Abstract
Motivation: Large-scale chemical cross-linking with mass spectrometry (XL-MS) analyses are quickly becoming a powerful means for high-throughput determination of protein structural information and protein–protein interactions. Recent studies have garnered thousands of cross-linked interactions, yet the field lacks an effective tool to compile experimental data or access the network and structural knowledge for these large scale analyses. We present XLinkDB 2.0 which integrates tools for network analysis, Protein Databank queries, modeling of predicted protein structures and modeling of docked protein structures. The novel, integrated approach of XLinkDB 2.0 enables the holistic analysis of XL-MS protein interaction data without limitation to the cross-linker or analytical system used for the analysis.
Availability and Implementation: XLinkDB 2.0 can be found here, including documentation and help: http://xlinkdb.gs.washington.edu/.
Contact: jimbruce@uw.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Large-scale structural characterization of interactions within proteins and protein complexes is a major goal of structural proteomics. Recent efforts have generated datasets featuring thousands of peptide-peptide links used to build networks of protein–protein interactions (PPIs). Yet, the full depth of these datasets is often unrealized as (i) the structural data inherent to chemical crosslinking-mass spectrometry (XL-MS) networks remains untapped on a network-wide scale, particularly for proteins lacking empirically derived structures and (ii) there is currently no centralized platform to host these large-scale networks. To address the lack of network level structural information we present XLinkDB 2.0 (hereafter XLinkDB), a web resource designed to integrate XL-MS data with databases of protein structures, enable generation of protein structure models for proteins lacking known structures, and generate automated protein–protein docking models. Protein structures are derived from empirical data when available (Protein Databank, PDB) or through high-quality structural prediction (Modeller). Similarly, structures of docked interacting proteins are queried from the PDB or docked with PatchDock. XLinkDB’s automation allows for the first, cross-platform, streamlined structural analysis of large XL-MS datasets and provides a curated, ever-growing reference database of cross-linked protein interactions for the general community.
2 Resource description
XLinkDB is implemented primarily within the PHP framework running on a Linux server (Apache 2.4.7, Ubuntu 14.0.2) supported by an SQL database system. For structural modeling and docking, XLinkDB uses the Integrative Modeling Platform (IMP; Russel, et al., 2012). The integration of the well-established, high-quality modeling (Modeller) and docking (PatchDock) modules within IMP, its invocation by Python (which provides flexibility for additional features), and relatively low runtime make IMP ideal for high-throughput analyses (Fig. 1) (Marti-Renom, et al., 2000; Schneidman-Duhovny, et al., 2005). The user input for XLinkDB is a tab-delimited text file with only six columns to identify cross-linked peptides and protein accessions: peptide sequence #1, protein UniProt accession #1, the relative position of the crosslinked residue within peptide #1, peptide sequence #2, protein UniProt accession #2 and the relative position of the crosslinked residue within peptide #2. XLinkDB has been preloaded with publicly-available, example XL-MS datasets from previous analyses. Importantly, as demonstrated by the publicly accessible datasets within XLinkDB, data from any XL-MS pipeline can be used as the input, including that generated by pLink (Yang, et al., 2012), XlinkX (Liu, et al., 2015), or ReACT (Chavez, et al., 2015; Chavez, et al., 2013; Navare, et al., 2015; Schweppe, et al., 2015; Weisbrod, et al., 2013).
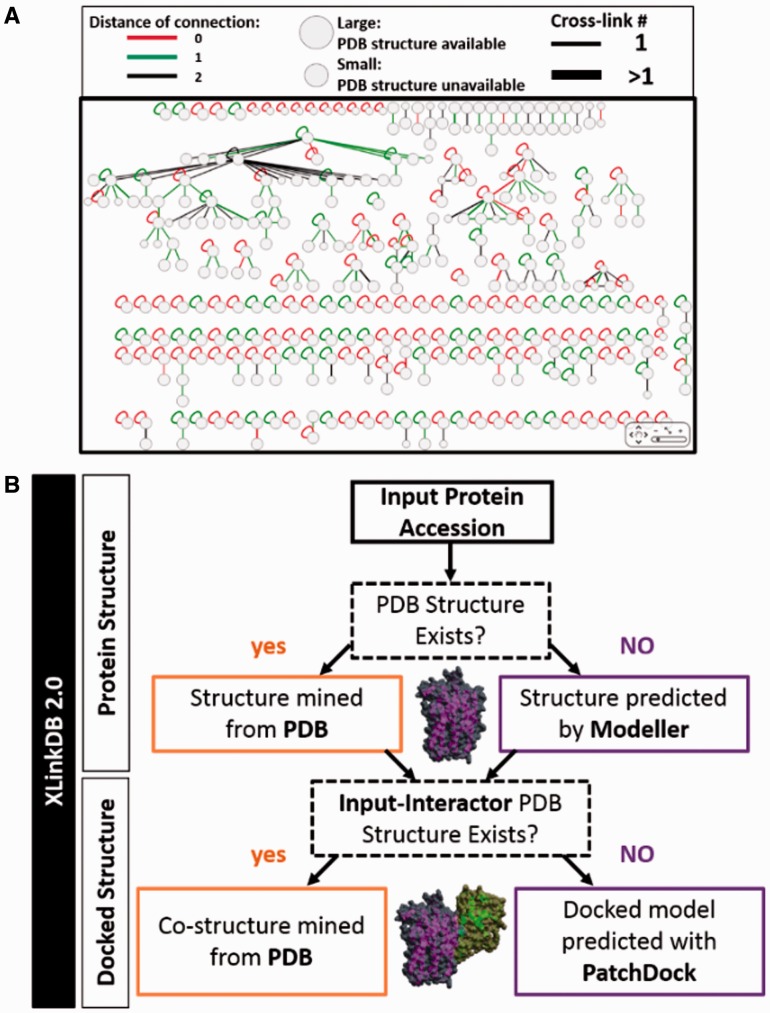
Fig. 1.
XlinkDB 2.0 Network and structural analysis. (A) Screenshot of cross-linked PPI network plot (Database: Ecoli2012_BingYang, (Yang, et al., 2012)). (B) Structure modeling workflow (Supplementary Fig. S2)
XLinkDB generates protein–protein interaction networks and queries the Uniprot database to determine the amino acid sequences for all input proteins (Supplementary Fig. S1). XLinkDB then queries the PDB to determine if a PDB structure exists for the protein and maps identified crosslinked residues to these structures. If no PDB structure exists XLinkDB generates a homology model using Modeller.
Through the PDB query or Modeller prediction, XLinkDB seeks to establish an associated PDB structure or structural model for each protein from the original input. For cross-linked relationships identified between two proteins (multi-protein complexes) XLinkDB will automatically query the PDB again, this time searching for co-protein structures that contain both input proteins. If no co-protein structures are identified, XLinkDB initiates IMP’s integrative docking to determine interaction interfaces of these proteins using sites of crosslinking as empirical distance constraints. Finally, XLinkDB retains the top-ranked docked model for each protein complex. Modeling and docking jobs are scheduled and initiated via a Jenkins server and job queueing is performed via a centralized Sun Grid Engine. In this manner, the docking and modeling platforms are maintained separately from network analysis. All associated structures, models, docked models and crosslinked residues therein can be visualized using Jmol directly within the user’s web browser.
3 Application and use cases
XLinkDB is able to generate large network plots and thousands of models based on minimal user input (Fig. 1). The automated, multi-dimensional data output allows for efficient analysis of XL-MS datasets. Considering both public and private data, XLinkDB currently contains 57 950 cross-linked peptide relationships and a total of 4872 protein structures. 1011 of these protein structures are from the PDB (including co-protein structures). Of the remaining 3861, XLinkDB 2.0 generated 1942 Modeller structures and 1919 docked protein models. Importantly, the database of structures will continue to grow with additional datasets being incorporated and initiating the structural workflow.
To further demonstrate the utility of XLinkDB, we uploaded datasets from three distinct XL-MS pipelines, all using different cross-linking chemistry, instrumentation and analytical tools. These included: pLink (Yang, et al., 2012), XlinkX (Liu, et al., 2015) and ReACT(Chavez, et al., 2015; Chavez, et al., 2013; Navare, et al., 2015; Schweppe, et al., 2015; Weisbrod, et al., 2013) (Supplementary Fig. S1). These datasets are available on the XLinkDB website as use cases, and for general data mining and network analysis.
4 Conclusions
The current XLinkDB server is the first integrated pipeline to analyze protein crosslinking data in the context of proteome scale structural knowledge, both from empirically evidenced structures and protein structural predictions. XLinkDB offers a platform that is flexible, adaptable and freely available. Moreover, because models and interactions can be input irrespective of cross-linking platform or linker chemistry, XLinkDB can integrate multiple data types. XLinkDB takes advantage of well-established modeling and docking protocols to automate time-intensive analytical bottlenecks. The prediction of almost 4000 protein structures and docked models highlights the broad utility of XLinkDB’s structural insights.
Finally, several large-scale protein cross-linking studies have been uploaded to XLinkDB from multiple species, including: Homo sapiens (Chavez, et al., 2013; Liu, et al., 2015; Schweppe, et al., 2015), Pseudomonas aeruginosa (Navare, et al., 2015), Escherichia coli (Weisbrod, et al., 2013; Yang, et al., 2012) and Acinetobacter baumannii (Schweppe, et al., 2015). These datasets, along with their structural and docking predictions, provide the first compendium of high-throughput XL-MS experiments and easy access to a wealth of network-level XL-MS information (Supplementary Figs S1–S4). This compendium is a novel, ever-growing resource for integrated structural and XL-MS data that will be constantly enriched by (i) further structure submissions to the PDB, (ii) integration of XL-MS knowledge for structure selection (Schweppe, et al., 2016) and (iii) future improvements to modeling/docking software.
Supplementary Material
Funding
This study was supported by: National Institutes of Health grants U19-AI107775-01, R01-AI101307-02, R01-HL110879-04, R01-GM086688-06, R01-GM097112-04; and in part by the University of Washington’s Proteomics Resource (UWPR95794).
Conflict of Interest: none declared.
References
- Chavez J.D. et al. (2015) Quantitative interactome analysis reveals a chemoresistant edgotype. Nat. Commun., 6, 7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez J.D. et al. (2013) Protein interactions, post-translational modifications and topologies in human cells. Mol. Cell. Proteomics, 12, 1451–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F. et al. (2015) Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat. Methods, 12, 1179–1184. [DOI] [PubMed] [Google Scholar]
- Marti-Renom M.A. et al. (2000) Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct., 29, 291–325. [DOI] [PubMed] [Google Scholar]
- Navare A.T. et al. (2015) Probing the protein interaction network of Pseudomonas aeruginosa cells by chemical cross-linking mass spectrometry. Structure, 23, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel D. et al. (2012) Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol., 10, e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D. et al. (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res., 33, W363–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweppe D.K. et al. (2015) Host-microbe protein interactions during bacterial infection. Cell Chem. Biol., 22, 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweppe D.K. et al. (2016) XLmap: an R package to visualize and score protein structure models based on sites of protein cross-linking. Bioinformatics, 32, 306–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrod C.R. et al. (2013) In vivo protein interaction network identified with a novel real-time cross-linked peptide identification strategy. J. Proteome Res., 12, 1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B. et al. (2012) Identification of cross-linked peptides from complex samples. Nat. Methods, 9, 904–906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.