) [32]. Branch 0 continues with extant populations from eastern and central Asia (0.ANT2-3), and culminates in a ‘big bang’ of explosive radiation [16], as witnessed by a polytomy that yielded extant branches 1, 2, 3 and 4. This radiation must have happened shortly before the Black Death in 1348, likely within decades, because the core genome from skeletons buried in 1348–1349 in London (1.PRE1) differ by only one informative SNP from that polytomous branch point. In turn, all subdivisions of populations within branches 1–4 and their transmissions to multiple geographic locations must have happened since the mid-fourteenth century.
(b). Patterns of global transmission since 1348
Little is yet known about branches 3 and 4, which encompass rare isolates from China and Mongolia. However, extant strains of branch 2 have been isolated from multiple geographically different sources, indicating extensive spread in the last 650 years since its origins. The 2.ANT3 population is common in northeastern China, and 2.MED3 in central China [16,30]. 2.ANT1 and 2.ANT3 were also found in China, and their geographical sources overlap with a branch of the Tea-Horse trade road connecting China with South Asia; 2.ANT1 has even been isolated in Nepal [16]. Similarly, 2.MED2 predominated along the southern route of the former Silk Roads. An even more dramatic correlation was found for 2.MED1, which was found in China along the northern route of the Silk Roads as well as throughout former Kurdistan. The Silk Roads followed oases through the deserts of central Asia, and passes through the high mountains in that region [49]. Finding Y. pestis along these former trade routes suggests that they might have also provided routes for the migrations of native rodents, and facilitated sequential plague infections between neighbouring rodent populations. Alternately, plague transmission may have been facilitated by human trade and migrations [16,30] or military manoeuvres, for example by the Mongols in the thirteenth century [50].
Our information on historical transmissions is greatest for branch 1. The reconstruction of ancient genomes have indicated that 1.PRE1 (figure 2a) caused the Black Death in London [33] as well as other medieval outbreaks in continental Europe [34,37]. A descendant of these bacteria, 1.PRE1B, which had accumulated 20 nucleotide substitutions (SNV), was isolated from Ellwangen, Germany (1485–1627) [34], and 1.PRE1A, a further descendant population with about 60 additional informative SNVs, caused plague in Marseille in 1722 [36], almost 400 years after the Black Death. Bacteria that are related to 1.PRE by low-resolution SNP genotyping were also isolated in Germany in the fourteenth and seventeenth centuries [35]. The 1.PRE1 set of populations forms a side-branch to branch 1. The direct descent along branch 1 is marked after one SNP by 1.PRE2. 1.PRE2 was isolated from London in the decades after the Black Death [36] and its close relatives, according to SNP typing, were isolated from plague deaths from the fourteenth century in The Netherlands [37]. 1.PRE3 designates a population that is one further SNP along branch 1; its genome was obtained from a mass grave in Bolgar City, Tatarstan, Russia (1362–1400) [34].
1.PRE3 is ancestral to the 1.ANT1–1.ANT3 populations, which have only been isolated in East Africa, followed along branch 1 by 1.IN1C–1.IN3, which have only been found in China [30,48]. In turn, 1.IN3 is the last known population that branched off before 1.ORI, which spread globally to cause the ongoing Third Pandemic from Hong Kong in 1894. The plague epidemic in Hong Kong is linked by epidemiological data from the nineteenth century to Yunnan province [51], which was the sole source of 1.IN3.
These data pose a conundrum, because the sequential SNPs along branch 1 can only have arisen in single cells, one at a time. The isolation of aDNA from 1.PRE1 populations from graves in Europe that differ in age by centuries is very suggestive that these bacteria established transmission chains in European rodents that lasted hundreds of years, and from whom waves of transmissions to humans occurred till the mid-eighteenth century. Historical records of repeated infections in remote Alpine villages suggest that one potential habitat for these rodents might have been in the Alps [52]. However, where did branch 1 itself continue to evolve? It has been suggested that a considerable part of that evolution, possibly for hundreds of years, was also in local reservoirs in Europe, with subsequent seeding of the 1.ANT populations in East and Central Africa followed by the 1.IN populations in China [32,34,36]. Alternately, that evolution might have continued in China and central Asia, with occasional epidemic sweeps that seeded Europe and East Africa [30]. The isolation of 1.PRE3 in the fourteenth century in Central Russia strongly supports this latter interpretation, but a definitive answer about the early epidemiological history of branch 1 must await further genomic studies on ancient DNA.
The third pandemic was unambiguously and exclusively caused by 1.ORI (figure 2d) [30,48]. Ten discrete subpopulations of ORI1–1.ORI3 are distinguished by informative SNPs that were fixed by bottlenecks during the spread from Hong Kong to other areas, including 1.ORI1 (Hawaii and continental USA), 1.ORI3 (sequentially to India, Madagascar and Turkey) and 1.ORI sublineages ii–ix which followed still other routes (figure 2d). Each of these lineages was apparently established within a few years after 1894, and the descendants of those clades have persisted to the present day in Madagascar, North America and Eastern Asia. These results provide a paradigm for the sparser results found with aDNA after the Black Death, which also showed rapid diversification during a pandemic. Rapid diversification may be caused by a combination of multiple bottlenecks, geographical isolation, large population sizes and an effectively high mutation rate per year owing to the demography of explosive transmission chains [16,30].
Our understanding of the detailed patterns of global transmissions of plague has increased very dramatically in the last 20 years owing to population genetic analyses. Bronze-age genomes of this pathogen were found in both Poland and Russia [31], and each of the last three pandemics has swept across large parts of Eurasia. Thus, plague caused by Y. pestis has swept across Asia on multiple occasions over the last 5000 years, or even longer.
4. How long have humans been afflicted by tuberculosis?
Tuberculosis is one of the most common infectious diseases of humans, and each year, an estimated nine million individuals develop clinical tuberculosis, and two million die of the disease. Tuberculosis is caused by the Mycobacterium tuberculosis complex of bacteria (MTBC), which all descend from a recent, common ancestor [53]. Human infection usually results in latent disease without clinical symptoms; it is thought that one-third of the global human population has been infected at some point in their lives [54].
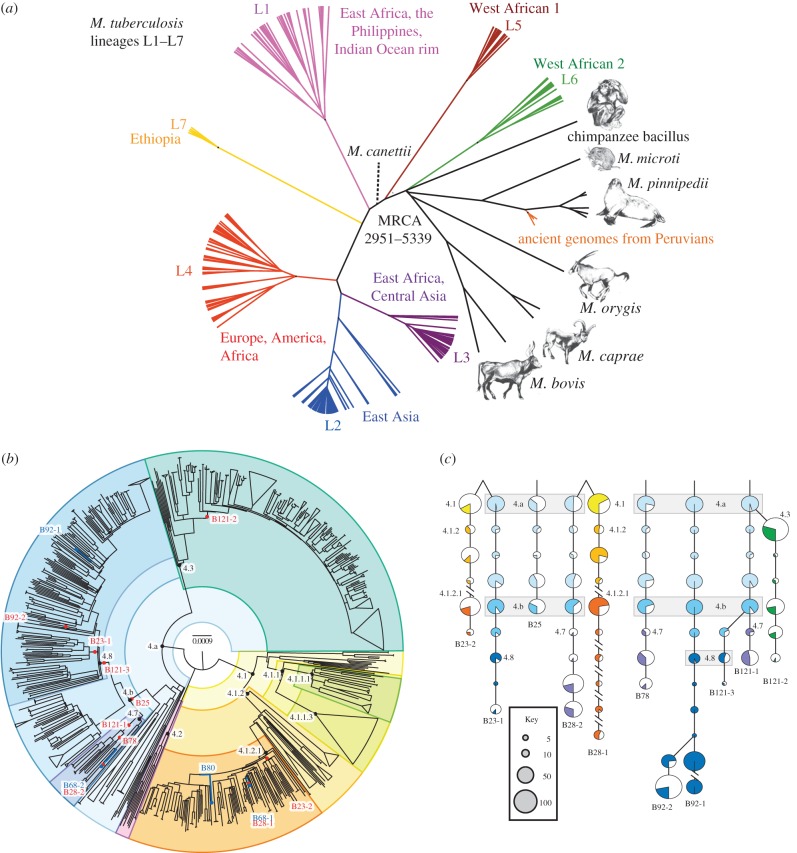
The MTBC consists of multiple genetic lineages (figure 3a) [57], some of which are specific for animals, but occasionally infect humans. Almost all of the human-specific lineages are found in Africa, and some (L5–L7) are only rarely isolated elsewhere. Similar to H. pylori, it has been proposed that the MTBC originated in Africa and first spread globally some 60 kyr ago during the out of Africa human migrations [57]. Lineages L2–L4 have shorter branches, and are interpreted as being ‘modern’, i.e. as having evolved after this migration, and then spread globally. The other lineages are interpreted as being ‘ancient’ (pre-60 kyr). In support of this interpretation, there is a statistically significant correlation (r2 = ∼0.3) in pairwise comparisons between the genetic and geographical distances between ‘ancient’ bacterial strains, and a weaker correlation (r2 = ∼0.1) for ‘modern’ strains [57]. Based on these observations, a single calibration date of 60 kyr for the out of Africa human migrations was recently used to estimate the tMRCA of the MTBC as 70 000 years ago [53]. However, none of the arguments summarized above are fully convincing, and this dating should be treated as a speculative hypothesis until it receives support from other sources.
Figure 3.
Estimated tMRCA of Mycobacterium tuberculosis. (a) Phylogenetic tree of genomes from lineages L1–L7 that infect humans, lineages that are associated with animal infections, and ancient genomes from Peruvians (modified from figure 3a from [55]). (b) Phylogenetic tree of 1582 genomes of lineage 4 of M. tuberculosis shows the location within the tree of 4 high coverage (blue lines and dots) and 10 low coverage genomes (red dots) from metagenomic sequences of mummified bodies in Hungary [56]. (c) Principles of phylogenetic placement with MGPlacer [56] for the mapping results in part (b). Polymorphisms that are identified by mapping short reads to a reference genome are traced down a pre-calculated phylogeny until no further SNPs match the branches leading to existing genotypes. The diameters of the pie charts reflect the number of known genotypes for each node, whereas the shaded slices indicate the proportion of characteristic SNPs within each pie that were found in the metagenomic reads. Parts (b) and (c) from figure 3 of [56].
The ancient history of M. tuberculosis has possibly been studied more intensively by palaeoarchaeologists than that of any other bacterial pathogen. The osteological signs of bone lesions that are specific to clinical tuberculosis have been found in multiple ancient skeletons. In some cases, PCR analyses of DNA extracts, coupled with limited sequencing and/or hybridization with CRISPR spacers, have provided presumptive genetic evidence that these lesions did arise from M. tuberculosis infections of humans and wild animals [58]. Those analyses dated human infections in Israel to 9000 years ago [59] and tuberculosis of wild bison in North America to 17 000 years ago [60]. It has been suggested that aDNA from Mycobacterium is less exposed to DNA damage than that of other bacterial taxa, because mycobacterial cell walls contain mycolic acid [61]. This explanation could explain an unusually high success rate for the PCR amplification of aDNA from M. tuberculosis. However, many older descriptions of aDNA do not satisfy current requirements for excluding environmental contamination [62], and a convincing case for age estimates of the MTBC of 17 000 years and older must await genome-based confirmation. Until now, I know only of two sites from which ancient genomes of M. tuberculosis have been elucidated.
The older of these two sites was near the coast of Peru, where three genomes were reconstructed from Peruvian Indians who died about 1000 years ago [55]. These genomes belonged to the MTBC, but surprisingly they were most closely related to M. pinnipedii, which infects seals (figure 3a). The authors suggest that ancient Peruvians were infected after eating seal meat, and the bacteria then made a successful host jump from seals to humans which resulted in interhuman transmissions. They further postulate the same bacteria became established among South American Indians and persisted for centuries until being replaced by ‘modern’ lineages from Europe and Africa after European colonization in 1492. Modern M. pinnipedii can also infect humans today, but those infections are not transmitted to other humans. It therefore remains possible that the Peruvian genomes represent human infections, but not interhuman transmissions, and genomic data from other ancient genomes of the MTBC in South America would be needed to substantiate a successful host jump from seals to humans.
The more recent of the two sites was a crypt in Hungary, in which corpses were interred in the eighteenth and early-nineteenth centuries, which was then bricked off from access until 1994 [56]. These bodies mummified spontaneously. Samples from 26 bodies revealed that eight of them contained MTBC, and allowed the reconstruction of a total of 14 genomes corresponding to 12 distinct genotypes [56]. (A mother and her daughter were each infected with the same two genotypes.) All genotypes belonged to lineage L4 (figure 3b), which is currently common throughout Europe. Because the MTBC is clonal, similar to Y. pestis, and homologous recombination is not known to occur, it was possible to identify the phylogenetic position of each genome, and to separate out distinct genomes from the bodies with dual or even triple infections using phylogenetic placement (MGPlacer, figure 3c).
Both studies [55,56] calculated a very similar molecular clock rate, approximately 5 × 10−8 (electronic supplementary material, table S1). If the clock rate is constant over longer time periods, then that calculation predicts a tMRCA of less than 6000 years for the whole MTBC. Because of lineage sorting and lineage extinction, tMRCAs need to be considered to represent only minimal estimates of the age of a taxon. Thus, it would not be terribly surprising if an archaeological sample that was older than 6000 years were to yield a genome of the MTBC, and a revised and older tMRCA. However, at the moment, the minimal age of the MTBC stands at approximately 5000 years, and the older estimates described above need substantiation by genomic data.
5. A vision for the future
I have summarized our current state of knowledge about the minimal age of three distinct bacterial pathogens: 100 kyr for H. pylori, less than 6 kyr for MTBC and more than 5 kyr for Y. pestis. All three estimates depended on a synthesis of very recent developments in understanding the population genetic structure of extant populations and in deciphering genomes from aDNA. Clearly, the combination of these approaches yielded synergies that supported exciting reconstructions of historical patterns of transmission and the evolutionary biology of the causes of gastritis, tuberculosis and plague. It can be anticipated that we will soon gain even more biological insights into these diseases, and hopefully also into other invasive diseases, especially if biologists and historians collaborate even more strongly.
Supplementary Material
Acknowledgements
The work I co-authored began at the Max Planck Institute for Molecular Genetics and then the Max Planck Institute for Infection Biology, Berlin, Germany. It continued at University College Cork, Ireland between 2007 and 2012, and at University of Warwick since then. I thank Monica Green for numerous suggestions, corrections, additional reading material and encouragement.
Competing interests
I declare I have no competing interests.
Funding
I received no funding for this study.
References
- 1.Holden MTG, et al. 2013. A genomic portrait of the emergence, evolution and global spread of a methicillin resistant Staphylococcus aureus pandemic. Genome Res 23, 653–664. ( 10.1101/gr.147710.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Z, et al. 2013. Neutral genomic microevolution of a recently emerged pathogen, Salmonella enterica serovar Agona. PLoS Genet. 9, e1003471 ( 10.1371/journal.pgen.1003471) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheppard SK, et al. 2013. Genome-wide association study identifies vitamin B5 biosynthesis as a host specificity factor in Campylobacter. Proc. Natl Acad. Sci. USA 110, 11 923–11 927. ( 10.1073/pnas.1305559110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chouikha I, Hinnebusch BJ. 2014. Silencing urease: a key evolutionary step that facilitated the adaptation of Yersinia pestis to the flea-borne transmission route. Proc. Natl Acad. Sci. USA 111, 18 709–18 714. ( 10.1073/pnas.1413209111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Z, McCann A, Weill FX, Blin C, Nair S, Wain J, Dougan G, Achtman M. 2014. Transient Darwinian selection in Salmonella enterica serovar Paratyphi A during 450 years of global spread of enteric fever. Proc. Natl Acad. Sci. USA 111, 12 199–12 204. ( 10.1073/pnas.1411012111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roumagnac P, et al. 2006. Evolutionary history of Salmonella typhi. Science 314, 1301–1304. ( 10.1126/science.1134933) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu P, et al. 2001. Fit genotypes and escape variants of subgroup III Neisseria meningitidis during three pandemics of epidemic meningitis. Proc. Natl Acad. Sci. USA 98, 5234–5239. ( 10.1073/pnas.061386098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ochman H, Wilson AC. 1987. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J. Mol. Evol. 26, 74–86. ( 10.1007/BF02111283) [DOI] [PubMed] [Google Scholar]
- 9.Wirth T, Morelli G, Kusecek B, Van Belkum A, van der Schee C, Meyer A, Achtman M. 2007. The rise and spread of a new pathogen: seroresistant Moraxella catarrhalis. Genome Res. 17, 1647–1656. ( 10.1101/gr.6122607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leopold SR, et al. 2009. A precise reconstruction of the emergence and constrained radiations of Escherichia coli O157 portrayed by backbone concatenomic analysis. Proc. Natl Acad. Sci. USA 106, 8713–8718. ( 10.1073/pnas.0812949106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ochman H, Elwyn S, Moran NA. 1999. Calibrating bacterial evolution. Proc. Natl Acad. Sci. USA 96, 12 638–12 643. ( 10.1073/pnas.96.22.12638) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morelli G, Didelot X, Kusecek B, Schwarz S, Falush D, Bahlawane C, Suerbaum S, Achtman M. 2010. Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families. PLoS Genet. 6, e1001036 ( 10.1371/journal.pgen.1001036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho SY, Shapiro B, Phillips MJ, Cooper A, Drummond AJ. 2007. Evidence for time dependency of molecular rate estimates. Syst. Biol. 56, 515–522. ( 10.1080/10635150701435401) [DOI] [PubMed] [Google Scholar]
- 14.Linz B, et al. 2014. A mutation burst during the acute phase of Helicobacter pylori infection in humans and rhesus macaques. Nat. Commun. 5, 4165 ( 10.1038/ncomms5165) [DOI] [PubMed] [Google Scholar]
- 15.Didelot X, Nell S, Yang I, Woltemate S, van der Merwe S, Suerbaum S. 2013. Genomic evolution and transmission of Helicobacter pylori in two South African families. Proc. Natl Acad. Sci. USA 110, 13 880–13 885. ( 10.1073/pnas.1304681110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui Y, et al. 2013. Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc. Natl Acad. Sci. USA 110, 577–582. ( 10.1073/pnas.1205750110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suerbaum S, Josenhans C. 2007. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat. Rev. Microbiol. 5, 441–452. ( 10.1038/nrmicro1658) [DOI] [PubMed] [Google Scholar]
- 18.Eppinger M, et al. 2006. Who ate whom? Adaptive Helicobacter genomic changes that accompanied a host jump from early humans to large felines. PLoS Genet. 2, e120 ( 10.1371/journal.pgen.0020120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Achtman M, et al. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32, 459–470. ( 10.1046/j.1365-2958.1999.01382.x) [DOI] [PubMed] [Google Scholar]
- 20.Moodley Y, et al. 2012. Age of the association between Helicobacter pylori and man. PLoS Pathog. 8, e1002693 ( 10.1371/journal.ppat.1002693) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nell S, et al. 2013. Recent acquisition of Helicobacter pylori by Baka pygmies. PLoS Genet. 9, e1003775 ( 10.1371/journal.pgen.1003775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moodley Y, et al. 2009. The peopling of the Pacific from a bacterial perspective. Science 323, 527–530. ( 10.1126/science.1166083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falush D, et al. 2003. Traces of human migrations in Helicobacter pylori populations. Science 299, 1582–1585. ( 10.1126/science.1080857) [DOI] [PubMed] [Google Scholar]
- 24.Wirth T, Wang X, Linz B, Novick RP, Lum JK, Blaser M, Morelli G, Falush D, Achtman M. 2004. Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: lessons from Ladakh. Proc. Natl Acad. Sci. USA 101, 4746–4751. ( 10.1073/pnas.0306629101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linz B, et al. 2007. An African origin for the intimate association between humans and Helicobacter pylori. Nature 445, 915–918. ( 10.1038/nature05562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prugnolle F, Manica A, Balloux F. 2005. Geography predicts neutral genetic diversity of human populations. Curr. Biol. 15, R159–R160. ( 10.1016/j.cub.2005.02.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maixner F, et al. 2016. The 5300-year-old Helicobacter pylori genome of the iceman. Science 351, 162–165. ( 10.1126/science.aad2545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cassidy LM, Martiniano R, Murphy EM, Teasdale MD, Mallory J, Hartwell B, Bradley DG. 2016. Neolithic and Bronze Age migration to Ireland and establishment of the insular Atlantic genome. Proc. Natl Acad. Sci. USA 113, 368–373. ( 10.1073/pnas.1518445113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollitzer R. 1951. Plague studies. 1. A summary of the history and survey of the present distribution of the disease. Bull. World Health Org. 4, 475–533. [PMC free article] [PubMed] [Google Scholar]
- 30.Morelli G, et al. 2010. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat. Genet. 42, 1140–1143. ( 10.1038/ng.705) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasmussen S, et al. 2015. Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago. Cell 163, 571–582. ( 10.1016/j.cell.2015.10.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner DM, et al. 2014. Yersinia pestis and the plague of Justinian 541–543 AD: a genomic analysis. Lancet Infect. Dis. 14, 319–326. ( 10.1016/S1473-3099(13)70323-2) [DOI] [PubMed] [Google Scholar]
- 33.Bos KI, et al. 2011. A draft genome of Yersinia pestis from victims of the Black Death. Nature 478, 506–510. ( 10.1038/nature10549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spyrou MA, et al. 2016. Historical Y. pestis genomes reveal the European black death as the source of ancient and modern plague pandemics. Cell Host Microbe 19, 874–881. ( 10.1016/j.chom.2016.05.012) [DOI] [PubMed] [Google Scholar]
- 35.Seifert L, Wiechmann I, Harbeck M, Thomas A, Grupe G, Projahn M, Scholz HC, Riehm JM. 2016. Genotyping Yersinia pestis in historical plague: evidence for long-term persistence of Y. pestis in Europe from the 14th to the 17th century. PLoS ONE 11, e0145194 ( 10.1371/journal.pone.0145194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bos KI, et al. 2016. Eighteenth century Yersinia pestis genomes reveal the long-term persistence of an historical plague focus. Elife 5, e12994 ( 10.7554/eLife.12994) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haensch S, et al. 2010. Distinct clones of Yersinia pestis caused the Black Death. PLoS Pathog. 6, e1001134 ( 10.1371/journal.ppat.1001134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Little LK. 2007. Plague and the end of antiquity. The pandemic of 541–750. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 39.Cohn SK., Jr 2002. The Black Death transformed: disease and culture in early renaissance Europe. London, UK: Arnold. [PubMed] [Google Scholar]
- 40.Little LK. 2011. Review article: plague historians in lab coats. Past & Present 213, 267–290. ( 10.1093/pastj/gtr014) [DOI] [PubMed] [Google Scholar]
- 41.Green MH. 2014. Pandemic disease in the medieval world: rethinking the black death. Kalamazoo, MI: Medieval Institute Publications. See http://scholarworks.wmich.edu/medieval_globe/1/. [Google Scholar]
- 42.Stenseth NC, Atshabar BB, Begon M, Belmain SR, Bertherat E, Carniel E, Gage KL, Leirs H, Rahalison L. 2008. Plague: past, present, and future. PLoS Med. 5, e3 ( 10.1371/journal.pmed.0050003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torrea G, Chenal-Francisque V, Leclercq A, Carniel E. 2006. Efficient tracing of global isolates of Yersinia pestis by restriction fragment length polymorphism analysis using three insertion sequences as probes. J. Clin. Microbiol. 44, 2084–2092. ( 10.1128/JCM.02618-05) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vogler AJ, et al. 2013. A decade of plague in Mahajanga, Madagascar: insights into the global maritime spread of pandemic plague. MBio 4, e00623-12. ( 10.1128/mBio.00623-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riehm JM, et al. 2012. Yersinia pestis lineages in Mongolia. PLoS ONE 7, e30624 ( 10.1371/journal.pone.0030624) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Achtman M. 2012. Insights from genomic comparisons of genetically monomorphic bacterial pathogens. Phil. Trans. R. Soc. B 367, 860–867. ( 10.1098/rstb.2011.0303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl Acad. Sci. USA 96, 14 043–14 048. ( 10.1073/pnas.96.24.14043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Achtman M, et al. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl Acad. Sci. USA 101, 17 837–17 842. ( 10.1073/pnas.0408026101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen V. 2012. The silk road: a new history. Oxford, UK: Oxford University Press. [Google Scholar]
- 50.Hymes R. 2014. Epilogue: a hypothesis on the East Asian beginnings of the Yersinia pestis polytomy. In Pandemic disease in the medieval world: rethinking the Black Death (ed. Green M.), pp. 285–308. Kalamazoo, MI: Medieval Institute Publications. See http://scholarworks.wmich.edu/medieval_globe/1/. [Google Scholar]
- 51.Xu L, et al. 2014. Wet climate and transportation routes accelerate spread of human plague. Proc. R. Soc. B 281, 20133159 ( 10.1098/rspb.2013.3159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carmichael AG. 2014. Plague persistence in Western Europe: a hypothesis. In Pandemic disease in the medieval world: rethinking the Black Death (ed. Green M.), pp. 157–191. Kalamazoo, MI: Medieval Institute Publications. See http://scholarworks.wmich.edu/medieval_globe/1/. [Google Scholar]
- 53.Comas I, et al. 2013. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet. 45, 1176–1182. ( 10.1038/ng.2744) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barry CE III, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, Schnappinger D, Wilkinson RJ, Young D. 2009. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 7, 845–855. ( 10.1038/nrmicro2236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bos KI, et al. 2014. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature 514, 494–497. ( 10.1038/nature13591) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kay GL, et al. 2015. Eighteenth-century genomes show that mixed infections were common at time of peak tuberculosis in Europe. Nat. Commun. 6, 6717 ( 10.1038/ncomms7717) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hershberg R, et al. 2008. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 6, e311 ( 10.1371/journal.pbio.0060311) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hershkovitz I, et al. 2015. Tuberculosis origin: the Neolithic scenario. Tuberculosis (Edinb.) 95(Suppl 1), S122–S126. ( 10.1016/j.tube.2015.02.021) [DOI] [PubMed] [Google Scholar]
- 59.Spigelman M, et al. 2015. Evolutionary changes in the genome of Mycobacterium tuberculosis and the human genome from 9000 years BP until modern times. Tuberculosis (Edinb.) 95(Suppl 1), S145–S149. ( 10.1016/j.tube.2015.02.022) [DOI] [PubMed] [Google Scholar]
- 60.Rothschild BM, Martin LD, Lev G, Bercovier H, Bar-Gal GK, Greenblatt C, Donoghue H, Spigelman M, Brittain D. 2001. Mycobacterium tuberculosis complex DNA from an extinct bison dated 17,000 years before the present. Clin. Infect. Dis. 33, 305–311. ( 10.1086/321886) [DOI] [PubMed] [Google Scholar]
- 61.Schuenemann VJ, et al. 2013. Genome-wide comparison of medieval and modern Mycobacterium leprae. Science 341, 179–183. ( 10.1126/science.1238286) [DOI] [PubMed] [Google Scholar]
- 62.Gilbert MT, Bandelt HJ, Hofreiter M, Barnes I. 2005. Assessing ancient DNA studies. Trends Ecol. Evol. 20, 541–544. ( 10.1016/j.tree.2005.07.005) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.