ABSTRACT
Many viruses contain RNA elements that modulate splicing and/or promote nuclear export of their RNAs. The RNAs of the major human pathogen, hepatitis B virus (HBV) contain a large (~600 bases) composite cis-acting 'post-transcriptional regulatory element' (PRE). This element promotes expression from these naturally intronless transcripts. Indeed, the related woodchuck hepadnavirus PRE (WPRE) is used to enhance expression in gene therapy and other expression vectors. These PRE are likely to act through a combination of mechanisms, including promotion of RNA nuclear export. Functional components of both the HBV PRE and WPRE are 2 conserved RNA cis-acting stem-loop (SL) structures, SLα and SLβ. They are within the coding regions of polymerase (P) gene, and both P and X genes, respectively. Based on previous studies using mutagenesis and/or nuclear magnetic resonance (NMR), here we propose 2 covariance models for SLα and SLβ. The model for the 30-nucleotide SLα contains a G-bulge and a CNGG(U) apical loop of which the first and the fourth loop residues form a CG pair and the fifth loop residue is bulged out, as observed in the NMR structure. The model for the 23-nucleotide SLβ contains a 7-base-pair stem and a 9-nucleotide loop. Comparison of the models with other RNA structural elements, as well as similarity searches of human transcriptome and viral genomes demonstrate that SLα and SLβ are specific to HBV transcripts. However, they are well conserved among the hepadnaviruses of non-human primates, the woodchuck and ground squirrel.
KEYWORDS: Covariance model, hepadnavirus, RNA secondary structure, stem-loop α, stem-loop β
Hepatitis B virus (HBV) RNAs
HBV is a widespread causative agent of human liver disease, with an estimated 240 million people currently infected. HBV is the smallest DNA virus but has characteristics of both DNA and RNA viruses. Because of the restrictions of its small genome size (~3.2 kb), HBV has extensive overlapping open reading frames (ORFs) encoding its proteins (Fig. 1).1 HBV is the type member of the mammalian hepadnaviridae. Related viruses of birds form the avihepadnaviridae. HBV RNAs are transcribed by cellular RNA polymerase II. They begin at different transcription start sites but end at a common non-canonical polyadenylation signal.2 Remarkably, HBV has 3 major greater-than-genome length intronless transcripts, the pregenomic RNA (pgRNA), preC RNA (pcRNA), and long X RNA (lxRNA).3 Despite the 3 transcripts containing all the HBV ORFs, they are not all translated into all the major HBV proteins. The pgRNA encodes the core (C) protein and polymerase (P) protein. This pgRNA also serves as the template for the synthesis of partial-double stranded HBV genomic DNA by the P protein. The longer pcRNA encodes the precore (also known as preC and PC) protein that forms the HBeAg (hepatitis B e antigen) after post-translational cleavage events. The lxRNA encodes a transcriptional trans-activator, the X protein.3 This X protein is also encoded by the shorter X mRNA. Other HBV subgenomic transcripts are preS1, preS2 and S mRNAs that encode 3 surface (S) proteins, the structural proteins of the virus particles and subparticles (HBsAg).
Figure 1.
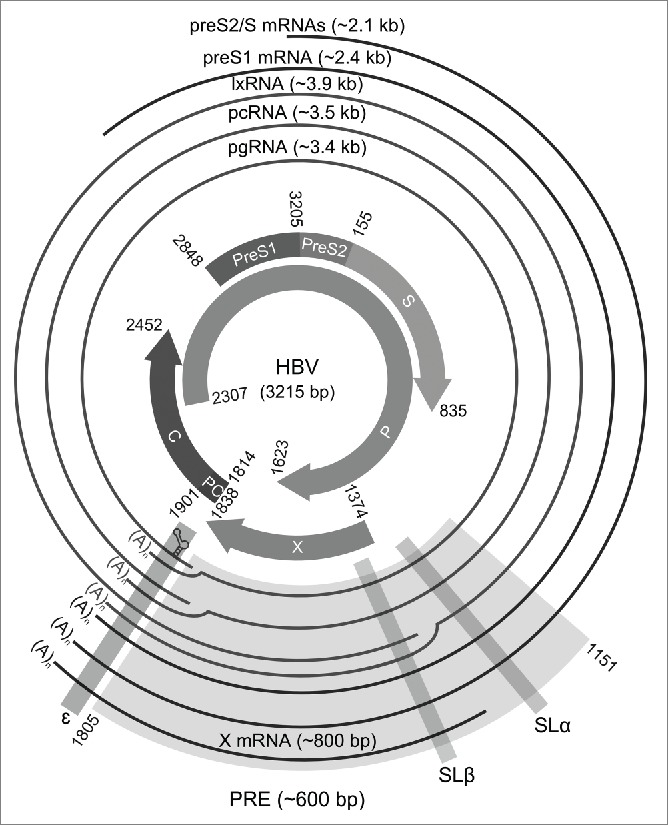
Map of the human HBV genome highlighting the multiple overlapping HBV ORFs and transcripts (RefSeq: NC_003977.1; GenBank:X04615.1). The inner ring represents the main HBV ORFs whereas the outer ring represents the main HBV transcripts. Most of the elements of the PRE (light gray shading) are present in all the main HBV transcripts. At least 2 of the 3 conserved RNA stem-loops (dark gray shadings), SLα, SLβ, and ɛ are present in all the main HBV transcripts.
The viral post-transcriptional regulatory element (PRE)
Notably, all the HBV transcripts contain most of the cis-acting PRE including 3 conserved RNA structures: the stem-loop α (SLα) and SLβ of the PRE at the 3′ end, and the epsilon element (ɛ) at the 3′ end or both ends (Fig. 1).4 The ɛ element located at the 5′ end of the pgRNA is necessary for the viral replication and packaging whereas, the ɛ element located at 3′ of the transcripts may have different functions and a slightly different conformation. The 5′ ɛ structure forms part of an RNA family ‘clan’.5 In contrast, SLα and SLβ are not present in avian hepadnavirus transcripts. The avian hepadnaviruses also lack the X ORF that harbors most of the PRE sequence.
The HBV PRE can enhance the expression of HBV transcripts and promote the nuclear export of unspliced HBV subgenomic RNAs via CRM1-independent pathway (see ref. 2 and 7 for reviews).2,4,6,7 In contrast, the PRE is not involved in the nuclear export of the full-length pgRNA that utilizes TAP/NXF1 dependent pathway.8 There are also spliced products of the pgRNA whose role is poorly understood.2,9 In the cytoplasm, HBV transcripts serve as mRNAs to produce HBV proteins.
The functions of SLα and SLβ
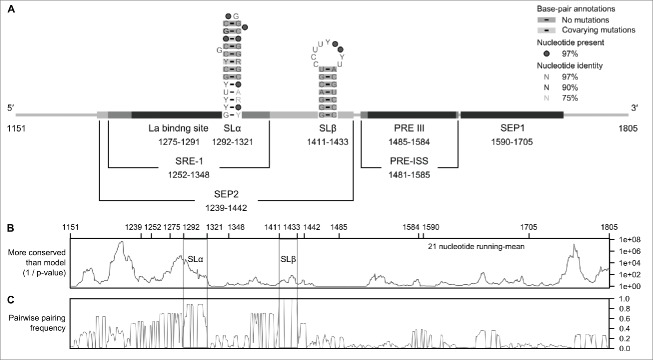
The HBV PRE (nucleotides 1151-1805; RefSeq: NC_003977.1; GenBank: X04615.1) contains 2 major nuclear export elements, recently named the sub-element of PRE 1 (SEP1) (nucleotides 1590-1705) and SEP2 (nucleotides 1239-1442) (Fig. 2A).10 The SEP1 element binds directly to the ZC3H18 protein and recruits the TREX complex and other cellular RNA export factors including ARS2, Acinus and Brr2 for nuclear export. The SEP2 can be further divided into 2 sub-elements, PREα and PREβ, these encompass SLα (nucleotides 1292-1321) and SLβ (nucleotides 1411-1433), respectively.4 However, SLα and SLβ can function independently in nuclear export. Minimum PRE fragments containing nucleotides 1278-1340 and 1347-1457 are sufficient to provide the nuclear export function of SLα and SLβ. Disrupting the base-pairs of the stem-loops using mutagenesis reduced their nuclear export activity significantly, which can be restored by compensatory mutations. In addition, SLα (that contains a weak 3′ splice site at position 1305) in conjunction with an upstream RNA stem-loop (that contains a weak 5′ splice site at position 458) can allow the preS2/S mRNAs to escape over-splicing.11 Thus, it has been postulated that cellular factors bind to SLα to prevent spliceosome assembly. However, the cellular splicing and nuclear export factors that bind to SLα and SLβ have yet to be determined. Additionally, the PRE promotes the splicing and stability of the pgRNA but not the nuclear export of this RNA.9,12 Intriguingly, the splicing regulatory element-1 (SRE-1) (nucleotides 1252-1348) of the PRE, which encompasses SLα, is an exonic splicing enhancer for the pgRNA. All of the observations indicate that SLα and SLβ are involved in the complex splicing and/or nuclear export of HBV transcripts.
Figure 2.
Functional elements and RNA secondary structures within the PRE. (A) A Map of the known functional elements and RNA secondary structures of HBV PRE. HBV (RefSeq: NC_003977.1; GenBank: X04615.1) PRE contains 2 major nuclear export elements, recently named SEP1 (nucleotides 1590-1705) and SEP2 (nucleotides 1239-1442).10 SEP1 contains the binding site for ZC3H18, a cellular factor for PRE-mediated nuclear export. SEP2 contains previously described SRE-1 (nucleotides 1252-1348),12 La binding site (nucleotides 1275-1291),34 SLα (nucleotides 1292-1321) and SLβ (nucleotides 1411-1433).4 The PRE intronic splicing silencer (PRE-ISS)35 region encompasses PRE III (nucleotides 1458-1584).36 The PRE III is a binding site for GAPDH and an alternative splicing regulator, polypyrimidine tract binding protein (PTB). PTB is also a cellular factor for a PRE-mediated nuclear export activity. The secondary structures of SLα and SLβ were inferred from NMR data (PDB: 2JYM)17 and/or previous mutation analyses.4,13 (B) Plot of conservation for an aligned, annotated, representative set of human HBV genotypes A-H (n = 32)16 and woodchuck hepadnavirus (n = 9) sequences. The p-value plot was produced by CDS-plotcon,30 in which the probability of the conservation in a 21 nucleotide running-mean is as great or greater than what is expected from a null model using a default "non-coding" setting. The gray boxes indicate the nucleotide positions for SLα and SLβ. (C) Plot of the frequencies of conserved paired sites (predicted RNA secondary structures) that was done using pairwise comparisons on the same set of aligned sequences (n = 41). The analysis was done using StructureDist in the SSE package,31 in which the maximum value of each frequency is equal to 1.
The woodchuck hepadnavirus PRE (WPRE) is more potent than HBV PRE, probably due to the presence of an additional third RNA element PREγ.13 Consequently, WPRE has been widely used in various vectors to promote expression of transgenes.14,15 The WPRE can functionally substitute for HBV PRE for the nuclear export of unspliced HBV subgenomic transcripts.13 As with HBV SLα, disrupting the base-pairs of the WPRE SLα using mutagenesis reduced the nuclear export activity significantly. However, there is insufficient evidence to support the presence of conserved RNA structure in the PREγ.
Covariance models for SLα and SLβ
Sequence conservation analyses of human HBV genotypes A-H (n = 32)16 and woodchuck hepadnavirus (n = 9) sequences show that SLα and SLβ are highly conserved RNA structures (Fig. 2B and 2C). Based on previous studies using nuclear magnetic resonance (NMR) (PDB: 2JYM)17 and mutagenesis,4,13 we created the seed alignments for SLα and SLβ (see the Stockholm files in the Supplemental Material). We then built the covariance models for SLα and SLβ using their seed alignments and compared the covariance models with other Rfam models using CMCompare.18 Relatively low pairwise-scores were obtained from the model comparisons, i.e., 12.1 or below, indicating that no similar RNA families were present in the database. However, it is notable that the comparison of SLα and SLβ models with other Rfam models returned a few stem-loop-like structures: human T-cell lymphotropic virus ribosomal frameshift site (RF01790) and retroviral readthrough element 2 (RF01092), respectively, with pairwise-scores of 9.8 and 10.7, respectively.
We next examined the sensitivity and specificity of the SLα and SLβ models using 41416 complete viral genomes (6495 mammalian hepadnaviruses and 34921 other viruses; retrieved from GenBank at 24 February 2016). Using the score thresholds obtained from 5-fold cross-validation, both SLα and SLβ models scored 100% specificity, with slightly lower sensitivities of 99.2% and 99.4%, respectively (see Table S1 and S2). Besides detecting the hepadnaviruses from human and woodchuck, the models also identified both the SLα and SLβ for non-human primate and ground squirrel hepadnaviruses. The searches did not return results above the score thresholds for the distantly related bat hepadnaviruses and some of the HBV (potentially non-viable) with deleted X gene or truncated and/or mutated P and/or X genes. Taken together, the results suggest that SLα and SLβ are evolutionary conserved RNA structures for most mammalian hepadnaviruses but not for bat hepadnaviruses, or the more distantly related avian hepadnaviruses.
On the other hand, searching human transcriptome (retrieved from RefSeq Release 74 at 1 March 2016) using the models did not return matches above the score thresholds, suggesting that similar RNA elements are not present in the host RNAs.
The constraints of P and X reading frames on SLα and SLβ
Compensating base-pair changes are common among the seed sequences of SLα, providing an independent support for the existence of the base-pairs. SLα is located within the P reading frame (Fig. 1 and Fig. S1A). There are a total of 3888 possible sequence variants that allow compensatory base-pair changes in SLα that preserve the P amino acid sequence. One hundred and three of the sequence variants (including missense variants/mutations and single base deletions) are found in mammalian hepadnaviruses in GenBank (n = 6450). Further, the most common sequence variant comprised 24% of the sequences, suggesting that the base-pairs of SLα are restricted by sequence in addition to P reading frame.
The RNA consensus diagram for the seed alignment of SLα (Fig. 2A) shows that the lower 5 base-pairs of the stem are less conserved. Indeed, this is in agreement with the consensus mammalian hepadnavirus sequences in GenBank (Fig. S2A). Interestingly, the apical loop of SLα for 99% of the mammalian hepadnavirus sequences have a U-bulge, which can be represented as CNGG(U). The first loop residue (C) pairs with the fourth loop residue (G), forming a tetraloop and bulging the fifth loop residue (U). Therefore, the minimum length of the 30-nucleotide stem-loop required for function is most likely the upper 8 base-pairs in conjunction with G- and U-bulge.
Based on mutagenesis, SLβ is likely to be a 23-nucleotide stem-loop which contains a 7-base-pair stem and a 9-residue loop containing mainly U and C with one G (Fig. 2A).4 However, no NMR or crystal structure is available. In contrast to SLα, there are no sequence variations for the stem of SLβ among the seed sequences. This region also encodes both the P and X proteins so its conservation is likely due to the constraints imposed by this (Fig. 1 and Fig. S1B). As a result, the stem of SLβ would be expected to lack compensatory base-pair changes. However, there are few changes that would change the protein sequences (2% in mammalian hepadnaviruses in GenBank (n = 6426); Fig. S1B).
Discussion
Unlike the HBV PRE, the nuclear export pathways mediated by retrovirus regulatory elements such as HIV-1 Rev response element (RRE) and the constitutive transport element (CTE) of simian retrovirus type 1 and Manson-pfizer monkey virus are well characterized.19 The lack of similarity of between the PRE and the retrovirus RRE and CTE suggests that the PRE utilizes a distinct nuclear export pathway(s) which is still poorly understood2,9 and recent evidence indicates that it may function differently in different cell types.20 The evidence for this distinction is, firstly, inhibiting leucine-rich NES-dependent export disrupted RRE-mediated RNA export (via CRM1-dependent pathway) but not PRE-mediated RNA export.21 Secondly, TAP/NXF1 binds directly to the CTE (in the TAP/NXF1-dependent pathway) but not the PRE.22 Thirdly, a RanBP1 mutant disrupted the nuclear export pathways for the PRE and RRE but not the CTE.21
Therefore, the proposed models for SLα and SLβ could serve as the basis for understanding the structural tolerance of the RNA stem-loops, which could be the binding sites for cellular splicing and nuclear export factors. Understanding the sequence variations of the RNA stem-loops is also important for antiviral siRNA (small interfering RNA) design, as siRNA targeting the SLα region has been effective in vitro.23-25
Methods
The representative set of HBV genotype A-H sequences (n = 32) were taken from a reference alignment used in HBVRegDB.5,16 The annotations on the HBV sequences were manually curated by comparing with the annotation analysis obtained from HBVdb.26 Representative woodchuck hepadnavirus sequences (n = 9) were retrieved from GenBank. The woodchuck hepadnavirus sequences were manually annotated. Multiple sequence alignment was done using the CLUSTALW 2.0.27,28 Phylogenetic analysis was done using HBVseq.29 Using the aligned sequences, CDS (coding sequence) annotations and phylogenetic data as input, sequence conservation analyses were done using CDS-plotcon webserver and StructureDist algorithm in SSE 1.1.30,31
Seed sequences of SLα and SLβ were extracted from the aligned sequences. Based on previous studies using NMR (PDB: 2JYM)17 and/or mutagenesis,4,13 Stockholm files of SLα and SLβ were prepared. RNA secondary structures were drawn using R2R 1.0.4.32 Covariance models were built and calibrated using Infernal 1.1.1.33 Comparison of the covariance models with other Rfam models were done using CMCompare webserver.18 Using cmsearch tool from Infernal, the covariance models were evaluated on 41416 complete viral genomes using 5-fold cross-validation (6495 Orthohepadnavirus and 34921 other viruses; retrieved from GenBank viral division at 24 February 2016). Similarity searches of human transcriptome (retrieved from RefSeq Release 74 at 1 March 2016) were done using the score thresholds obtained from the cross-validation step. The covariance models have been submitted to Rfam database.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
CSL is a recipient of a Dr Sulaiman Daud 125th Jubilee Postgraduate Scholarship.
References
- 1.Chen A, Kao YF, Brown CM. Translation of the first upstream ORF in the hepatitis B virus pregenomic RNA modulates translation at the core and polymerase initiation codons. Nucleic Acids Res 2005; 33:1169-81; PMID:15731337; http://dx.doi.org/ 10.1093/nar/gki251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen A, Panjaworayan T-Thienprasert N, Brown CM. Prospects for inhibiting the post-transcriptional regulation of gene expression in hepatitis B virus. World J Gastroenterol WJG 2014; 20:7993-8004; PMID:25009369; http://dx.doi.org/ 10.3748/wjg.v20.i25.7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doitsh G, Shaul Y. A long HBV transcript encoding pX is inefficiently exported from the nucleus. Virology 2003; 309:339-49; PMID:12758180; http://dx.doi.org/ 10.1016/S0042-6822(03)00156-9 [DOI] [PubMed] [Google Scholar]
- 4.Smith GJ, Donello JE, Hope TJ, Lück R, Steger G. The hepatitis B virus post-transcriptional regulatory element contains two conserved RNA stem-loops which are required for function. Nucleic Acids Res 1998; 26:4818-27; PMID:9776740; http://dx.doi.org/ 10.1093/nar/26.21.4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen A, Brown CM. Distinct families of cis-acting RNA replication elements epsilon from hepatitis B viruses. RNA Biol 2012; 9:130-6; PMID:22418844; http://dx.doi.org/ 10.4161/rna.18649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang ZM, Yen TS. Role of the hepatitis B virus posttranscriptional regulatory element in export of intronless transcripts. Mol Cell Biol 1995; 15:3864-9; PMID:7791793; http://dx.doi.org/ 10.1128/MCB.15.7.3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sommer G, Heise T. Posttranscriptional control of HBV gene expression. Front Biosci 2008; 13:5533-47; PMID:18508603; http://dx.doi.org/ 10.2741/3097 [DOI] [PubMed] [Google Scholar]
- 8.Yang C-C, Huang E-Y, Li H-C, Su P-Y, Shih C. Nuclear export of human hepatitis B virus core protein and pregenomic RNA depends on the cellular NXF1-p15 machinery. PLoS ONE 2014; 9:e106683; PMID:25360769; http://dx.doi.org/ 10.1371/journal.pone.0106683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee GH, Wasser S, Lim SG. Hepatitis B pregenomic RNA splicing—the products, the regulatory mechanisms and its biological significance. Virus Res 2008; 136:1-7; PMID:18579251; http://dx.doi.org/ 10.1016/j.virusres.2008.05.007 [DOI] [PubMed] [Google Scholar]
- 10.Chi B, Wang K, Du Y, Gui B, Chang X, Wang L, Fan J, Chen S, Wu X, Li G, et al. A sub-element in PRE enhances nuclear export of intronless mRNAs by recruiting the TREX complex via ZC3H18. Nucleic Acids Res 2014; 42:7305-18; PMID:24782531; http://dx.doi.org/ 10.1093/nar/gku350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hass M, Hannoun C, Kalinina T, Sommer G, Manegold C, Günther S. Functional analysis of hepatitis B virus reactivating in hepatitis B surface antigen-negative individuals. Hepatology 2005; 42:93-103; PMID:15962285; http://dx.doi.org/ 10.1002/hep.20748 [DOI] [PubMed] [Google Scholar]
- 12.Heise T, Sommer G, Reumann K, Meyer I, Will H, Schaal H. The hepatitis B virus PRE contains a splicing regulatory element. Nucleic Acids Res 2006; 34:353-63; PMID:16410615; http://dx.doi.org/ 10.1093/nar/gkj440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donello JE, Loeb JE, Hope TJ. Woodchuck hepatitis virus contains a tripartite posttranscriptional regulatory element. J Virol 1998; 72:5085-92; PMID:9573279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zufferey R, Donello JE, Trono D, Hope TJ. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol 1999; 73:2886-92; PMID:10074136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zanta-Boussif MA, Charrier S, Brice-Ouzet A, Martin S, Opolon P, Thrasher AJ, Hope TJ, Galy A. Validation of a mutated PRE sequence allowing high and sustained transgene expression while abrogating WHV-X protein synthesis: application to the gene therapy of WAS. Gene Ther 2009; 16:605-19; PMID:19262615; http://dx.doi.org/ 10.1038/gt.2009.3 [DOI] [PubMed] [Google Scholar]
- 16.Panjaworayan N, Roessner SK, Firth AE, Brown CM. HBVRegDB: annotation, comparison, detection and visualization of regulatory elements in hepatitis B virus sequences. Virol J 2007; 4:136; PMID:18086305; http://dx.doi.org/ 10.1186/1743-422X-4-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwalbe M, Ohlenschläger O, Marchanka A, Ramachandran R, Häfner S, Heise T, Görlach M. Solution structure of stem-loop α of the hepatitis B virus post-transcriptional regulatory element. Nucleic Acids Res 2008; 36:1681-9; PMID:18263618; http://dx.doi.org/ 10.1093/nar/gkn006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eggenhofer F, Hofacker IL, Siederdissen CHZU. CMCompare webserver: comparing RNA families via covariance models. Nucleic Acids Res 2013; 41:W499-503; PMID:23640335; http://dx.doi.org/ 10.1093/nar/gkt329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandes J, Jayaraman B, Frankel A. The HIV-1 Rev response element. RNA Biol 2012; 9:6-11; PMID:22258145; http://dx.doi.org/ 10.4161/rna.9.1.18178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Visootsat A, Payungporn S, T-Thienprasert NP. A conserved RNA structural element within the hepatitis B virus post-transcriptional regulatory element enhance nuclear export of intronless transcripts and repress the splicing mechanism. Mol Biol Rep 2015; 42:1603-14; PMID:26514143; http://dx.doi.org/ 10.1007/s11033-015-3928-0 [DOI] [PubMed] [Google Scholar]
- 21.Zang W-Q, Benedict Yen TS. Distinct export pathway utilized by the hepatitis B virus posttranscriptional regulatory element. Virology 1999; 259:299-304; PMID:10388654; http://dx.doi.org/ 10.1006/viro.1999.9777 [DOI] [PubMed] [Google Scholar]
- 22.Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber BK, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell 1998; 1:649-59; PMID:9660949; http://dx.doi.org/ 10.1016/S1097-2765(00)80065-9 [DOI] [PubMed] [Google Scholar]
- 23.Panjaworayan N, Payungporn S, Poovorawan Y, Brown CM. Identification of an effective siRNA target site and functional regulatory elements, within the hepatitis B virus posttranscriptional regulatory element. Virol J 2010; 7:216; PMID:20822550; http://dx.doi.org/ 10.1186/1743-422X-7-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panjaworayan N, Brown CM. Effects of HBV genetic variability on RNAi strategies. Hepat Res Treat 2011; 2011:e367908; PMID:21760994; http://dx.doi.org/ 10.1155/2011/367908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thongthae N, Payungporn S, Poovorawan Y, T-Thienprasert NP. A rational study for identification of highly effective siRNAs against hepatitis B virus. Exp Mol Pathol 2014; 97:120-7; PMID:24953337; http://dx.doi.org/ 10.1016/j.yexmp.2014.06.006 [DOI] [PubMed] [Google Scholar]
- 26.Hayer J, Jadeau F, Deléage G, Kay A, Zoulim F, Combet C. HBVdb: a knowledge database for Hepatitis B Virus. Nucleic Acids Res 2013; 41:D566-70; PMID:23125365; http://dx.doi.org/ 10.1093/nar/gks1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23:2947-8; PMID:17846036; http://dx.doi.org/ 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 28.Okonechnikov K, Golosova O, Fursov M. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 2012; 28:1166-7; PMID:22368248; http://dx.doi.org/ 10.1093/bioinformatics/bts091 [DOI] [PubMed] [Google Scholar]
- 29.Rhee S-Y, Margeridon-Thermet S, Nguyen MH, Liu TF, Kagan RM, Beggel B, Verheyen J, Kaiser R, Shafer RW. Hepatitis B virus reverse transcriptase sequence variant database for sequence analysis and mutation discovery. Antiviral Res 2010; 88:269-75; PMID:20875460; http://dx.doi.org/ 10.1016/j.antiviral.2010.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Firth AE, Brown CM. Detecting overlapping coding sequences with pairwise alignments. Bioinformatics 2005; 21:282-92; PMID:15347574; http://dx.doi.org/ 10.1093/bioinformatics/bti007 [DOI] [PubMed] [Google Scholar]
- 31.Simmonds P. SSE: a nucleotide and amino acid sequence analysis platform. BMC Res Notes 2012; 5:50; PMID:22264264; http://dx.doi.org/ 10.1186/1756-0500-5-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinberg Z, Breaker RR. R2R - software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics 2011; 12:3; PMID:21205310; http://dx.doi.org/ 10.1186/1471-2105-12-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013; 29:2933-5; PMID:24008419; http://dx.doi.org/ 10.1093/bioinformatics/btt509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horke S, Reumann K, Rang A, Heise T. Molecular characterization of the human La protein·hepatitis B virus RNA.B interaction in vitro. J Biol Chem 2002; 277:34949-58; PMID:12121976; http://dx.doi.org/ 10.1074/jbc.M201911200 [DOI] [PubMed] [Google Scholar]
- 35.Huang C, Xie M-H, Liu W, Yang B, Yang F, Huang J, Huang J, Wu Q, Fu X-D, Zhang Y. A structured RNA in hepatitis B virus post-transcriptional regulatory element represses alternative splicing in a sequence-independent and position-dependent manner. FEBS J 2011; 278:1533-46; PMID:21371260; http://dx.doi.org/ 10.1111/j.1742-4658.2011.08077.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zang W-Q, Li B, Huang P-Y, Lai MMC, Yen TSB. Role of polypyrimidine tract binding protein in the function of the hepatitis B virus posttranscriptional regulatory element. J Virol 2001; 75:10779-86; PMID:11602719; http://dx.doi.org/ 10.1128/JVI.75.22.10779-10786.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.