In addition to the hallmark translocations involving the MYC oncogene and immunoglobulin loci,1 Burkitt lymphomas (BL) frequently carry mutations in the inhibitor of DNA binding 3 (ID3) gene.2,3 Genetic aberrations of ID3 in BL comprise a spectrum of mono- and bi-allelic structural and point mutations.3 ID3 acts as negative transcriptional regulator by sequestering transcription factors with basic helix-loop-helix motifs. Mutated ID3 attenuates this regulatory interaction.4,5 ID3 and its interaction partner, TCF3, are involved in controlling cell cycle progression and survival pathways through tonic B-cell signaling.6,7
ID3 mutations occur in 34–68% of BL but are rare in diffuse large B-cell lymphomas (DLBCL).2,3,7 Interestingly, the incidence of ID3 mutations was reported to be higher in B-cell lymphomas, unclassifiable, with features intermediate between DLBCL and BL,8 than in DLBCL. However, in the quoted study a molecular diagnosis was not available and the diagnosis of an “intermediate” lymphoma was based on histopathological features only.8,9
Mutation-specific immunohistochemistry is a valuable diagnostic tool,10 and we used it to test six anti-ID3 antibodies for their ability to detect ID3 mutational variants in molecularly defined BL (mBL), “intermediate” lymphomas and non-mBL lymphomas11 (Online Supplementary Table S1).
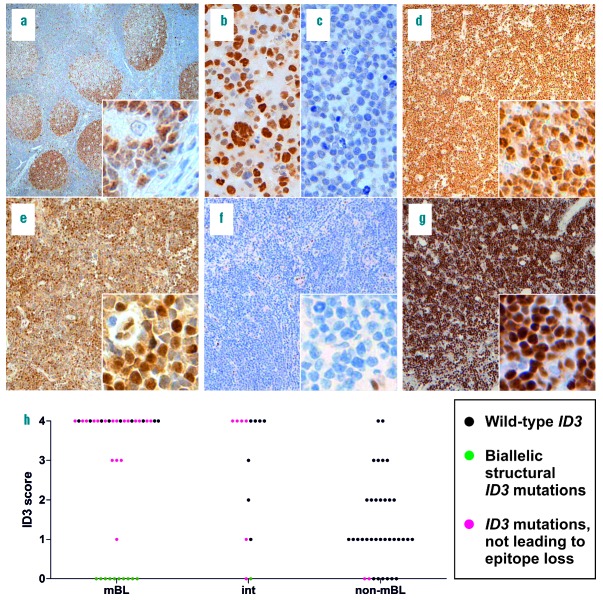
First, we tested all six antibodies on formalin-fixed and paraffin-embedded tonsil tissue using immunohistochemistry. ID3 has been reported to be strongly expressed in the dark zone and less intensively in the light zone of germinal centers.12 The expected staining pattern was only observed with clone 17-3 (BioCheck Inc., Foster City, USA) (Figure 1), but not for the other antibodies tested (Online Supplementary Figure S1). To determine whether clone 17-3 shows a mutation-specific staining pattern, selected wild-type (wt) and point-mutated ID3 cell lines and lymphoma specimens were tested by immunohistochemistry (Online Supplementary Figure S1) and by western blotting (Online Supplementary Figure S2). As expected, clone 17-3 showed no reactivity in BL cell lines or mBL biopsies with homozygous loss of ID3 (Figure 1; Online Supplementary Figures S1 and S2; Online Supplementary Table S2). The other five antibodies positively stained cell lines and/or biopsies by immunohistochemistry despite a homozygous deletion of the ID3 locus and were not, therefore, used further (Online Supplementary Figure S1; Online Supplementary Table S2).
Figure 1.
Immunohistochemistry for ID3 and ID3 mutation distribution among mBL, “intermediate” lymphomas, and non-mBL. (a–g). ID3 immunohistochemistry of formalin-fixed, paraffin-embedded sections of tonsil, BL and BL cell lines; original magnification of tonsil 50×, inlet 400×, cell lines 400×, cases 100×, inlets 400×. (a–g) were stained with clone 17-3. (a) Reactive tonsil, pronounced ID3 distribution in the dark zone of germinal centers; (b) BL cell line EB-1, wt ID3; (c) BL cell line BL-41, biallelic stop gain, loss of ID3 amino acids 69-109; (d) case 34, mBL, wt ID3; (e) case 17; mBL, two ID3 point mutations; (f) case 2, mBL, homozygous loss of ID3 C-terminal domains; (g) case 15, mBL, stop gain and splice site mutation, the latter without structural consequences; (h) Scatter plot of the ID3 immunohistochemical scoring based on percentages of ID3-positive tumor cells: 0=0%, 1=1–25%, 2=26–50%, 3=51–75%, and 4=76–100%. Each point is a case and the color codes illustrate the mutational status of ID3. Interpretable ID3 staining failed in two cases which are not included in the plot. BL: Burkitt lymphoma; int: intermediate; mBL: molecular Burkitt lymphoma; non-mBL: non-molecular Burkitt lymphoma; wt: wild-type.
The Online Supplementary Data contains more detailed information on the materials and methods, cell lines and cases.
Mutation-sensitive ID3 immunohistochemistry using clone 17-3 was performed on 89 formalin-fixed, paraffin-embedded lymphoma biopsies. The conventionally assigned diagnoses based on histomorphological and immunophenotypic features according to the current World Health Organization classification8 were as follows: BL (23/89), Burkitt leukemia (1/89), atypical BL (15/89), DLBCL (27/89), high-grade B-cell non-Hodgkin lymphoma (3/89), B-cell lymphomas, unclassifiable (2/89), follicular lymphoma grade 1-3a (10/89), transformed follicular lymphoma grade 3a/b/DLBCL (4/89), primary mediastinal B-cell lymphoma (2/89), primary central nervous system DLBCL (1/89), and post-transplant lymphoproliferative disease with features of DLBCL (1/89) (Online Supplementary Table S3). All cases were molecularly studied in either the MMML (n=41) or ICGC MMML-Seq (n=43) projects or both (n=5). Thus, the molecular classification based on gene expression analysis as well as the ID3 mutation status based on whole genome and/or Sanger sequencing were available.3,11,13,14 In detail, according to a defined gene expression signature, the so-called mBL signature index, which reflects the probability that a case resembles a BL, all cases were assigned their specific molecular diagnosis. In accordance with Hummel et al.11 cases with a mBL signature index score higher than 0.95 were classified as mBL (38/89), cases with an intermediate mBL signature index score between 0.05 and 0.95 as “intermediate” lymphomas (14/89), and cases with a mBL signature index score lower than 0.05 as non-mBL (36/89); one nodal manifestation of BL leukemia was not assigned a molecular diagnosis (Online Supplementary Table S3; Online Supplementary Methods).
ID3 expression in mBL showed a biphasic pattern. Almost all BL displayed either high ID3 expression scores (>50% positive lymphoma cells) or no expression (Figure 1h and Online Supplementary Table S3). mBL with wt, monoallelic point or monoallelic structural ID3 mutations (deletions, insertions or frameshifts) displayed ID3 immunoreactivity by immunohistochemistry (27/27, 100%; interpretable staining failed in 2 mBL). All mBL in our series with lack of ID3 immunoreactivity (10/10, 100%) harbored complex biallelic structural ID3 mutations (e.g. biallelic frameshifts). Sequence analyses predicted a loss of the C-terminal ID3 epitope of clone 17-3 (Figure 1; Online Supplementary Table S3).
We detected a broad spectrum of ID3 immunoreactivity in non-mBL, ranging from no expression to high expression (Figure 1h and Online Supplementary Table S3). However, a high level of ID3 expression (>50% positive lymphoma cells) was rare in non-mBL (6/36, 17%). ID3 expression in non-mBL seems to be independent of the mutational status, since none of the ID3-negative non-mBL harbored biallelic structural ID3 variants (0/8, 0%). Monoallelic mutations of ID3 were detected in only two cases of non-mBL (2/36, 6%) and neither of these lymphomas showed ID3 immunoreactivity (Online Supplementary Table S3).
Like non-mBL, molecularly defined “intermediate” lymphomas showed a broad spectrum of ID3 expression (Figure 1 and Online Supplementary Table S3). High ID3 immunoreactivity (>50% positive lymphoma cells) was as frequent as in mBL (9/14, 64% versus 27/37, 73%, respectively) and more frequent than in non-mBL (6/36, 17%). Complete lack of ID3 immunoreactivity was observed in only 2/14 “intermediate” lymphomas which both harbored ID3 structural mutations. One was a pediatric case with a biallelic ID3 frameshift insertion and a splice site alteration. Unfortunately, we were not able to assess whether the lesion is biallelic in the second case (Online Supplementary Table S3). Interestingly, none of the four double- or triple-hit lymphomas with MYC and either BCL2 and/or BCL6 translocations in our series had mutated ID3 nor lacked ID3 immunoreactivity (Online Supplementary Table S3).
Here we describe an anti-ID3 antibody which was the only one of six antibodies tested with high specificity for ID3 in immunohistochemistry and western blots. Clone 17-3 showed highly specific immunoreactivity for wt and point-mutated ID3 in mature aggressive B-cell lymphomas. We found that ID3 is highly expressed in mBL and the “intermediate” group of lymphomas, whereas it is not expressed, or only moderately expressed in non-mBL. Furthermore, both mBL and “intermediate” lymphomas are characterized by a high frequency of ID3 mutations whereas non-mBL are not. Moreover, mBL and “intermediate” lymphomas,11 mainly resembling BL, atypical BL and B-cell lymphomas, unclassifiable,8 show a complete lack of ID3 staining only when biallelic structural aberrations causing a loss of C-terminal domains of ID3 are present. In contrast, none of the non-mBL cases in this study, mainly resembling DLBCL, harbored biallelic structural ID3 mutations. ID3-negative non-mBL had either wt ID3 or harbored monoallelic ID3 locus deletions, so the lack of ID3 expression was not associated with a genetic loss of ID3 domains but probably due to transcriptional regulation. Thus, lack of ID3 staining in a mature aggressive B-cell lymphoma with features of BL can be regarded as an indicator of biallelic loss of ID3. Since lack of ID3 immunoreactivity also occurs in a small subset of non-mBL with wt or monoallelic structural ID3 aberrations, staining for ID3 currently seems to be of limited value in the differential diagnosis of lymphoma. A potential use in combination with other biomarkers needs to be determined in future studies.
Acknowledgments
The authors gratefully acknowledge O. Batic, C. Botz-von Drathen, T. Engel, C. Becher, and U. Schnaidt for their excellent technical assistance.
Footnotes
Funding: this research was supported by Deutsche Krebshilfe in the framework of the joint project Molecular Mechanisms in Malignant Lymphoma (MMML, 70-3173-Tr3) and the German Ministry for Education and Science (BMBF) in the framework of the International Cancer Genome Consortium MMML by Sequencing (ICGC MMML-Seq, 01KU1002A-J). RS and WK were supported by the BMBF within the framework of e:Bio Molecular Mechanisms in Malignant Lymphoma with MYC Deregulation project (MMML-MYC-SYS, 0316166B) and the BMBF within the framework of e:Med Molecular Mechanisms in Malignant Lymphoma – Demonstrator of the Personalized Medicine (MMML Demonstrator, 031A428D). RS and WK were also supported by the KinderKrebsInitiative (KKI), Buchholz, Holm-Seppensen e.V. RW is a recipient of a Christoph-Schubert-Award of the KKI, Buchholz, Holm-Seppensen. JR is supported by the Dr. Werner Jackstädt Foundation in the framework of a Junior Excellence Research Group (S134 - 10.100).
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA. 1982;79(24):7824–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmitz R, Young RM, Ceribelli M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richter J, Schlesner M, Hoffmann S, et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet. 2012;44(12):1316–1320. [DOI] [PubMed] [Google Scholar]
- 4.Murre C, McCaw PS, Vaessin H, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58(3):537–544. [DOI] [PubMed] [Google Scholar]
- 5.Benezra R, Davis RL, Lassar A, et al. Id: a negative regulator of helix-loop-helix DNA binding proteins. Control of terminal myogenic differentiation. Ann N Y Acad Sci. 1990;599:1–11. [DOI] [PubMed] [Google Scholar]
- 6.Dave SS, Fu K, Wright GW, et al. Molecular diagnosis of Burkitt’s lymphoma. N Engl J Med. 2006;354(23):2431–2442. [DOI] [PubMed] [Google Scholar]
- 7.Love C, Sun Z, Jima D, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44(12):1321–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swerdlow SH. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed World Health Organization classification of tumours Lyon, France: International Agency for Research on Cancer; 2008. [Google Scholar]
- 9.Momose S, Weißbach S, Pischimarov J, et al. The diagnostic gray zone between Burkitt lymphoma and diffuse large B-cell lymphoma is also a gray zone of the mutational spectrum. Leukemia. 2015;29(8):1789–1791. [DOI] [PubMed] [Google Scholar]
- 10.Capper D, Preusser M, Habel A, et al. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011;122(1):11–19. [DOI] [PubMed] [Google Scholar]
- 11.Hummel M, Bentink S, Berger H, et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Eng J Med. 2006;354(23):2419–2430. [DOI] [PubMed] [Google Scholar]
- 12.Schmitz R, Ceribelli M, Pittaluga S, Wright G, Staudt LM. Oncogenic mechanisms in Burkitt lymphoma. Cold Spring Harb Perspect Med. 2014;4(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kretzmer H, Bernhart SH, Wang W, et al. DNA methylome analysis in Burkitt and follicular lymphomas identifies differentially methylated regions linked to somatic mutation and transcriptional control. Nat Genet. 2015;47(11):1316–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]