Abstract
Migration of hosts and parasites can have a profound impact on host–parasite ecological and evolutionary interactions. Using the bacterium Pseudomonas aeruginosa UCBPP-PA14 and its phage DMS3vir, we here show that immigration of naive hosts into coevolving populations of hosts and parasites can influence the mechanistic basis underlying host defence evolution. Specifically, we found that at high levels of bacterial immigration, bacteria switched from clustered regularly interspaced short palindromic repeats (CRISPR-Cas) to surface modification-mediated defence. This effect emerges from an increase in the force of infection, which tips the balance from CRISPR to surface modification-based defence owing to the induced and fixed fitness costs associated with these mechanisms, respectively.
Keywords: clustered regularly interspaced short palindromic repeats, phage, migration, induced defence, constitutive defence
1. Introduction
It is becoming increasingly clear that migration of hosts and parasites in spatially structured environments has important epidemiological and coevolutionary consequences [1,2], affecting, for example, host–parasite coexistence, local adaptation and coevolutionary dynamics [3–5]. Migration may also affect the type of host defence that is favoured by selection [6,7]. One reason for this is that host migration may affect the temporal variation in exposure to parasites, as well as the mean force of infection (the frequency at which infections take place), both of which are predicted to influence the relative benefits of defences that are associated with fixed versus parasite-dependent (induced) fitness costs [8–10]. This is because induced fitness costs increase as infection frequencies go up, while fixed costs per definition remain the same [8–10].
In response to phage infection, the bacterium Pseudomonas aeruginosa strain UCBPP-PA14 can evolve either surface modification (sm)- or CRISPR-Cas (clustered regularly interspaced short palindromic repeats; CRISPR-associated)-mediated defence [10]. We have previously shown that sm through the loss of the phage receptor is associated with a fixed fitness cost [10–12], whereas CRISPR-Cas-mediated defence is associated with an induced fitness cost [10,13]. The induced fitness cost of P. aeruginosa CRISPR-Cas may either be because the system is induced upon infection, as is the case in other species [14–16], or because of phage-induced damage to the host before it is eliminated. Regardless of the underlying mechanism, the way in which the fitness costs and benefits are manifested has profound impact on the evolution of CRISPR-Cas defence, as the level of phage exposure can tip the balance from CRISPR-Cas to sm [10].
The mechanism of CRISPR-Cas has been well studied over the past decade. CRISPR-Cas is an adaptive immune system of prokaryotes that integrates sequences from phage and other mobile genetic elements into CRISPR loci on the host genome ([17], reviewed in [18,19]). The P. aeruginosa UCBPP-PA14 Type IF CRISPR-Cas system consists of a single cas operon that is flanked by two CRISPR loci (CRISPR1 and CRISPR2), both of which rapidly acquire novel sequences in response to infection by its phage DMS3vir [10]. CRISPR loci are transcribed and processed into short crRNA (CRISPR RNA) molecules, which associate with Cas proteins to form a surveillance immune complex [20]. Upon re-infection, crRNA guides Cas proteins to bind and cleave complementary phage DNA, resulting in phage resistance ([21], reviewed in [22,23]).
Recently, we demonstrated that evolution of CRISPR-Cas-mediated immunity in P. aeruginosa results in rapid phage extinction [24]. In this study, we examine the impact of naive host immigration to a population of P. aeruginosa that use CRISPR-Cas to protect against phage infection. We demonstrate that high immigration levels prevent phage extinction, thus increasing the mean force of infection. As a consequence, hosts switch from CRISPR-mediated defence to sm-mediated defence at increasing immigration levels.
2. Material and methods
(a). Bacterial strains and phages
The bacterial strains used in this study are P. aeruginosa UCBPP-PA14 (wild-type, WT), UCBPP-PA14 csy3::LacZ (CRISPR-Cas KO) and the CRISPR-Cas KO-derived surface mutant sm1, which have been described previously [10,25]. The CRISPR-Cas KO strain has a non-functional CRISPR-Cas system due to disruption of the essential csy3 gene. The sm1 strain is deficient for production of the pilus, which is the receptor for phage DMS3vir. In addition, BIM-4sp was used, which is a WT-derived strain that acquired four spacers against phage DMS3vir. Phage DMS3vir is a previously described mutant of phage DMS3 that cannot enter the lysogenic life cycle owing to mutation of the repressor gene [10,25].
(b). Evolution experiments
Evolution experiments were performed in six replicates by inoculating 6 ml M9 growth medium (22 mM Na2HPO4; 22 mM KH2PO4; 8.6 mM NaCl; 20 mM NH4Cl; 1 mM MgSO4; 0.1 mM CaCl2) supplemented with 0.2% glucose with approximately 106 colony forming units (cfu) of WT or CRISPR-Cas KO strains from fresh overnight cultures and adding 104 plaque forming units (pfu) of DMS3vir, followed by incubation at 37°C while shaking at 180 r.p.m. Cultures were transferred daily 1 : 100 into fresh growth medium. Migration was performed as follows: WT bacteria were grown in M9, as above, and transferred daily in parallel to the phage-infected cultures. During each daily transfer these uninfected WT bacteria were mixed with the co-evolving bacteria-phage community by adding 10 µl uninfected WT bacterial culture (corresponding to approx. 106 cfu) to 990 µl co-evolving culture (1%; low immigration) or 100 µl WT bacterial culture (corresponding to approximately 107 cfu) to 900 µl co-evolving culture (10%; high immigration). From these mixtures, 60 µl was transferred to 6 ml fresh M9 growth medium (1 : 100 transfer). Bacterial immunity against the ancestral phage was determined at the time points indicated by streaking individual clones (typically either 16 or 24 individual clones per replicate) through ancestral phage on Luria-Bertani (LB) agar. Resistance was scored as the ability to grow through the phage. To confirm CRISPR-Cas-mediated immunity, the same clones were also streaked through phage carrying an anti-CRISPR gene (DMS3mvir + acrF1 [26]; bacterial clones with CRISPR-Cas-mediated immunity are sensitive to this phage) and to monitor acquisition of novel spacers, PCRs were performed using primers CTAAGCCTTGTACGAAGTCTC and CGCCGAAGGCCAGCGCGCCGGTG (for CRISPR 1) and primers GCCGTCCAGAAGTCACCACCCG and TCAGCAAGTTACGAGACCTCG (for CRISPR 2). Surface modification was confirmed based on all of the following traits: (i) a distinct, smooth, colony morphology, (ii) broad-range immunity to both DMS3vir and DMS3mvir + acrF1, and (iii) an absence of newly acquired CRISPR spacers.
(c). Competition experiments
Competition experiments were performed in glass microcosms in a total volume of 6 ml M9 growth medium supplemented with 0.2% glucose. Competition experiments were initiated by inoculating with approximately 106 cfu (i.e. 1 : 100 from a 1 : 1 mixture of overnight cultures that were spun down and resuspended in the same volume of M9 salts) of sm1 and BIM-4sp. Phage was added at the start of the experiment using 104 pfu. Cultures were transferred daily into fresh M9 medium, and immigration was performed as described above. Cells were serially diluted in M9 salts and plated at 5 days post-infection (dpi) on LB agar supplemented with 50 µg ml−1 X-gal. Each competition experiment was performed in 12 replicates.
(d). Time-shift analysis
From the coevolution experiment with 10% daily immigration, 24 individual bacterial clones were isolated from each replicate experiment from time point 3 and 11 dpi. Phage was isolated by chloroform extraction from 3 and 11 dpi and subjected to a single round of amplification on the P. aeruginosa csy3::lacZ (CRISPR-Cas KO) strain. This amplification was performed by infecting CRISPR-KO bacteria (108 cfu) with phage (103 pfu) in a 0.5% agar overlay in a square 14 × 14 cm polypropylene Petri dish. This plate was incubated overnight at 37°C, and the following day phage was harvested from these plates by washing the plate with 10 ml M9 salt buffer and performing a chloroform extraction. Bacterial clones were grown overnight in LB at 37°C and streaked through lines of phage on LB agar (25 µl phage at 108 pfu ml−1 for a single streak on a square 14 × 14 cm polypropylene Petri dish). Resistance was scored as the ability to grow through the phage.
(e). Statistical analyses
In the evolution experiments, the effect of immigration treatment and time on the angular transformed proportion of surface modification colonies was analysed as an REML mixed model, with replicate fitted as a random effect. To examine whether host immigration increased phage persistence, a one-tailed Fisher's test was performed. To examine how the various treatments impact fitness associated with CRISPR in the competition experiments, the relative frequencies of the CRISPR-resistant BIM-4sp strain and sm1 strain were determined for the time points indicated and used to calculate the relative fitness (rel. fitness = [(fraction strain A at t = x) × (1 − (fraction strain A at t = 0))]/[(fraction strain A at t = 0) × (1 − (fraction strain A at t = x)]). Relative fitness values were used for Student's t-test, ANOVA if data were normal distributed or Kruskal–Wallis test if residuals were not normal distributed. For t-test and ANOVA, data were transformed as appropriate to normalize residuals. All subsequent statistical analyses were carried out using JMP (v. 10) software. In the time-shift experiments, the effect of phage time point and bacteria time point and their interaction on angular transformed proportion of resistant colonies was analysed using a linear mixed model, with replicate fitted as a random effect, using JMP (v. 12) software. Effects were considered significant at p < 0.05.
3. Results
We have previously shown that P. aeruginosa strain UCBPP-PA14 rapidly drives phage DMS3vir extinct following the evolution of CRISPR-Cas-mediated immunity [24]. While parasites and hosts often go extinct locally, it is frequently observed that they can persist in structured environments where hosts and parasites migrate between subpopulations [3]. We, therefore, tested whether naive host immigration would allow for phage persistence in this experimental system and examined whether phage persistence would influence the type of evolved defence and coevolutionary interaction with the virus.
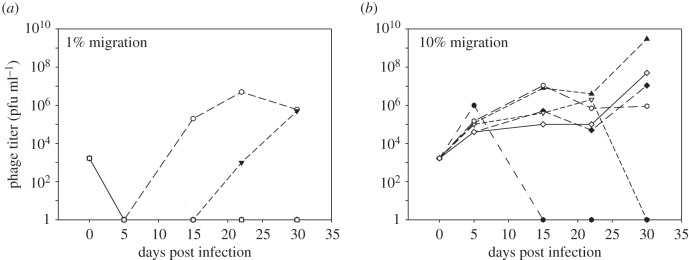
To this end, we performed long-term co-culture experiments between the WT strain and phage DMS3vir, with immigration of WT bacteria from a phage-free environment at each daily transfer. We included two migration treatments: low immigration of bacteria from a phage-free environment (1% migrants at each transfer), and high immigration from a phage-free environment (10% migrants at each transfer). In the low-migration treatment, phage persisted in two out of six replicate experiments (figure 1a), and at high immigration (10% immigration), phage persisted in four out of six replicates (figure 1b). In the parallel experiment that lacked naive host immigration, phage went extinct in all replicates [24]. Hence, high naive host immigration significantly reduced phage extinction (one-tailed Fisher's exact test, p = 0.03), causing an increase in the force of infection.
Figure 1.
Phage persistence during long-term coevolution between phage DMS3vir and WT bacteria at low or high immigration of sensitive hosts. Phage titers over time upon infection of WT bacteria with phage DMS3vir and (a) low (1%) immigration (n = 6) or (b) high (10%) immigration of sensitive WT bacteria at each transfer (n = 6).
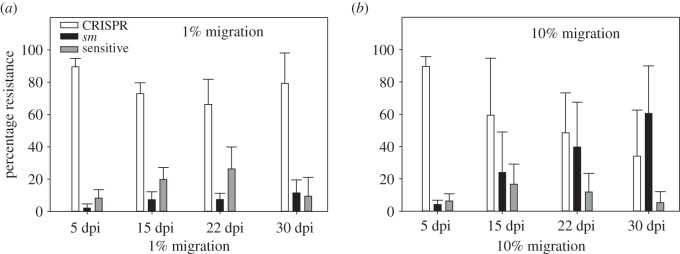
Next, we examined the impact of naive host immigration on the evolution of P. aeruginosa immune mechanisms. Pseudomonas aeruginosa can evolve phage resistance using two distinct immune mechanisms: CRISPR-Cas or surface modification. At both low and high levels of immigration, bacteria initially evolved almost exclusively CRISPR-Cas-mediated phage resistance (figure 2). While an increase in sm-mediated immunity was observed over time, this increase was much more rapid under high levels of immigration (figure 2; F1,34 = 12.9, p = 0.001). Collectively, these data show that immigration of naive hosts leads to an increase in the evolution of sm immunity.
Figure 2.
Types of evolved immunity during long-term coevolution between phage DMS3vir and WT bacteria at low or high immigration. (a) Immunity profile at low immigration (n = 6). (b) Immunity profile at high immigration (n = 6). Error bars correspond to 95% confidence intervals (CI).
We envisaged two, not mutually exclusive, reasons why immigration may cause selection against CRISPR-Cas-mediated immunity: (i) sm-mediated phage resistance is favoured at high immigration because of the increase in phage exposure, and (ii) phage can evolve to overcome CRISPR-Cas-mediated resistance because of the increased evolutionary potential associated with immigration of phage-sensitive bacteria.
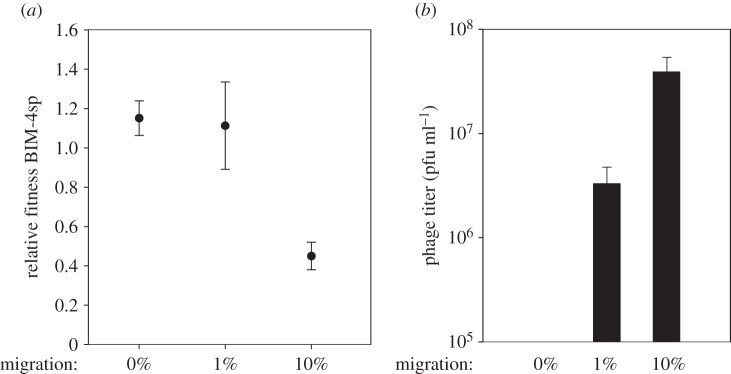
To test the first possibility, we performed competition experiments between a CRISPR-Cas-resistant clone (BIM-4sp) and a previously described surface modification-resistant clone (sm1) [10]. BIM-4sp carries multiple spacers that target phage DMS3vir and the phage can, therefore, not evolve to overcome CRISPR immunity [24,27], hence excluding the second, coevolutionary explanation as a confounding factor during this experiment. Competition was performed in the presence of phage DMS3vir and in the presence or absence of naive host immigration. In accordance with the experiments described above, an increase in immigration of phage-sensitive WT bacteria was associated with a decrease in the fitness of BIM-4sp relative to sm1 (F2,33 = 29, p < 0.0001; figure 3a). As expected, CRISPR escape phage could not be detected, but importantly, phage could persist during naive host immigration, which indicates that escape phage is not required for phage persistence under these conditions (figure 3b). This suggests that immigration of susceptible hosts selects against CRISPR immunity, by allowing higher phage densities and thus increasing the force of infection, which is in agreement with a previous study which shows that fitness of CRISPR-resistant bacteria decreases relative to surface mutants when the frequency of infection increases [10].
Figure 3.
Fitness of CRISPR-Cas- and sm-mediated defences at different levels of immigration. (a) Relative fitness of BIM-4sp (four spacers against DMS3vir) at different levels of immigration at 5 days post infection. (b) Total phage (pfu ml−1) at different levels of immigration. In the absence of migration, no phage could be detected. CRISPR escape phage was absent in all treatments. Error bars correspond to 95% CI.
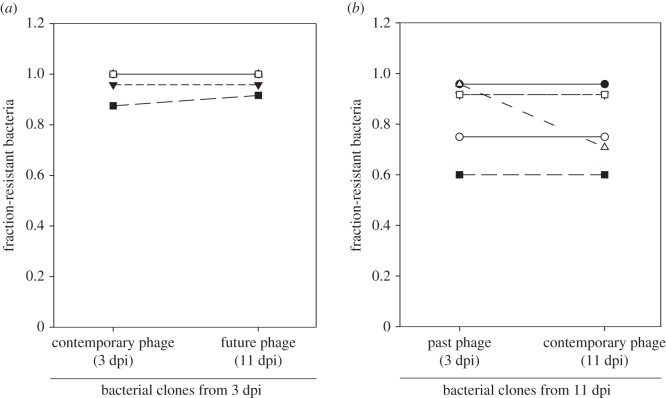
To test the second possibility, we performed time-shift experiments [28] to determine whether significant amounts of coevolution occurred during our first experiment. We isolated phage and individual bacterial clones from time points 3 and 11 dpi of the evolution experiment with high immigration. If phage evolved to overcome CRISPR-Cas-mediated resistance under high immigration, bacterial clones from 3 dpi would be expected to be more resistant to phage from 3 dpi compared with phage from 11 dpi. However, we detected no difference in resistance of individual clones from 3 dpi against the contemporary or future phage population (figure 4a), and bacteria from 11 dpi were equally resistant to phage from 3 and 11 dpi (figure 4b) (F1,19 = 0.1312, p = 0.7212). The lack of detectable coevolution is presumably because of the high level of CRISPR spacer diversity that naturally evolves in P. aeruginosa upon infection with DMS3vir, which has previously been shown to constrain the evolution of escape phage in the absence of immigration [24].
Figure 4.
Time-shift assays reveal no detectable coevolution at high immigration. (a) Resistance of bacterial clones from 3 dpi against contemporary and future (11 dpi) phage populations. (b) Resistance of bacterial clones from 11 dpi against contemporary and past (3 dpi) phage populations. Each symbol corresponds to an individual replicate experiment. Error bars correspond to 95% CI.
4. Discussion
Bacteria have a range of immune mechanisms to protect against their phages and other parasites [29,30], but it is unclear why this diversity in immune strategies evolved in the first place [31]. One hypothesis, which is supported by theory and data, is that different immune strategies are favoured under different ecological conditions. For example, previous studies have shown that spatial structure and migration can impact the evolution of altruistic immune strategies, such as abortive infection (Abi) in bacteria [32–34], as they are subject to kin selection and their benefit, therefore, depends on spatial structure [35]. The evolution of Abi is, therefore, constrained by ecological factors that impact relatedness. Other bacterial immune systems may also be favoured under different ecological conditions, but the way ecology drives the evolution of distinct immune strategies is largely unknown.
In this study, we have examined how ecology can impact the evolution of CRISPR-Cas adaptive immunity in bacteria. We and others have previously shown that the CRISPR-Cas adaptive immune system of bacteria is associated with an induced cost of resistance [10,13]. This was observed both in P. aeruginosa strain UCBPP-PA14 and in Streptococcus thermophilus strain DGCC7710 [10,13], although in the latter species a constitutive cost of carrying the system was also detected [13]. The induced cost of CRISPR may be because of toxic effects from increased Cas protein production, including autoimmunity [36–38], or because of phage-induced damage to the host before the phage is destroyed, or both. Even though we did not find any evidence for this previously [10], we also cannot exclude the possibility that CRISPR-Cas systems can be overwhelmed at high frequencies of infection. Understanding the mechanistic basis underlying the observed induced cost of CRISPR-Cas-mediated defence will require further investigation.
Regardless of the underlying mechanism, the way fitness costs are manifested has clear implications for the conditions under which they are favoured [8,10,39]. As the induced cost of CRISPR resistance increases with increasing infection rates, sm, which is associated with a fixed cost, will be favoured at a high force of infection [10]. This effect can emerge in different ways, such as through increased population densities of hosts and parasites as a result of increased resource availability [10]. In addition, as we show here, the same effect can emerge from immigration of naive hosts into subpopulations that contain virus. In the absence of such immigration, phage is driven to extinction rapidly when the bacterial population evolves CRISPR immunity with high spacer diversity [24]. The rapid reduction in the force of infection thus provides a clear benefit to CRISPR clones over surface mutants, which pay a fixed cost of resistance even after the phage has gone extinct. In the presence of high levels of naive host immigration, however, surface mutants are favoured over CRISPR-resistant clones because of the increase in the infection risk. This study on the crucial role of spatial structure on defence evolution may help to explain the observed variation in bacterial defence mechanisms in natural populations [40,41].
Data accessibility
Raw data files available on Dryad: http://dx.doi.org/10.5061/dryad.k7cr5.
Authors' contributions
A.B. and E.R.W. designed the study. H.C. and S.v.H. carried out the experiments. H.C., S.v.H., N.M.H.-K., A.B. and E.R.W. interpreted the data. E.R.W. wrote the manuscript.
Competing interests
We declare we have no competing interests.
Funding
H.C. was funded by the European Union (Erasmus + program), the CROUS (French State) and the Région Rhône Alpes (Explora'Sup program). S.v.H. received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement no. 660039. N.M.H.-K. received funding from the Danish Council for Independent Research, Postdoctoral Research Fellowship DFF-4090-00265. We also acknowledge NERC, BBSRC, Wellcome Trust, the Royal Society and the AXA research funds for funding.
References
- 1.Lion S, Gandon S. 2015. Evolution of spatially structured host-parasite interactions. J. Evol. Biol. 28, 10–28. ( 10.1111/jeb.12551) [DOI] [PubMed] [Google Scholar]
- 2.Brockhurst MA, Buckling A, Poullain V, Hochberg ME. 2007. The impact of migration from parasite-free patches on antagonistic host-parasite coevolution. Evolution 61, 1238–1243. ( 10.1111/j.1558-5646.2007.00087.x) [DOI] [PubMed] [Google Scholar]
- 3.Holyoak M, Lawler SP. 1996. Persistence of an extinction-prone predator-prey interaction through metapopulation dynamics. Ecology 77, 1867–1879. ( 10.2307/2265790) [DOI] [Google Scholar]
- 4.Morgan AD, Gandon S, Buckling A. 2005. The effect of migration on local adaptation in a coevolving host-parasite system. Nature 437, 253–256. ( 10.1038/nature03913) [DOI] [PubMed] [Google Scholar]
- 5.Schrag SJ, Mittler JE. 1996. Host-parasite coexistence: the role of spatial refuges in stabilizing bacteria-phage interactions. Am. Nat. 148, 348–377. ( 10.1086/285929) [DOI] [Google Scholar]
- 6.Cipollini D, Mbagwu J, Barto K, Hillstrom C, Enright S. 2005. Expression of constitutive and inducible chemical defenses in native and invasive populations of Alliaria petiolata. J. Chem. Ecol. 31, 1255–1267. ( 10.1007/s10886-005-5284-3) [DOI] [PubMed] [Google Scholar]
- 7.Beaton LL, Van Zandt PA, Esselman EJ, Knight TM. 2011. Comparison of the herbivore defense and competitive ability of ancestral and modern genotypes of an invasive plant, Lespedeza cuneata. Oikos 120, 1413–1419. ( 10.1111/j.1600-0706.2011.18893.x) [DOI] [Google Scholar]
- 8.Hamilton R, Siva-Jothy M, Boots M. 2008. Two arms are better than one: parasite variation leads to combined inducible and constitutive innate immune responses. Proc. R. Soc. B 275, 937–945. ( 10.1098/rspb.2007.1574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shudo E, Iwasa Y. 2001. Inducible defense against pathogens and parasites: optimal choice among multiple options. J. Theor. Biol. 209, 233–247. ( 10.1006/jtbi.2000.2259) [DOI] [PubMed] [Google Scholar]
- 10.Westra ER, et al. 2015. Parasite exposure drives selective evolution of constitutive versus inducible defense. Curr. Biol. 25, 1043–1049. ( 10.1016/j.cub.2015.01.065) [DOI] [PubMed] [Google Scholar]
- 11.Lenski RE. 1988. Experimental studies of pleiotropy and epistasis in Escherichia coli. I. Variation in competitive fitness among mutants resistant to virus-T4. Evolution 42, 425–432. ( 10.2307/2409028) [DOI] [PubMed] [Google Scholar]
- 12.Brockhurst MA, Buckling A, Rainey PB. 2005. The effect of a bacteriophage on diversification of the opportunistic bacterial pathogen, Pseudomonas aeruginosa. Proc. R. Soc. B 272, 1385–1391. ( 10.1098/rspb.2005.3086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vale PF, Lafforgue G, Gatchitch F, Gardan R, Moineau S, Gandon S. 2015. Costs of CRISPR-Cas-mediated resistance in Streptococcus thermophilus. Proc. R. Soc. B 282, 20151270 ( 10.1098/rspb.2015.1270) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agari Y, Sakamoto K, Tamakoshi M, Oshima T, Kuramitsu S, Shinkai A. 2010. Transcription profile of Thermus thermophilus CRISPR systems after phage infection. J. Mol. Biol. 395, 270–281. ( 10.1016/j.jmb.2009.10.057) [DOI] [PubMed] [Google Scholar]
- 15.Quax TE, et al. 2013. Massive activation of archaeal defense genes during viral infection. J. Virol. 87, 8419–8428. ( 10.1128/JVI.01020-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young JC, et al. 2012. Phage-induced expression of CRISPR-associated proteins is revealed by shotgun proteomics in Streptococcus thermophilus. PLoS ONE 7, e38077 ( 10.1371/journal.pone.0038077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. ( 10.1126/science.1138140) [DOI] [PubMed] [Google Scholar]
- 18.Amitai G, Sorek R. 2016. CRISPR-Cas adaptation: insights into the mechanism of action. Nat. Rev. Microbiol. 14, 67–76. ( 10.1038/nrmicro.2015.14) [DOI] [PubMed] [Google Scholar]
- 19.Sternberg SH, Richter H, Charpentier E, Qimron U. 2016. Adaptation in CRISPR-Cas Systems. Mol. Cell 61, 797–808. ( 10.1016/j.molcel.2016.01.030) [DOI] [PubMed] [Google Scholar]
- 20.Brouns SJ, et al. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964. ( 10.1126/science.1159689) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garneau JE, et al. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71. ( 10.1038/nature09523) [DOI] [PubMed] [Google Scholar]
- 22.van der Oost J, Westra ER, Jackson RN, Wiedenheft B. 2014. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 12, 479–492. ( 10.1038/nrmicro3279) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marraffini LA. 2015. CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61. ( 10.1038/nature15386) [DOI] [PubMed] [Google Scholar]
- 24.van Houte S, et al. 2016. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532, 385–388. ( 10.1038/nature17436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O'Toole GA. 2012. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J. Bacteriol. 194, 5728–5738. ( 10.1128/JB.01184-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. 2013. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432. ( 10.1038/nature11723) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin BR, Moineau S, Bushman M, Barrangou R. 2013. The population and evolutionary dynamics of phage and bacteria with CRISPR-mediated immunity. PLoS Genet. 9, e1003312 ( 10.1371/journal.pgen.1003312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gandon S, Buckling A, Decaestecker E, Day T. 2008. Host-parasite coevolution and patterns of adaptation across time and space. J. Evol. Biol. 21, 1861–1866. ( 10.1111/j.1420-9101.2008.01598.x) [DOI] [PubMed] [Google Scholar]
- 29.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327. ( 10.1038/nrmicro2315) [DOI] [PubMed] [Google Scholar]
- 30.Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. 2012. The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 46, 311–339. ( 10.1146/annurev-genet-110711-155447) [DOI] [PubMed] [Google Scholar]
- 31.van Houte S, Buckling A, Westra ER. 2016. Evolutionary ecology of prokaryotic immune mechanisms. Microbiol. Mol. Biol. Rev. 80, 745–763. ( 10.1128/MMBR.00011-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berngruber TW, Lion S, Gandon S. 2013. Evolution of suicide as a defence strategy against pathogens in a spatially structured environment. Ecol. Lett. 16, 446–453. ( 10.1111/ele.12064) [DOI] [PubMed] [Google Scholar]
- 33.Fukuyo M, Sasaki A, Kobayashi I. 2012. Success of a suicidal defense strategy against infection in a structured habitat. Sci. Rep. 2, 238 ( 10.1038/srep00238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Refardt D, Bergmiller T, Kümmerli R. 2013. Altruism can evolve when relatedness is low: evidence from bacteria committing suicide upon phage infection. Proc. R. Soc. B 280, 20123035 ( 10.1098/rspb.2012.3035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Debarre F, Lion S, Van Baalen M, Gandon S. 2012. Evolution of host life-history traits in a spatially structured host-parasite system. Am. Nat. 179, 52–63. ( 10.1086/663199) [DOI] [PubMed] [Google Scholar]
- 36.Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. 2010. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 26, 335–340. ( 10.1016/j.tig.2010.05.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vercoe RB, et al. 2013. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 9, e1003454 ( 10.1371/journal.pgen.1003454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei Y, Terns RM, Terns MP. 2015. Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Genes Dev. 29, 356–361. ( 10.1101/gad.257550.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tollrian R, Harvell D. 1999. The ecology and evolution of inducible defenses. Princeton, NJ: Princeton University Press. [Google Scholar]
- 40.Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D. 2011. Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature 474, 604–608. ( 10.1038/nature10172) [DOI] [PubMed] [Google Scholar]
- 41.Held NL, Whitaker RJ. 2009. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ. Microbiol. 11, 457–466. ( 10.1111/j.1462-2920.2008.01784.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data files available on Dryad: http://dx.doi.org/10.5061/dryad.k7cr5.