We read with interest the comments raised by Gong et al.1 on our recently published study,2 and appreciate these important and critical remarks.
We agree that contamination is a potential important problem with polymerase chain reaction (PCR)-based work. To avoid the possibility of contamination, we confirmed our data before completion of our manuscript. We performed the PCR experiment using new experimental reagents at a different laboratory. We also repeated the PCR to detect the fusion using two different set of primers, the new primer set producing longer amplicons than the old primer set. All these efforts produced identical results, confirming that our samples were not contaminated by PCR amplicons.
As the direct fusion of one exon of CTLA4 to an exon of CD28 is less likely than fusion breakpoints within the introns, we confirmed direct fusion of the two exons at the genomic DNA level with other experiments before submitting our manuscript. As shown in Figure 1, direct fusion of exon 3 of CTLA4 to exon 4 of CD28 was confirmed by PCR using primers from intron 2 of CTLA4 and the 3’ region of CD28 using genomic DNA from formalin-fixed, paraffin-embedded samples. PCR products were validated by Sanger sequencing. DNA from normal blood was used as a control as well as no template controls. These data confirm that the fusion is present at the DNA level and is not the result of contamination.
Figure 1.
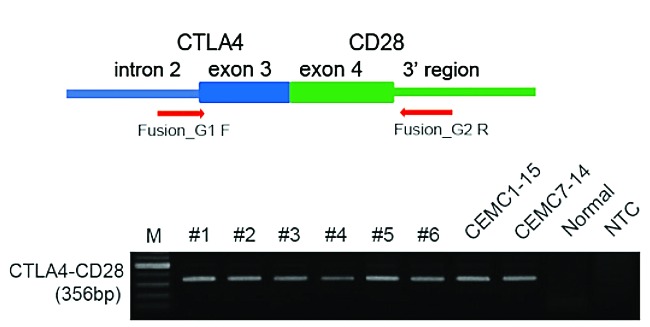
Validation of the direct CTLA4-CD28 fusion cases. Direct fusion of 3rd exon of CTLA4 and 4th exon of CD28 were confirmed by PCR using primers from intron2 of CTLA4 and 3’ region of CD28 using genomic DNA from FFPE samples. Arrows indicate the approximate positions of oligonucleotide primers on the CTLA4-CD28 fusion gene. PCR products were validated by Sanger sequencing. DNA from normal blood was used as control and NTC indicates the no template control.
We also tried to perform fluorescent in situ hybridization experiments. However it was difficult to design probes which can distinguish the fusion segment because the CD28 and CTLA4 genes are located very close to each other. As shown in our manuscript2 in Online Supplementary Figure S5, the result was not helpful for delineating the fusion signal from the normal signal.
Footnotes
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Gong Q, Wang C, Rohr J, Feldman A, Chan WC, McKeithan TW. Comment on: Frequent CTLA4-CD28 gene fusion in diverse types of T-cell lymphoma by Yoo et al. Haematologica. 2016;101(6)000–000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoo HY, Kim P, Kim WS, et al. Frequent CTLA4-CD28 gene fusion in diverse types of T-cell lymphoma. Haematologica. 2016;101(6)000–000. [DOI] [PMC free article] [PubMed] [Google Scholar]


