Abstract
Motivation: How chromatin folds in three-dimensional (3D) space is closely related to transcription regulation. As powerful tools to study such 3D chromatin conformation, the recently developed Hi-C technologies enable a genome-wide measurement of pair-wise chromatin interaction. However, methods for the detection of biologically meaningful chromatin interactions, i.e. peak calling, from Hi-C data, are still under development. In our previous work, we have developed a novel hidden Markov random field (HMRF) based Bayesian method, which through explicitly modeling the non-negligible spatial dependency among adjacent pairs of loci manifesting in high resolution Hi-C data, achieves substantially improved robustness and enhanced statistical power in peak calling. Superior to peak callers that ignore spatial dependency both methodologically and in performance, our previous Bayesian framework suffers from heavy computational costs due to intensive computation incurred by modeling the correlated peak status of neighboring loci pairs and the inference of hidden dependency structure.
Results: In this work, we have developed FastHiC, a novel approach based on simulated field approximation, which approximates the joint distribution of the hidden peak status by a set of independent random variables, leading to more tractable computation. Performance comparisons in real data analysis showed that FastHiC not only speeds up our original Bayesian method by more than five times, bus also achieves higher peak calling accuracy.
Availability and Implementation: FastHiC is freely accessible at:http://www.unc.edu/∼yunmli/FastHiC/
Contacts: yunli@med.unc.edu or ming.hu@nyumc.org
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
The spatial organizations of chromosomes play a critical role in transcription regulation. In particular, regulatory elements such as enhancers, often contact with the promoters of targeted genes by forming long-range DNA looping. Understanding such 3D chromatin conformation provides novel insights into the regulation mechanisms of gene expression (Gorkin et al., 2014). The recently developed Hi-C technologies enable a genome-wide measurement of chromatin interaction (Lieberman-Aiden et al., 2009). In the Hi-C data, the contact frequency between any two chromatin loci is measured by the number of paired-end reads spanning across them. Higher read count indicates more frequent chromatin interaction and closer spatial proximity.
The rapid accumulation of Hi-C data with high sequencing depth enables the study of chromatin interaction at the unprecedented resolution (Jin et al., 2013; Rao et al., 2014). However, the detection of biologically meaningful long-range chromatin interactions poses great challenges. Most existing methods (Ay et al., 2014; Jin et al., 2013; Rao et al., 2014) assume that the chromatin interaction frequencies for different loci pairs are independent, which is clearly invalid. If two loci show high interaction frequency, their adjacent loci will be more likely to interact with each other. To fill in this gap, we have recently developed a novel hidden Markov random field based Bayesian (HMRFBayes) method to explicitly model the spatial dependency among adjacent loci (Xu et al., 2016). By borrowing information across neighborhood loci, HMRF achieves substantially improved robustness and enhanced statistical power.
While extremely promising, HMRFBayes is based on a Bayesian framework which requires intensive computation. The key challenge is to efficiently and effectively infer the dependency structure underlying the hidden peak status. To address this challenge, we developed a novel algorithm named FastHiC, which approximates the joint distribution of the hidden peak status by a set of independent random variables. Under such approximation, we adopted an EM algorithm for statistical inference. Compared with HMRFBayes, FastHiC not only speeds up by more than five times, but also achieves higher peak calling accuracy.
2 Statistical model
We use the same HMRF model as in our previous work (Xu et al., 2016). Our goal is to detect biologically meaningful chromatin interactions among loci. Let and represent the observed and expected chromatin contact frequency between loci and , respectively . Here, is known based on the pre-specified background model (Ay et al., 2014; Jin et al., 2013). Let be the hidden peak status: indicates a biologically meaningful interaction, while indicates a random collision. We further assume that follows a negative binomial distribution with mean and over-dispersion . Here, is the signal-noise-ratio of the peak over the background. In the HMRF model, we assume that ’s are conditionally independent given the hidden peak status , where follows an Ising distribution (Besag, 1974):
Here, is the parameter accounting for spatial dependency. Larger indicates higher spatial dependency. is the normalization constant without explicit expression form.
The key challenge is the efficient and effective inference of the spatial dependency parameter . In our previous work (Xu et al., 2016), we used the pseudo-likelihood (PL) to approximate the Ising distribution, and devised a Gibbs sampler for statistical inference.
However, in the PL approximation, the neighborhood of each loci pair can still fluctuate, resulting in intractable computation of . To solve this problem, we developed a novel algorithm named FastHiC, which uses the simulated field to approximate the joint distribution of the peak status implicated by the Ising distribution (Celeux et al., 2003), leading to a modified pseudo-likelihood (MPL):
This MPL approximates the Ising distribution by a set of independent random variables, enabling tractable computation of . FastHiC then adopts an EM algorithm for inference. Details of the simulated field approximation and EM algorithm can be found in Supplementary Material Section S1.
3 Results
We first conducted simulation study to compare the performance of FastHiC with HMRFBayes (Supplementary Material Section S2). These two methods achieved comparable statistical efficiency in parameter estimations (Supplementary Table S1) and peak calling accuracy (Supplementary Fig. S1 and Table S2). Noticeably, FastHiC ran more than five times faster than HMRFBayes (Supplementary Table S3), due to the novel implementation of simulated field approximation.
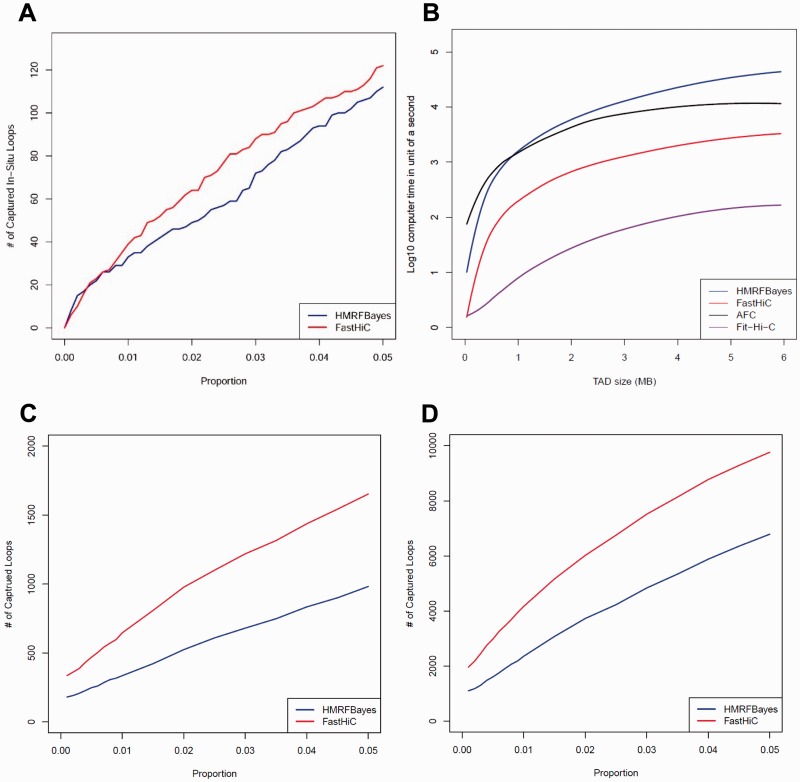
Next, we re-analyzed the Hi-C data in human IMR90 cells (Jin et al., 2013) where 2262 topological associated domains (TADs) were identified (Dixon et al., 2012). We analyzed each TAD separately to detect intra-TAD chromatin interactions at 4Kb resolution. We didnot analyze inter-TAD chromatin interactions because low sequencing depth for inter-TAD reads (the average number of intra-TAD and inter-TAD reads are 58.91 and 1.57, respectively). Since we have shown that HMRFBayes (Xu et al., 2016) outperforms AFC (Jin et al., 2013) and Fit-Hi-C (Ay et al., 2014), in this work, we only compared the performance of FastHiC and HMRFBayes (Supplementary Table S4). We did not compare with HiCCUPS (Rao et al., 2014) since its software is not publicly available. Overall, FastHiC and HMRFBayes obtained highly similar peak calling results. The Spearman correlation coefficient of peak probabilities between FastHiC and HMRFBayes within each TAD has median 0.934 and standard deviation 0.121. In addition, we compared the peak calling results from FastHiC and HMRFBayes with the chromatin loops identified from the in situ Hi-C data (Rao et al., 2014). Figure 1A shows that peaks identified by FastHiC have slightly higher overlap with chromatin loops than peaks identified by HMRFBayes. Overall, FastHiC and HMRFBayes achieved highly similar peak calling accuracy in real Hi-C data in human IMR90 cells.
Fig. 1.
(A) The overlap between in-situ Hi-C loops (Rao et al., 2014) and peaks identified by FastHiC and HMRFBayes. (B) Computational time of four peak callers. (C) The overlap between ChIA-PET loops (Ji et al., 2015) and peaks identified by FastHiC and HMRFBayes in primed H1 cells. (D) The overlap between ChIA-PET loops (Tang et al., 2015) and peaks identified by FastHiC and HMRFBayes in GM12878 cells
Next, we compared the computational time of four different peak callers with different TAD sizes (Fig. 1B). In general, the running time increases quadratically with the TAD size. For all 2262 TADs, the average running time for FastHiC, HMRFBayes, AFC and Fit-Hi-C are 428, 4134, 2605 and 19 s, respectively. FastHiC runs much faster than HMRFBayes and AFC. Although Fit-Hi-C runs faster than FastHiC, we have showed that the HMRF-based approach outperformed Fit-Hi-C in peak calling accuracy (Xu et al., 2016). Compared with Fit-Hi-C, we believe that FastHiC achieves a reasonable balance between speed and accuracy.
To further compare the performance of FastHiC and HMRFBayes, we analyzed two additional Hi-C datasets in human H1 embryonic stem cells (Dixon et al., 2015) and human GM12878 lymphoblastic cells (Selvaraj et al., 2013). We detected intra-TAD chromatin interactions at 40 kb resolution. To evaluate peak calling accuracy, we compared the peak calling results from FastHiC and HMRFBayes with the recently published ChIA-PET data on H1 (Ji et al., 2015) and GM12878 cells (Tang et al., 2015). We found that peaks identified from FastHiC show much higher overlap with chromatin loops detected from ChIA-PET data than those identified from HMRFBayes, in both H1 (Fig. 1C and Supplementary Fig. S2) and GM12878 cells (Fig. 1D). These results demonstrate that FastHiC achieves higher peak calling accuracy than HMRFBayes.
In addition, we explored modeling the dependency structure with broader neighborhood, but neither simulation studies nor real data analysis showed performance improvement (Supplementary Material Section S4).
4 Conclusion
In summary, we have developed FastHiC, a fast and accurate algorithm to detect long-range chromatin interactions from Hi-C data. FastHiC utilized the same hidden Markov random field model as in our previous work (Xu et al., 2016), but with a novel implementation of the simulated field approximation, leading to more than five times speed-up and higher peak calling accuracy. We believe that FastHiC has potential to become a useful tool in studying chromatin spatial organization.
Supplementary Material
Funding
This research was supported by the National Institute of Health grants R01-HG006292 and R01-HG006703 (awarded to YL), and 1U54DK107977-01 (awarded to MH).
Conflict of Interest: none declared.
References
- Ay F. et al. (2014) Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res., 24, 999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besag J. (1974) Spatial interaction and the statistical analysis of lattice systems (with Discussion). J. R. Stat. Soc. Ser. B, 36, 192–236. [Google Scholar]
- Celeux G. et al. (2003) EM procedures using mean field-like approximations for Markov model-based image segmentation. Pattern Recognit., 36, 131–144. [Google Scholar]
- Dixon J.R. et al. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature, 485, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J.R. et al. (2015) Chromatin architecture reorganization during stem cell differentiation. Nature, 518, 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorkin D.U. et al. (2014) The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell, 14, 771–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X. et al. (2015) 3D chromosome regulatory landscape of human pluripotent cells. Cell Stem Cell, 18, 262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F. et al. (2013) A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature, 503, 290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E. et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science, 326, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S.P. et al. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell, 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj S. et al. (2013) Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nat. Biotechnol., 31, 1111–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z. et al. (2015) CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell, 163, 1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. et al. (2016) A hidden Markov random field based Bayesian method for the detection of long-range chromosomal interactions in Hi-C Data. Bioinformatics, 32, 650–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.