Abstract
Blockade of the renin–angiotensin–aldosterone system (RAAS) with ACE inhibitors has been a cornerstone of heart failure therapy for over 15 years. More recently, further blockade of RAAS with aldosterone antagonists and angiotensin receptor blockers (ARBs) has been studied. While these therapies have certainly improved outcomes in the treatment of heart failure, morbidity and mortality remain extremely high. Furthermore, polypharmacy and complex regimens of seven medications on average is the norm for management of heart failure. This results in increased costs, patient burden, and uncertainty as to the best course of therapy. The ability to personalize patients’ therapeutic regimens using pharmacogenomics has the potential of providing more effective and efficient use of RAAS-modulating medications. This review highlights the implications of major RAAS pharmacogenetic studies, while outlining future directions for translation to practice.
Keywords: ACE inhibitors, Angiotensin receptor blockers, Aldosterone antagonists, Pharmacogenetics, Pharmacogenomics, Renin–angiotensin–aldosterone system
Introduction
Blockade of the renin–angiotensin–aldosterone system (RAAS) with ACE inhibitors has been a cornerstone of heart failure therapy for over 15 years. During that time, indications for ACE inhibitor use have been broadened to include treatment in patients with left ventricular dysfunction, including asymptomatic patients. Additionally, further blockade of RAAS with aldosterone antagonists and angiotensin receptor blockers (ARBs) has been studied in combination with ACE inhibitors and other standard heart failure therapies. While these therapies have certainly improved outcomes in the treatment of heart failure, morbidity and mortality remain extremely high, with heart failure being the leading cause of hospitalizations among people on Medicare [1].
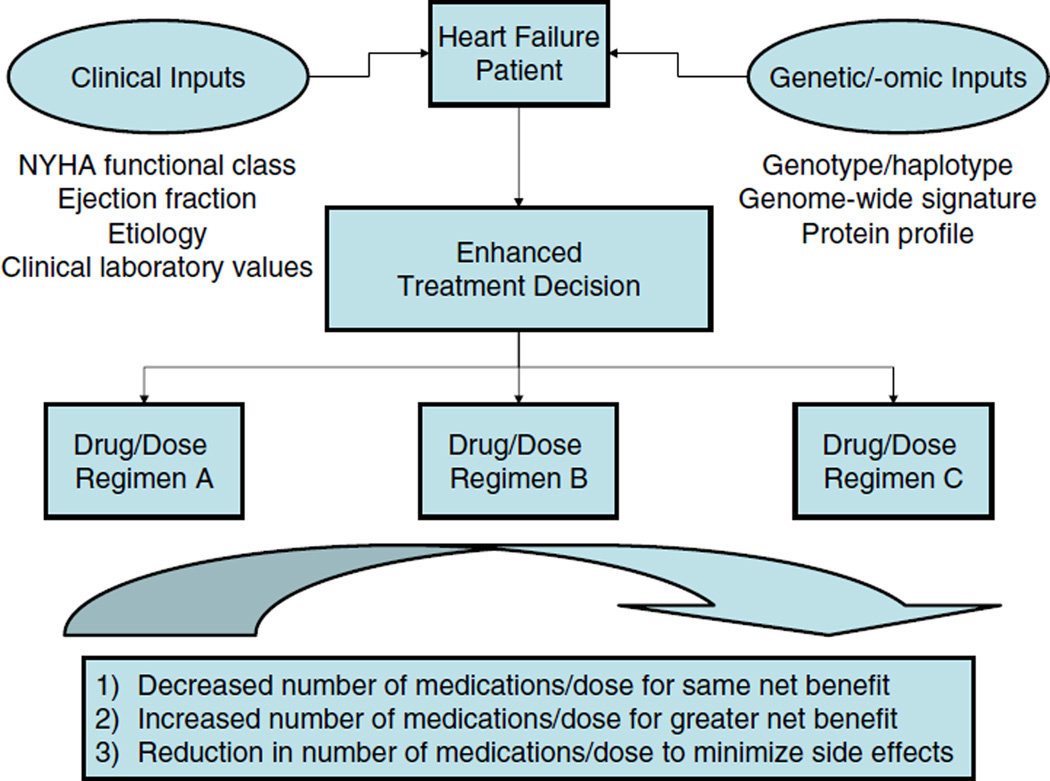
On the basis of treatment algorithms set forth by the American Heart Association, American College of Cardiology, and the Heart Failure Society of America, a patient may be treated with multiple medications for their heart failure alone, and a great deal of variability to each individual medication is likely to exist, resulting in variable incremental benefit for each medication added to a given regimen. Furthermore, it remains unclear exactly which patients may benefit from more aggressive RAAS blockade (e.g., ARB in addition to β-blocker, ACE inhibitor, and aldosterone antagonist), given the diverse nature of heart failure patients [2–4]. Therefore, the ability to personalize patients’ therapeutic regimens could theoretically result in (1) decreased total number of necessary medications for the same net benefit or (2) maintaining maximized number of medications but with increased benefit:risk ratio. Pharmacogenomics is one method of personalizing heart failure regimens for individual patients in order to achieve the most effective and efficient use of medications (Fig. 1). There are few pharmacogenomic studies in heart failure to date. However, consideration of RAAS pharmacogenomics in the context of other cardiovascular diseases with phenotypes relevant to heart failure can begin to help operationalize pharmacogenomics in the heart failure patient. This review will focus on our knowledge to date with regard to the genetic contribution to ACE inhibitor, ARB, and aldosterone antagonist response and outline future steps for implementation of pharmacogenomics in practice.
Fig. 1.
Pharmacogenomics-based treatment decisions in heart failure. Simultaneous consideration of traditional clinical prognostic variables (e.g., functional class, ejection fraction), genetic variants, and protein biomarkers (e.g., cytokines) can lead to more refined patient stratification. An enhanced treatment decision would result in a specific drug or dose regimen aimed at simplifying drug regimens, optimizing medication use, and minimizing side effects. NYHA, New York Heart Association
Candidate RAAS genes
The primary methodological approach taken to date with regard to heart failure pharmacogenomics has been the candidate gene design. In this hypothesis-driven approach, genes in drug targets, drug metabolizing enzymes, or relevant to drug target physiology (i.e., RAAS-mediated pathogenesis) are chosen a priori to be studied [5]. An advantage of this method is that it is based in principle on biological plausibility or previously demonstrated functional effects of the genetic polymorphisms to be studied. Additionally, the candidate gene approach greatly reduces the number of variants tested, saving cost and multiple statistical testing compared to an approach without preexisting hypotheses (e.g., genome-wide association study).
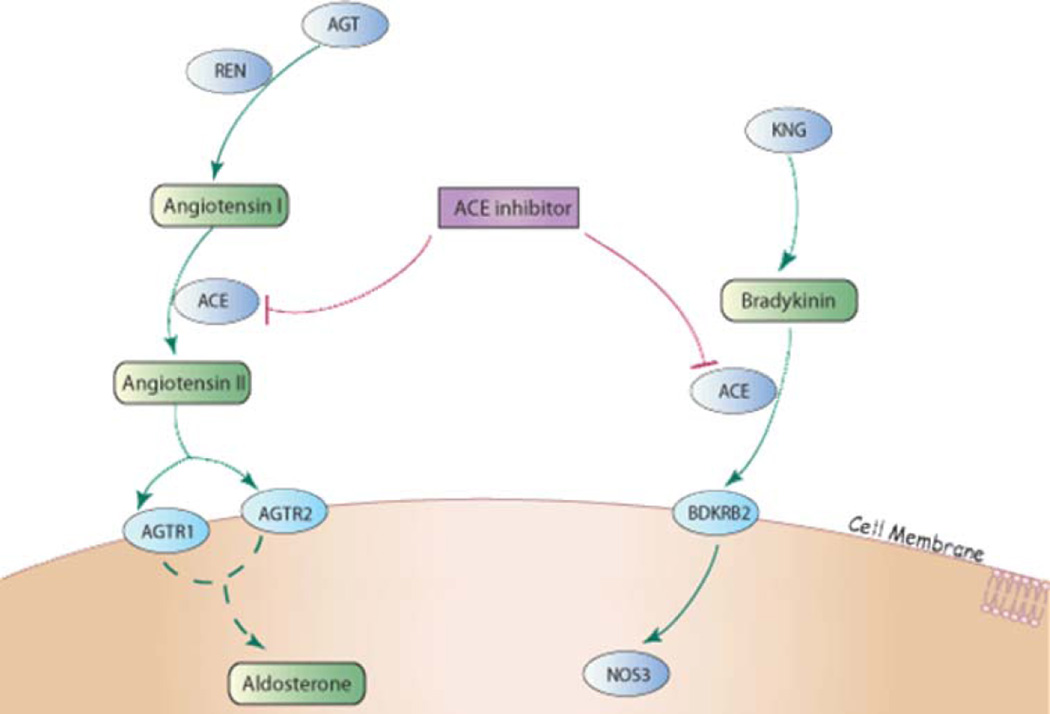
The success of the candidate gene approach in the area of RAAS pharmacogenomics is not surprising given that the target gene products and related downstream pathways of ACE inhibitors, ARBs, and aldosterone antagonists are relatively well described (Fig. 2). Angiotensinogen (encoded by AGT) is cleaved by renin (REN) to angiotensin I. ACE inhibitors then block angiotensin-converting enzyme (ACE)-mediated conversion of angiotensin I to angiotensin II, a potent vasoconstrictor and stimulator of aldosterone and norepinephrine release. Additionally, ACE inhibitors prevent the breakdown of vasodilatory bradykinin, the cardiovascular effects of which are mediated through the bradykinin B2 receptor (BDKRB2). In contrast, ARBs antagonize the angiotensin II type 1 receptor (AGTR1), thereby blunting adverse angiotensin II effects at the myocardial and adrenal levels. In addition, AGTR1 blockade allows for greater binding of angiotensin II to the angiotensin II type 2 receptor (AGTR2), which has further beneficial vasodilatory and antigrowth effects. Aldosterone synthase (CYP11B2) regulates aldosterone secretion and even in the presence of ACE inhibitors and/or ARBs, aldosterone secretion is increased in patients with heart failure. Spironolactone and eplerenone are aldosterone antagonists which exert their effects through blockade of the mineralocorticoid receptor (NR3C2). When further considering genes in related pathways (e.g., adrenergic) and those that might be important in drug disposition (i.e., pharmacokinetics), a list of candidates can be generated that will likely be the focus of broader candidate gene studies examining greater numbers of variants and larger, more complex pathways (Table 1).
Fig. 2.
Renin angiontensin aldosterone system candidate genes. ACE inhibitor/ARB pathway in a non-tissue specific cell type. ACE inhibitors target the ACE gene product and ARBs antagonize the angiotensin II type 1 receptor (AGTR1). (From PharmGKB: http://www.pharmgkb.org)
Table 1.
Sample candidate genes/polymorphisms in RAAS pharmacogenetics
| Gene symbol |
Gene name | OMIM No. |
Protein function | Candidate polymorphisms |
Reported functional impact of variant(s) |
|---|---|---|---|---|---|
| ACE | Angiotensin I- converting enzyme |
106180 | Converts angiotensin I to angiotensin II |
287 bp I/D | Unknown |
| −3892 T>C | Unknown | ||||
| ACE2 | Angiotensin I- converting enzyme-2 |
300335 | Converts angiotensin I to angiotensin 1–9 |
Rs2106809 | Unknown |
| AGT | Angiotensinogen | 106150 | Precursor to angiotensin I | −6 A>G | A allele has increased transcription |
| M235T | In LD with −6 A>G | ||||
| AGTR1 | Angiotensin II type 1 receptor |
106165 | Receptor for angiotensin II; mediates detrimental effects of angiotensin II in heart failure |
A1166>C | C allele results in decreased expression |
| AGTR2 | Angiotensin II type I receptor |
300034 | Receptor for angiotensin II; thought to mediate beneficial effects of angiotensin II in heart failure |
A190>G | Unknown |
| G1675>A | G allele has increased luciferase reporter gene activity |
||||
| G3726>C | Unknown | ||||
| BDKRB2 | Bradykinin receptor B2 | 113503 | Receptor for bradykinin; mediates bradykinin vasodilation |
−58C>T | C allele has reduced transcriptional activity |
| +9/−9 | −9 allele has increased expression | ||||
| CYP11B2 | Aldosterone synthase | 124080 | Converts 11-deoxycorticosterone to aldosterone |
−344 C>T | C allele has greater affinity for putative SF1 site |
| A6547>G | Unknown | ||||
| NR3C2 | Mineralocorticoid receptor |
600983 | Receptor for cortisol and aldosterone |
Ile180Val | Val allele has decreased cortisol responsiveness |
| REN | Renin | 179820 | Converts angiotensinogen to angiotensin I |
−4021 C>T | Unknown |
| −5312 C>T | T allele has increased transcriptional activity |
||||
| A4280>C | Unknown |
LD, linkage disequilibrium
Angiotensin I-converting enzyme (ACE)
The 45 kb ACE gene is located on chromosome 17q23.3 and contains 21 exons. There are at least three alternative transcripts of the gene. The longest transcript is known as the somatic or endothelial form, designated isoform 1, and is expressed widely. The germinal or testicular form (expressed only in sperm), designated isoform 2, contains an inframe alternate exon in the 5′ region and differs from isoform 1 in both the 5′ and 3′ regions. A third transcript, isoform 3, also contains the alternate in-frame exon in the 5′ coding region and has an alternate splice site in the 3′ coding region.
By far the most widely studied polymorphism in ACE has been the 287 bp insertion/deletion (I/D) polymorphism located in intron 16. Interestingly, this polymorphism was originally studied as a linkage marker to help define the role of the ACE gene on circulating ACE concentrations. The D allele was found to be associated with higher circulating plasma ACE levels in an additive manner (with I/D individuals having intermediate levels compared to I/I and D/D individuals), explaining 47% of the variability in circulating ACE [6]. The D allele was also found to associate with higher cellular ACE levels (with D/D individuals having higher levels than I/I individuals) [6, 7]. Soon thereafter, the association between the I/D polymorphism and risk of myocardial infarction in low risk individuals was made [8] and, since that time, several hundred (often conflicting) papers assessing the I/D polymorphism have been published [9].
Since the ACE I/D polymorphism is thought to be a marker polymorphism in linkage with a functional variant, additional work in trying to identify the functional polymorphisms in ACE has yielded two potential quantitative trait loci (QTLs) on chromosome 17 linked to ACE concentrations— one in tight linkage disequilibrium with the I/D polymorphism and one in the 50 region of the ACE gene. Interestingly, a third potential QTL for ACE concentrations in Mexican–Americans has been identified on chromosome 4 using data from the San Antonio Heart Study. This QTL, which is on another chromosome entirely from the ACE gene, suggests that another gene(s) may contribute substantially to variability in ACE concentrations and might provide an explanation for the lack of consistency among association studies in the literature for this gene [10].
Angiotensin I-converting enzyme-2 (ACE2)
ACE2 is located on chromosome Xp22. It has considerable homology to ACE and may counteract some of the effects of ACE. ACE2 has 18 exons, is 41 kb in size, and has a coding region made up of 805 amino acids.
Angiotensinogen (AGT)
AGT is located on chromosome 1q42–q43, spans 12 kb, and contains 5 exons. Two commonly studied SNPs are −6 A>G located in the promoter region and M235>T. The 235T allele has been associated with hypertension and elevated angiotensinogen concentrations. However, these single nucleotide polymorphisms (SNPs) are in complete linkage disequilibrium in most populations and it is the −6 A>G SNP that appears to have functional activity. The −6 A allele was found to have approximately 20–40% higher basal transcription levels compared to the G allele [11]. Additionally, vectors containing the −6 G allele were found to preferentially bind a trans-acting factor resulting in decreased transcriptional activity.
Angiotensin receptors 1 and 2 (AGTR1 and AGTR2)
AGTR1 is located on chromosome 3q21-q25 and spans 60 kb, while AGTR2 is located on chromosome Xq21-q23. AGTR1 contains at least four transcript variants. Transcript A contains exons 1 and 5; transcript B contains exons 1, 3, and 5; transcript C contains exons 1, 2, and 5; and transcript D contains exons 1, 2, 3, and 5 [12]. The coding region is contained entirely in the last exon of all variants. AGTR2 contains 3 exons, the first two of which are untranslated. Alternative splicing of both AGTR1 and AGTR2 appears to be important in modulating translation efficiency and protein expression in various tissues [13].
The most widely studied polymorphism in AGTR1 is the A1166>C polymorphism, which is located in the 3′ UTR. Until recently, the functional relevance of the A1166>C SNP was unknown. However, investigators have recently found a reduced abundance of mRNA transcripts with the 1166C [14]. The same investigators also found CC individuals to have reduced placental mRNA level compared to AC and AA individuals in a dose-response manner [14]. AGTR2 has been much less widely studied to date than AGTR1, likely because its function is less well understood than AGTR1.
Bradykinin receptor B2 (BDKRB2)
BDKRB2 is a 46 kb gene located on 14q32.1-q32.2 and contains two isoforms with differing start codons. BDKRB2 has three exons, the first of which is not translated. A promoter polymorphism, −58 C>T and a 9 bp I/D (+9/−9) in exon 1 have been the most widely studied variants. The +9/−9 polymorphism has been found to affect transcription such that the −9 allele had increased mRNA compared to the +9 allele [15].
Aldosterone synthase (CYP11B2)
Aldosterone synthase, encoded by CYP11B2, is located on chromosome 8q21–q22. CYP11B2 has 9 exons and a coding region approximately 3 kb in size. The two most commonly studied polymorphisms in CYP11B2 are −344 C>T and A6547>G. The −344 C[T SNP is located in the promoter region at a putative steroidogenic factor-1 (SF1) binding site, with the C allele having greater affinity for the site than the T allele [16]. The A6547>G SNP has unknown functional significance. However, its location in the 3′-UTR lends the possibility of it altering mRNA stability.
Renin (REN)
Renin is encoded by REN and is located on chromosome 1q32. REN is approximately 11.5 kb in size and contains 10 exons. Two isoforms of REN exist which have differing first exons and distinctive expression patterns [17]. Mutations in REN have been found to cause familial hyperproreninemia.
RAAS pharmacogenetics
ACE inhibitors
It is somewhat surprising to note that while numerous pharmacogenetic studies of ACE inhibitors exist in hypertension, they are fairly limited in the heart failure population. One potential reason for the paucity of pharmacogenetic studies is that the phenotypes of interest that determine drug response in heart failure are less readily available compared to in hypertension. For example, while blood pressure is relatively easy to obtain, routinely measured, and well correlated with outcomes, serial echocardiograms are not routinely obtained in clinical practice and are less well correlated with outcomes. Pharmacogenomic studies to assess ACE inhibitor response are difficult to conduct, given that ACE inhibitor are a well established therapy in the treatment of heart failure and that the vast majority of patients are already being treated with them. In addition, an ideal dataset for analyses would include genetic samples and hard clinical endpoints related mortality and morbidity, and such datasets are rare.
McNamara and colleagues have established a prospective cohort of heart failure patients in whom approximately 85% were on ACE inhibitors at baseline, 10% were on ARBs, and 40% were on β-blockers; genetic samples have been banked on these individuals [18]. In this population, the ACE I/D polymorphism was associated with event-free survival with the (D/D) genotype having the poorest event-free survival. These findings are consistent with the above-described functional data in which the D allele is associated with higher ACE levels, and presumably higher RAAS activity. Interestingly, the association with ACE I/D genotype was most apparent in those on low-dose ACE inhibitor therapy or not on β-blocker therapy, suggesting a genotype-drug-disease interaction in which variant homozygotes (D/D) represent a genetic subgroup that could derive greatest benefit from drugs that interfere with RAAS (i.e., ACE inhibitors and β-blockers). In support of this relationship, the negative effects of the D/D genotype on survival were attenuated with β-blocker therapy, suggesting that the increased risk associated with a particular genotype may be masked with contemporary drug therapy. In fact, additional studies have corroborated the observations that adequate use of RAAS and adrenergic system antagonists in a heart failure setting can attenuate the detrimental effects of RAAS and related system gene polymorphisms on prognosis [19]. While ACE pharmacogenetic studies have not necessarily identified which patients derive the most benefit from ACE inhibitors, they have provided insight into disease pathogenesis and the importance of RAAS blockade in individuals with ACE D alleles.
Angiotensin receptor blockers
The majority of pharmacogenetic association studies with ARBs have evaluated the gene that encodes their binding site, the angiotensin II receptor type 1 gene, AGTR1. The A1166>C polymorphism in AGTR1 has been associated with blood pressure, renal blood flow, and renal vascular resistance responses to ARBs with C allele carriers having an attenuated response [20, 21]. In addition to the AGTR1 associations, a small study has evaluated changes in left ventricular hypertrophy (LVH) in response to losartan compared by ACE insertion/deletion (I/D) genotype [22]. In 32 patients on chronic hemodialysis, individuals carrying the D allele had significantly greater reductions in left ventricular mass index (LVMI) than I homozygotes, again consistent with the functional data regarding this variant [22].
In a genetic substudy of the Swedish Irbesartan Left Ventricular Hypertrophy Investigation versus Atenolol (SILVIA) trial, bradykinin receptor BDKRB2 −9/+9 genotype was determined in a group of 90 patients with left ventricular hypertrophy who were randomized to irbesartan or atenolol [23]. Although the investigators did not identify a treatment by genotype interaction, they did find that −9 allele carriers had twice the regression in left ventricular mass compared to those with the +9/+9 genotype, regardless of treatment assignment.
Pharmacogenomics studies to date with ARBs have found some interesting associations. Moreover, given the lack of firm recommendations regarding when to add an ARB (versus aldosterone antagonist or nitrates/hydralazine) to existing therapy in symptomatic patients [24], pharmacogenomics would seem to be a useful modality for elucidating which patients could benefit most from their addition. Although at this time, the populations investigated have generally been small and without replication. Therefore, whether using pharmacogenomics for these agents has clinical utility will become clearer as the body of evidence grows and more comprehensive studies are designed.
Aldosterone antagonists
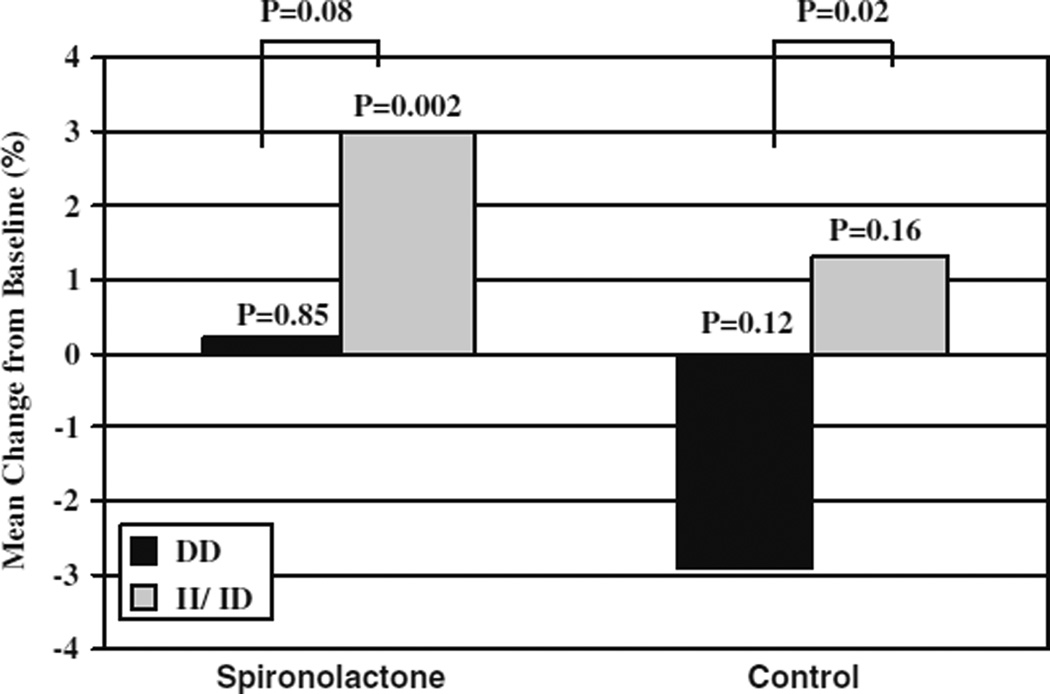
Aldosterone antagonism represents a newer modality for neurohormonal blockade in heart failure relative to ACE inhibition. It has been promulgated that circulating aldosterone concentrations increase in the setting of continual ACE inhibition in heart failure, and that this hormone contributes to detrimental remodeling and volume expansion. Cicoira and colleagues have assessed the “aldosterone escape” phenomenon with regard to the ACE I/D polymorphism in a population of heart failure patients treated with maximally tolerated ACE inhibitor therapy. They found the DD genotype to be nearly three times more prevalent among patients with aldosterone escape (defined as plasma aldosterone concentrations above the upper limit of the reference range) than I carriers (62% versus 24%, respectively, P < 0.005) [25]. However, aldosterone escape was only present in 13 of the 132 patients which somewhat limits the conclusions which could be drawn from the study. Nonetheless, a 10% prevalence of aldosterone escape among a general heart failure population represents a large portion; in this sample ACE genotype might serve as a biomarker to guide whether to enhance a heart failure regimen with aldosterone antagonists upstream. In a follow-up study, the same investigators evaluated the ACE I/D genotype and spironolactone response in 93 patients with heart failure randomized to standard therapy with or without spironolactone 25–50 mg/day for 12 months [26]. All patients were on maximum tolerated ACE inhibitor treatment and 69% of patients were treated with β-blockers. Among patients treated with spironolactone, only those who carried at least one I allele experienced significant increases in ejection fraction relative to baseline (Fig. 2) [26]. End diastolic volume and end systolic volume were also improved in this group. Also of note, this small study lends further support that individuals with the DD genotype may be predisposed to worsening disease, as individuals with this genotype treated with standard therapies (without spironolactone) experienced decline in EF over 12 months compared with I allele carriers (Fig. 3). It is somewhat surprising that DD individuals, who were more likely to have aldosterone escape in the previous study, would respond less favorably to spironolactone than I carriers; therefore, the exact genetic subgroup that receives the greatest benefit from spironolactone has yet to be defined. Consideration of other genetic variants in RAAS could perhaps refine the approach to genotype-tailored spironolactone use and should be explored further.
Fig. 3.
Change in left ventricular ejection fraction by ACE I/D genotype. Change in ejection fraction after 12 months in spironolactone group and control group compared by ACE I/D genotype. Black bars indicate the D/D genotype group and gray bars indicate the I/D + I/I genotype groups. Adapted from Cicoira et al. [26]
The blood pressure phenotype
Theoretically, the mechanisms by which RAAS antagonists exert their antihypertensive effects are, in part, the same mechanisms by which benefit is conferred in heart failure. Although it is unclear whether genetic associations with drug response will translate across phenotypes, in this section, we present data from other disease states in which ACE inhibitor and ARB therapy are used. Early studies evaluating the ACE I/D polymorphism and blood pressure response to ACE inhibitors were small and often inconsistent in their results [27–29]. More recently, two much larger studies, the Genetics of Hypertension-Associated Treatment (GenHAT) study and the Perindopril Protection Against Recurrent Stroke Study (PROGRESS), which included nearly 38,000 and 6,000 participants, respectively, found no association between the ACE I/D polymorphism and antihypertensive response to ACE inhibitors [30, 31]. While these studies fairly decisively indicate that the ACE I/D polymorphism does not influence ACE inhibitor response in a clinically meaningful way, it is still possible that further ACE gene variability (e.g., comprehensive tagSNPs or haplotypes) might influence drug response.
Recently, Fan and colleagues used a comprehensive tagSNP approach to capture variation in the ACE2 gene as well as the ACE I/D polymorphism [32]. They identified one SNP in ACE2, rs2106809, that was associated with increased risk of hypertension that was further increased when the ACE DD genotype was also present. The same ACE2 SNP was also associated with an approximately 3 mmHg greater blood pressure response to captopril in homozygous wild type women compared to variant carrier women. No association was seen in men, which might not be surprising given ACE2s location on the X chromosome.
Using a postmarketing surveillance trial in a Chinese population, investigators found several SNPs/haplotypes in RAAS genes (other than ACE) to influence antihypertensive response to benazepril. They genotyped 1,447 subjects (randomly divided into two groups in an effort to provide a sample set and a validation set from the same population) for four SNPs in AGT, seven in AGTR1, and three in AGTR2 in order to evaluate the common variation in these genes [33]. One SNP in AGT, rs7079 (C11537>A) was associated with diastolic blood pressure (DBP) response in both random subsets of patients. Individuals with the CC genotype had a slightly smaller reduction in DBP with benazepril compared to CA or AA individuals, 7.1 ± 8.9 mmHg compared to 8.8 ± 9.5 and 8.3 ± 6.2 mmHg, respectively (P = 0.028). Using haplotype analysis, two haplotypes (H2- CCGTGCA and H3- TTAAGCA for SNPs rs1492078, rs2638362, rs2640543, rs389566, rs275649, rs5182, and rs5186) in AGTR1 were associated with systolic blood pressure (SBP) response to benazepril in both subsets. In the total sample, individuals carrying both haplotypes H2 and H3 had a 13.6 ± 16.7 mmHg reduction in SBP, those with neither H2 nor H3 had a 10.9 ± 15.4 mmHg reduction, and those with H3 but not H2 had a 6.6 ± 16 mmHg reductions (P = 0.004). None of the AGTR2 SNPs/haplotypes were associated with differences in BP response to benazepril in both subsets. These associations accounted for approximately 13% of the variability in SBP response and 10% of the variability in DBP response. Interestingly, the two most commonly studied SNPs from AGT and AGTR1 (−6 A/G and A1166C, respectively) were not associated with BP response to ACE inhibitor in this study. However, the minor allele frequencies of both of these SNPs are much lower in Chinese individuals than in Caucasians, which is the population in which the majority of the previous associations have been based, suggesting that limited statistical power may have contributed to their inability to replicate the previous findings.
One study to date has assessed the role of CYP11B2 and ACE inhibitor response. The −344 C>T SNP in CYP11B2 was associated with DBP response to ACE inhibitor treatment with benazepril or imidapril in a Chinese population [34]. Individuals with the C/C genotype had an approximately 5 mmHg fall in DBP after 6 weeks of treatment, whereas C/T and T/T individuals had an approximately 9 mmHg decline [34]. The investigators did not find an association with the A6547>G variant and blood pressure response, nor did they find an interaction between the two SNPs.
Many pharmacogenomic studies in hypertension have evaluated the association between RAAS candidate genes and ACE inhibitor or ARB responses. The largest of these studies have been negative [30, 31]; however, both of these studies evaluated the ACE I/D polymorphism, which may not adequately tag the truly functional SNP(s) in the ACE gene. Future studies investigating larger numbers of patients and other RAAS genes/SNPs will likely provide evidence for whether pharmacogenomics will be clinically useful in this setting and for guiding heart failure pharmacogenomics studies as well.
Racial/ethnic differences in ACE inhibitor response and pharmacogenetics
It is a well-recognized phenomenon that variability exists in antihypertensive response to ACE inhibitors by race/ethnicity because of differences in renin levels. Variability also exists by race/ethnicity in the allele frequencies of many SNPs associated with differential response to ACE inhibitors. Therefore, it is tempting to conclude that the racial/ethnic differences in medication response are a result of genetic differences. Unfortunately, this issue is quite complex because of the difficulty in disentangling all of the environmental, cultural, and socioeconomic factors also captured by a race term that may not be directly measured [35]. Therefore, it is difficult to determine whether racial differences in drug response are due to genetic differences or some other unmeasured differences between populations. Associations that are assessed in multiple populations and persist within each population may have a genetic basis to them.
Implications and future directions
The heart failure syndrome represents a challenging disease state in which to establish pharmacogenomic associations, and yet a disease in desperate need of individualized therapy. The underlying molecular derangements in heart failure are currently innumerable, and include myocardial energy dysregulation, inflammation, neurohormonal activation, and oxidative stress. Given the complex relationship between these processes, RAAS pharmacogenomic studies should more comprehensively consider candidate genes in proteins involved in myocardial contractility, cytoskeletal integrity, cellular calcium homeostasis, and various cell signaling pathways. Until then, RAAS pharmacogenomics is likely to remain limited in scope.
In terms of current utility, the promise of heart failure pharmacogenomics is twofold: (1) to identify the most appropriate “add-on” therapies in increasingly complex polypharmacy patient regimens and (2) to identify likely responders to new heart failure treatments in drug development. With respect to the first promise, instead of being classified with an ischemic, non-ischemic, or idiopathic etiology, heart failure patients could be given a more molecularly based diagnosis. Using this approach, clinicians would have a better idea of whether adding an aldosterone antagonist or a nitrate/hydralazine combination would best treat the underlying pathogenesis for a particular patient who remained symptomatic with ACE inhibitor and β-blocker treatment. In terms of drug development, several new treatment modalities including sympathetic modulators (moxonidine), TNF-alpha blockers (etanercept), and vasopressin receptor blockers (tolvaptan) have failed to show incremental benefits in heart failure populations, costing millions of dollars and in some cases harming patients [36–40]. Pharmacogenomics has been advocated as a tool to streamline the development process and minimize the likelihood for failure in the drug product pipeline in late-phase clinical trials [41].
Because of the mortality and/or morbidity benefits already conferred by such agents as ACE inhibitors, beta-blockers, spironolactone, digoxin, and nitrates/hydralazine, the likelihood for demonstrating robust, incremental benefit for a first-in-class agent on top of these therapies in a general heart failure population is becoming progressively more difficult. Rather an enrichment strategy whereby clinical studies enroll patients with “at-risk” genotypes is more likely to allow for drug benefits to be seen. This concept is not new to heart failure. The first ACE inhibitor studies demonstrated mortality benefits in heart failure with less than 300 patients [42] by enriching the sample with an at-risk group (in this case, class III or IV heart failure). A pharmacogenomic approach would be analogous to the early ACE inhibitor studies, by identifying patient niches in which these novel therapeutics can be applied. In order for these promises of pharmacogenomics to be fulfilled in heart failure, focused candidate pathway and genome-wide efforts will be needed in drug discovery, development, and post-approval patient care settings.
Acknowledgments
This work was supported in part by National Institutes of Health P50 HL077113 (ALB), GM74492 (IZ), American Heart Association Heartland Affiliate 0655496Z (ALB) and Florida/Puerto Rico Affiliate 0435278B (IZ).
References
- 1.Hospitalization in the United States. Rockville, MD: Agency for Healthcare Research and Quality; 1997. [May 2000]. HCUP Fact Book No 1. http://www.ahrq.gov/data/hcup/factbk1. AHRQ Publication No. 00-0031. [Google Scholar]
- 2.Cohn JN, Tognoni G. A randomized trial of the angiotensin-receptor blocker valsartan in chronic heart failure. N Engl J Med. 2001;345(23):1667–1675. doi: 10.1056/NEJMoa010713. [DOI] [PubMed] [Google Scholar]
- 3.McMurray JJ, Ostergren J, Swedberg K, Granger CB, Held P, Michelson EL, et al. Effects of candesartan in patients with chronic heart failure and reduced left-ventricular systolic function taking angiotensin-converting-enzyme inhibitors: the CHARM-Added trial. Lancet. 2003;362(9386):767–771. doi: 10.1016/S0140-6736(03)14283-3. [DOI] [PubMed] [Google Scholar]
- 4.Pfeffer MA, McMurray JJ, Velazquez EJ, Rouleau JL, Kober L, Maggioni AP, et al. Valsartan, captopril, or both in myocardial infarction complicated by heart failure, left ventricular dysfunction, or both. N Engl J Med. 2003;349(20):1893–1906. doi: 10.1056/NEJMoa032292. [DOI] [PubMed] [Google Scholar]
- 5.Zineh I, Johnson JA. Pharmacogenetics of chronic cardiovascular drugs: applications and implications. Expert Opin Pharmacother. 2006;7(11):1417–1427. doi: 10.1517/14656566.7.11.1417. [DOI] [PubMed] [Google Scholar]
- 6.Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86(4):1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costerousse O, Allegrini J, Lopez M, Alhenc-Gelas F. Angiotensin I-converting enzyme in human circulating mononuclear cells: genetic polymorphism of expression in T-lymphocytes. Biochem J. 1993;290(Pt 1):33–40. doi: 10.1042/bj2900033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cambien F, Poirier O, Lecerf L, Evans A, Cambou JP, Arveiler D, et al. Deletion polymorphism in the gene for angiotensin-converting enzyme is a potent risk factor for myocardial infarction. Nature. 1992;359(6396):641–644. doi: 10.1038/359641a0. [DOI] [PubMed] [Google Scholar]
- 9.Sayed-Tabatabaei FA, Oostra BA, Isaacs A, van Duijn CM, Witteman JC. ACE polymorphisms. Circ Res. 2006;98(9):1123–1133. doi: 10.1161/01.RES.0000223145.74217.e7. [DOI] [PubMed] [Google Scholar]
- 10.Kammerer CM, Gouin N, Samollow PB, VandeBerg JF, Hixson JE, Cole SA, et al. Two quantitative trait loci affect ACE activities in Mexican–Americans. Hypertension. 2004;43(2):466–470. doi: 10.1161/01.HYP.0000111830.36999.94. [DOI] [PubMed] [Google Scholar]
- 11.Inoue I, Nakajima T, Williams CS, Quackenbush J, Puryear R, Powers M, et al. A nucleotide substitution in the promoter of human angiotensinogen is associated with essential hypertension and affects basal transcription in vitro. J Clin Invest. 1997;99(7):1786–1797. doi: 10.1172/JCI119343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin MM, Willardson BM, Burton GF, White CR, McLaughlin JN, Bray SM, et al. Human angiotensin II type 1 receptor isoforms encoded by messenger RNA splice variants are functionally distinct. Mol Endocrinol. 2001;15(2):281–293. doi: 10.1210/mend.15.2.0598. [DOI] [PubMed] [Google Scholar]
- 13.Warnecke C, Surder D, Curth R, Fleck E, Regitz-Zagrosek V. Analysis and functional characterization of alternatively spliced angiotensin II type 1 and 2 receptor transcripts in the human heart. J Mol Med. 1999;77(10):718–727. doi: 10.1007/s001099900049. [DOI] [PubMed] [Google Scholar]
- 14.Abdollahi MR, Lewis RM, Gaunt TR, Cumming DV, Rodriguez S, Rose-Zerilli M, et al. Quantitated transcript haplotypes (QTH) of AGTR1, reduced abundance of mRNA haplotypes containing 1166C (rs5186:A>C), and relevance to metabolic syndrome traits. Hum Mutat. 2007;28(4):365–373. doi: 10.1002/humu.20454. [DOI] [PubMed] [Google Scholar]
- 15.Lung CC, Chan EK, Zuraw BL. Analysis of an exon 1 polymorphism of the B2 bradykinin receptor gene and its transcript in normal subjects and patients with C1 inhibitor deficiency. J Allergy Clin Immunol. 1997;99(1 Pt 1):134–146. doi: 10.1016/s0091-6749(97)70310-5. [DOI] [PubMed] [Google Scholar]
- 16.Bassett MH, Zhang Y, Clyne C, White PC, Rainey WE. Differential regulation of aldosterone synthase and 11beta-hydroxylase transcription by steroidogenic factor-1. J Mol Endocrinol. 2002;28(2):125–135. doi: 10.1677/jme.0.0280125. [DOI] [PubMed] [Google Scholar]
- 17.Lee-Kirsch MA, Gaudet F, Cardoso MC, Lindpaintner K. Distinct renin isoforms generated by tissue-specific transcription initiation and alternative splicing. Circ Res. 1999;84(2):240–246. doi: 10.1161/01.res.84.2.240. [DOI] [PubMed] [Google Scholar]
- 18.McNamara DM, Holubkov R, Postava L, Janosko K, MacGowan GA, Mathier M, et al. Pharmacogenetic interactions between angiotensin-converting enzyme inhibitor therapy and the angiotensin-converting enzyme deletion polymorphism in patients with congestive heart failure. J Am Coll Cardiol. 2004;44(10):2019–2026. doi: 10.1016/j.jacc.2004.08.048. [DOI] [PubMed] [Google Scholar]
- 19.Shin J, Lobmeyer MT, Gong Y, Zineh I, Langaee TY, Yarandi H, et al. Relation of beta(2)-adrenoceptor haplotype to risk of death and heart transplantation in patients with heart failure. Am J Cardiol. 2007;99(2):250–255. doi: 10.1016/j.amjcard.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 20.Spiering W, Kroon AA, Fuss-Lejeune MJ, de Leeuw PW. Genetic contribution to the acute effects of angiotensin II type 1 receptor blockade. J Hypertens. 2005;23(4):753–758. doi: 10.1097/01.hjh.0000163143.66965.06. [DOI] [PubMed] [Google Scholar]
- 21.Sookoian S, Castano G, Garcia SI, Viudez P, Gonzalez C, Pirola CJ. A1166C angiotensin II type 1 receptor gene polymorphism may predict hemodynamic response to losartan in patients with cirrhosis and portal hypertension. Am J Gastroenterol. 2005;100(3):636–642. doi: 10.1111/j.1572-0241.2005.41168.x. [DOI] [PubMed] [Google Scholar]
- 22.Nakayama M, Nakano H, Tsuboi N, Kurosawa T, Tsuruta Y, Iwasaki Y, et al. The effect of angiotensin receptor blockade ARB on the regression of left ventricular hypertrophy in hemodialysis patients: comparison between patients with D allele and non-D allele ACE gene polymorphism. Clin Nephrol. 2005;64(5):358–363. doi: 10.5414/cnp64358. [DOI] [PubMed] [Google Scholar]
- 23.Hallberg P, Lind L, Michaelsson K, Karlsson J, Kurland L, Kahan T, et al. B2 bradykinin receptor (B2BKR) polymorphism and change in left ventricular mass in response to antihypertensive treatment: results from the Swedish Irbesartan Left Ventricular Hypertrophy Investigation versus Atenolol (SILVHIA) trial. J Hypertens. 2003;21(3):621–624. doi: 10.1097/00004872-200303000-00029. [DOI] [PubMed] [Google Scholar]
- 24.Heart Failure Society Of A. HFSA 2006. Comprehensive Heart Failure Practice Guideline. J Card Fail. 2006;12(1):e1–e2. doi: 10.1016/j.cardfail.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Cicoira M, Zanolla L, Rossi A, Golia G, Franceschini L, Cabrini G, et al. Failure of aldosterone suppression despite angio-tensin-converting enzyme (ACE) inhibitor administration in chronic heart failure is associated with ACE DD genotype. J Am Coll Cardiol. 2001;37(7):1808–1812. doi: 10.1016/s0735-1097(01)01237-2. [DOI] [PubMed] [Google Scholar]
- 26.Cicoira M, Rossi A, Bonapace S, Zanolla L, Perrot A, Francis DP, et al. Effects of ACE gene insertion/deletion polymorphism on response to spironolactone in patients with chronic heart failure. Am J Med. 2004;116(10):657–661. doi: 10.1016/j.amjmed.2003.12.033. [DOI] [PubMed] [Google Scholar]
- 27.Ohmichi N, Iwai N, Uchida Y, Shichiri G, Nakamura Y, Kinoshita M. Relationship between the response to the angiotensin converting enzyme inhibitor imidapril and the angiotensin converting enzyme genotype. Am J Hypertens. 1997;10(8):951–955. doi: 10.1016/s0895-7061(97)00121-0. [DOI] [PubMed] [Google Scholar]
- 28.Stavroulakis GA, Makris TK, Krespi PG, Hatzizacharias AN, Gialeraki AE, Anastasiadis G, et al. Predicting response to chronic antihypertensive treatment with fosinopril: the role of angiotensin-converting enzyme gene polymorphism. Cardiovasc Drugs Ther. 2000;14(4):427–432. doi: 10.1023/a:1007820401377. [DOI] [PubMed] [Google Scholar]
- 29.Ueda S, Meredith PA, Morton JJ, Connell JM, Elliott HL. ACE (I/D) genotype as a predictor of the magnitude and duration of the response to an ACE inhibitor drug (enalaprilat) in humans. Circulation. 1998;98(20):2148–2153. doi: 10.1161/01.cir.98.20.2148. [DOI] [PubMed] [Google Scholar]
- 30.Arnett DK, Davis BR, Ford CE, Boerwinkle E, Leiendecker-Foster C, Miller MB, et al. Pharmacogenetic association of the angiotensin-converting enzyme insertion/deletion polymorphism on blood pressure and cardiovascular risk in relation to antihypertensive treatment: the Genetics of Hypertension-Associated Treatment (GenHAT) study. Circulation. 2005;111(25):3374–3383. doi: 10.1161/CIRCULATIONAHA.104.504639. [DOI] [PubMed] [Google Scholar]
- 31.Harrap SB, Tzourio C, Cambien F, Poirier O, Raoux S, Chalmers J, et al. The ACE gene I/D polymorphism is not associated with the blood pressure and cardiovascular benefits of ACE inhibition. Hypertension. 2003;42(3):297–303. doi: 10.1161/01.HYP.0000088322.85804.96. [DOI] [PubMed] [Google Scholar]
- 32.Fan X, Wang Y, Sun K, Zhang W, Yang X, Wang S, et al. Polymorphisms of ACE2 gene are associated with essential hypertension and antihypertensive effects of Captopril in women. Clin Pharmacol Ther. 2007;82(2):187–196. doi: 10.1038/sj.clpt.6100214. [DOI] [PubMed] [Google Scholar]
- 33.Su X, Lee L, Li X, Lv J, Hu Y, Zhan S, et al. Association between angiotensinogen, angiotensin II receptor genes, and blood pressure response to an angiotensin-converting enzyme inhibitor. Circulation. 2007;115(6):725–732. doi: 10.1161/CIRCULATIONAHA.106.642058. [DOI] [PubMed] [Google Scholar]
- 34.Yu HM, Lin SG, Liu GZ, Zhang YQ, Ma WJ, Deng CY. Associations between CYP11B2 gene polymorphisms and the response to angiotensin-converting enzyme inhibitors. Clin Pharmacol Ther. 2006;79(6):581–589. doi: 10.1016/j.clpt.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Risch N, Burchard E, Ziv E, Tang H. Categorization of humans in biomedical research: genes, race and disease. Genome Biol. 2002;3(7) doi: 10.1186/gb-2002-3-7-comment2007. comment 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohn JN, Pfeffer MA, Rouleau J, Sharpe N, Swedberg K, Straub M, et al. Adverse mortality effect of central sympathetic inhibition with sustained-release moxonidine in patients with heart failure (MOXCON) Eur J Heart Fail. 2003;5(5):659–667. doi: 10.1016/s1388-9842(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 37.Bozkurt B, Torre-Amione G, Warren MS, Whitmore J, Soran OZ, Feldman AM, et al. Results of targeted anti-tumor necrosis factor therapy with etanercept (ENBREL) in patients with advanced heart failure. Circulation. 2001;103(8):1044–1047. doi: 10.1161/01.cir.103.8.1044. [DOI] [PubMed] [Google Scholar]
- 38.Gheorghiade M, Gattis WA, O’Connor CM, Adams KF, Jr, Elkayam U, Barbagelata A, et al. Effects of tolvaptan, a vasopressin antagonist, in patients hospitalized with worsening heart failure: a randomized controlled trial. JAMA. 2004;291(16):1963–1971. doi: 10.1001/jama.291.16.1963. [DOI] [PubMed] [Google Scholar]
- 39.Gheorghiade M, Konstam MA, Burnett JC, Jr, Grinfeld L, Maggioni AP, Swedberg K, et al. Short-term clinical effects of tolvaptan, an oral vasopressin antagonist, in patients hospitalized for heart failure: the EVEREST Clinical Status Trials. JAMA. 2007;297(12):1332–1343. doi: 10.1001/jama.297.12.1332. [DOI] [PubMed] [Google Scholar]
- 40.Kaye DM, Krum H. Drug discovery for heart failure: a new era or the end of the pipeline? Nat Rev Drug Discov. 2007;6(2):127–139. doi: 10.1038/nrd2219. [DOI] [PubMed] [Google Scholar]
- 41.Roses AD. Pharmacogenetics and drug development: the path to safer and more effective drugs. Nat Rev Genet. 2004;5(9):645–656. doi: 10.1038/nrg1432. [DOI] [PubMed] [Google Scholar]
- 42.Effects of enalapril on mortality in severe congestive heart failure. Results of the Cooperative North Scandinavian Enalapril Survival Study (CONSENSUS) (1987) The CONSENSUS Trial Study Group. N Engl J Med. 316(23):1429–1435. doi: 10.1056/NEJM198706043162301. [DOI] [PubMed] [Google Scholar]