Abstract
Summary: Modern biotechnical research is becoming increasingly reliant on computational structural modeling programs to develop novel solutions to scientific questions. Rosetta is one such protein modeling suite that has already demonstrated wide applicability to a number of diverse research projects. Unfortunately, Rosetta is largely a command-line-driven software package which restricts its use among non-computational researchers. Some graphical interfaces for Rosetta exist, but typically are not as sophisticated as commercial software. Here, we present InteractiveROSETTA, a graphical interface for the PyRosetta framework that presents easy-to-use controls for several of the most widely used Rosetta protocols alongside a sophisticated selection system utilizing PyMOL as a visualizer. InteractiveROSETTA is also capable of interacting with remote Rosetta servers, facilitating sophisticated protocols that are not accessible in PyRosetta or which require greater computational resources.
Availability and implementation: InteractiveROSETTA is freely available at https://github.com/schenc3/InteractiveROSETTA/releases and relies upon a separate download of PyRosetta which is available at http://www.pyrosetta.org after obtaining a license (free for academic use).
Contact: bystrc@rpi.edu
1 Introduction
Computational molecular modeling has exhibited significant advances over the past several years, leading to its more widespread use in academic and commercial research for novel structural engineering applications. One significant software package is the Rosetta protein modeling suite (Leaver-Fay, 2011), which provides access to a wide array of structural modeling tools which have been successfully used for protein engineering, including protein design (Kuhlman, 2003), structure prediction (Raman et al., 2009), loop modeling (Mandell et al., 2009) and protein docking (Chaudhury, 2008), among other tools. Unfortunately, the main Rosetta software package has a command-line interface, intended for use on Unix operating systems, which limits its usability mainly to researchers with extensive C++ and Unix operating system experience. One significant improvement in Rosetta accessibility was the development of the PyRosetta package (Chaudhury et al., 2010), which provides Python bindings to several of the main Rosetta protocols and permits the user to design their own customized protocols using these bindings in Python scripts. Later, visualization capabilities were added to PyRosetta by synchronizing Rosetta “pose” structures with the PyMOL Molecular Visualization System (www.pymol.org) via the so-called “PyMOL movers” (Baugh et al., 2011; DeLano, 2002) However, a central graphical interface that provides access to PyRosetta for researchers who are uncomfortable with Python scripting is still a major concern. Two notable graphical interfaces exist for PyRosetta: FoldIT and the PyRosetta Toolkit. FoldIT provides an easy-to-use visualization and selection system, but there only exist a couple of basic Rosetta protocols such as minimization and basic protein design (Khatib et al., 2011). The PyRosetta Toolkit provides a graphical interface designed with Python’s standard Tkinter GUI library (Adolf-Bryfogle and Dunbrack, 2013), but a sophisticated visualization system is still missing. Recently, RosettaDiagrams (www.rosettadiagrams.org) provides a user-friendly interface for RosettaScripts (Fleishman et al., 2011). Rosie (Rosetta Online Server that Includes Everyone) provides access to computational resources for big Rosetta jobs (Lyskov et al., 2013).
We have developed a novel Rosetta GUI called ‘InteractiveROSETTA’. InteractiveROSETTA provides several user-friendly controls for interacting with many of the common PyRosetta protocols, including superimposition, energy minimization, fixed backbone protein design, kinematic closure (KIC) loop modeling, protein–protein docking, point mutations and parameterization tools for unrecognized PDB types. InteractiveROSETTA does not require a familiarity with the structure and organization of Rosetta and provides easy access to its structural modeling applications.
2 InteractiveROSETTA features
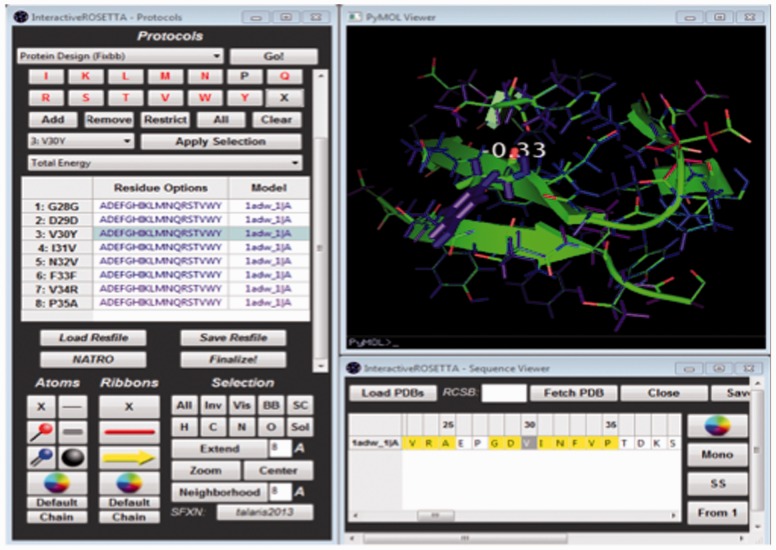
The InteractiveROSETTA GUI consists of three main windows: a PyMOL window for structure visualization and selection, a sequence viewer for displaying and selecting the primary sequences of loaded structures, and a protocol window that permits the user to execute various Rosetta protocols (Fig. 1). InteractiveROSETTA also makes extensive use of BioPython (Cock et al., 2009) for storing and manipulating structures efficiently and quickly. The GUI controls are generated using the wxPython library, which provides Python bindings for the C++ wxWidgets library. InteractiveROSETTA is a cross-platform and runs efficiently in 64-bit architectures running Windows 7/8 and Debian/RedHat-based Linux distributions. The Mac OSX version is still being optimized.
Fig. 1.
A hypothetical InteractiveROSETTA fixed backbone design session in Windows 7. InteractiveROSETTA simplifies setting up and viewing the results of a design by coloring residues according to Rosetta scores
2.1 Structure visualization and manipulation
As previously stated, structures are visualized using a PyMOL viewer which communicates with the underlying InteractiveROSETTA Python code. Segments of primary sequence can be selected in the sequence viewer which synchronously selects the equivalent tertiary structures in PyMOL. Selections in PyMOL are also synchronized to select the corresponding residue positions in the sequence viewer. Furthermore, the sequences can be colored according to secondary structure, as determined by DSSP, (Kabsch and Sander, 1983) or B-factor. InteractiveROSETTA also provides a number of tools to facilitate customizing atomic display and extending selections or views.
2.2 Running Rosetta protocols
The principal strength of InteractiveROSETTA lies in its ability to utilize the visualization and selection tools mentioned earlier to greatly simplify the process of designing Rosetta protocols. The protocols window contains a dropdown menu listing all of the currently supported Rosetta/PyRosetta protocols. Several proteins may be superimposed using one loaded structure as an immobile base model, either by Cα or backbone atoms. Superimposition also calls a MUSCLE executable (Edgar, 2004) to align the primary sequences into a multiple sequence alignment for the user’s convenience. Energy minimization allows the user to select regions of the structures to minimize along backbone and/or sidechain atoms. Fixed backbone design permits the user redesign part or all of a structure’s primary sequence and provides several controls for setting up the design parameters. Loop modeling with KIC is also supported which allows the researcher to remodel existing loops or create new loops de novo given a desired primary sequence of the novel loop. Basic protein–protein docking is also supported, which will accept various structural chains as static and movable structures that will be docked together given a specified number of coarse and refined models to generate. A simple comparative structure prediction protocol is provided, although this protocol currently relies upon the Robetta server (Chivian et al., 2003) to generate fragment files for the structure prediction. A researcher may also design proteins using the multi-state design (MSD) protocol. MSD typically requires enumeration of several different files so the controls provided by InteractiveROSETTA greatly simplify setting up MSD jobs. Finally, a residue and ligand creator is provided to easily parameterize unrecognized residue types into a Rosetta “params” file.
3 Outlook
InteractiveROSETTA should provide a simple to use interface for non-computational researchers to utilize Rosetta more fully or for the first time for their own applications. In addition, InteractiveROSETTA is extremely amenable to community development of novel protocols. Separate Python scripts which extend the “ScrolledPanel” widget can be added to the “scripts” directory to add new protocols to InteractiveROSETTA’s database.
Acknowledgements
We acknowledge all the contributions of the Rosetta developers on RosettaCommons for all of their tireless work to put all of these tools in Rosetta, and Christoph Gohlke, University of California, Irvine, for providing many useful Python modules for Windows.
Funding
InteractiveROSETTA was funded through the National Institute of Health Grant R01 GM099827.
Conflicts of Interest: none declared.
References
- Adolf-Bryfogle J., Dunbrack R.L., Jr. (2013) The PyRosetta toolkit: a graphical user interface for the Rosetta software suite. PLoS One, 8, e66856–e66856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh E.H., et al. (2011) Real-time PyMOL visualization for Rosetta and PyRosetta. PLoS One, 6, e21931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury S., Gray J.J. (2008) Conformer selection and induced fit in flexible backbone protein–protein docking using computational and NMR ensembles. J. Mol. Biol., 381, 1068–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury S., et al. (2010) PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics, 26, 689–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivian D., et al. (2003) Automated prediction of CASP-5 structures using the Robetta server. Proteins, 53, 524–533. [DOI] [PubMed] [Google Scholar]
- Cock P.J.A., et al. (2009) Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics, 25, 1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W.L. (2002) Pymol: An open-source molecular graphics tool. CCP4 Newsletter On Protein Crystallography, 40, 82–92. [Google Scholar]
- Edgar R.C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics, 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishman S.J., et al. (2011). RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS One, 6, e20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Sander C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Khatib F., et al. (2011) Algorithm discovery by protein folding game players. Proc. Natl Acad. Sci. U. S. A., 108, 18949–18953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman B., et al. (2003) Design of a novel globular protein fold with atomic-level accuracy. Science, 302, 1364–1368. [DOI] [PubMed] [Google Scholar]
- Leaver-Fay A., et al. (2011) Rosetta3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol., 487, 545–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyskov S., et al. (2013) Serverification of molecular modeling applications: the Rosetta Online Server that Includes Everyone (ROSIE). PLoS One, 8, e63906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell D.J., et al. (2009) Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat. Methods, 6, 551–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S., et al. (2009) Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins, 77, 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]