ABSTRACT
Pre-mRNA splicing is a key post-transcriptional regulation process in which introns are excised and exons are ligated together. A novel class of structured intron was recently discovered in fish. Simple expansions of complementary AC and GT dimers at opposite boundaries of an intron were found to form a bridging structure, thereby enforcing correct splice site pairing across the intron. In some fish introns, the RNA structures are strong enough to bypass the need of regulatory protein factors for splicing. Here, we discuss the prevalence and potential functions of highly structured introns. In humans, structured introns usually arise through the co-occurrence of C and G-rich repeats at intron boundaries. We explore the potentially instructive example of the HLA receptor genes. In HLA pre-mRNA, structured introns flank the exons that encode the highly polymorphic β sheet cleft, making the processing of the transcript robust to variants that disrupt splicing factor binding. While selective forces that have shaped HLA receptor are fairly atypical, numerous other highly polymorphic genes that encode receptors contain structured introns. Finally, we discuss how the elevated mutation rate associated with the simple repeats that often compose structured intron can make structured introns themselves rapidly evolving elements.
KEYWORDS: Dinucleotide repeats, intron secondary structure, pre-mRNA splicing, single nucleotide polymorphism, U2AF2
Introduction
Pre-mRNA splicing is, perhaps, the most surprising step in gene expression where intervening intronic sequence are removed from nascent transcript and the remaining coding exonic sequence are ligated to make mRNA which is then translated into protein. The intron boundary is defined by 5′ splice site (5ss) and 3′ splice site (3ss). For the eukaryotic nuclear pre-mRNA, the precise recognition of splice sites and the splicing reaction is catalyzed by a large small nuclear ribonucleoprotein (snRNP) complex, called the spliceosome, which is de novo assembled onto each intron. The spliceosome is composed with several snRNP particles, namely U1, U2, U4, U5 and U6, each containing a small RNA (typically 100 to 300 nucleotides [nts]) and many other proteins.1-3 Splicing takes place in 2 steps. In the first transesterification reaction, U1 small RNAs basepair with the 5ss and U2 small RNAs basepair with the branch site sequence. Interactions between U1 and U2 bring the splice sites together to promote splicing. The initial basepairing between the 5ss and U1 is disrupted and replaced by U6 interaction with the 5ss after the engagement of the U4/U6/U5 tri-snRNP complex. U2 and U6 basepairing displaces the original U4 and U6 basepairing, forming a catalytically active site that carries out the first transesterification reaction. The 2′-OH group of the branchpoint nucleotide is bulged out to perform nucleophilic attack on the 3′ end of the upstream exon, resulting in a lariat intermediate and a free upstream exon. Following rearrangement of snRNPs, the second transesterification step occurs. In this step, the 3′-OH group of the upstream exon ligates to the 5′ end of the downstream exon (3ss), producing an excised intronic lariat and spliced exons.4,5
The recognition of the 3ss is distinct from 5ss recognition by the nature of the signal. While U1snRNP recognizes the 9nt 5ss, the 3ss is a bipartite signal that comprises a polypyrimidine tract about −4 to −20 nt upstream of the 3ss6 and the 3ss AG dinucleotide. Many proteins in the U2 snRNP complex promote 3ss recognition and defects in these proteins are associated with diseases.7-11 The U2 auxiliary factors U2AF1 recognizes 3ss AG and U2AF2 binds to the polypyrimidine tract upstream from the 3ss (Fig. 1A). The affinity of these elements to spliceosomal small RNAs or auxiliary protein factors varies among transcripts and thus lead to variable level of exon inclusion. In depletion and add-back studies in vitro, U2AF2 has been suggested to play an essential role in splicing.12-14 It is an essential gene for viability in fly, worms and yeast.15-19 However, genome-wide U2AF2 binding studies show that ∼12% of human introns are not bound by U2AF2, and knockdown of U2AF2 only affects the inclusion level of ∼6% of the exons analyzed, suggesting that some introns are more dependent on U2AF2 than others.20 Similarly, studies utilizing human cell lines show that knockdown of U2AF2 shifts the splicing toward introns harboring strong polypyrimidine tracts,21 and that U2AF2 dependency can be eliminated by replacement of a stronger 5ss.22 In addition, in fission yeast S. pombe, a large number of introns remain efficiently spliced upon the inactivation of U2AF,59 the U2AF2 homolog. In this case stronger 5ss and A/U-rich sequence in the 5′ intronic region are associated with U2AF59-insensitive introns.23 Taken together, these studies suggest that there exist compensatory mechanisms to bypass the need of essential splicing factors for splice site recognition.
Figure 1.
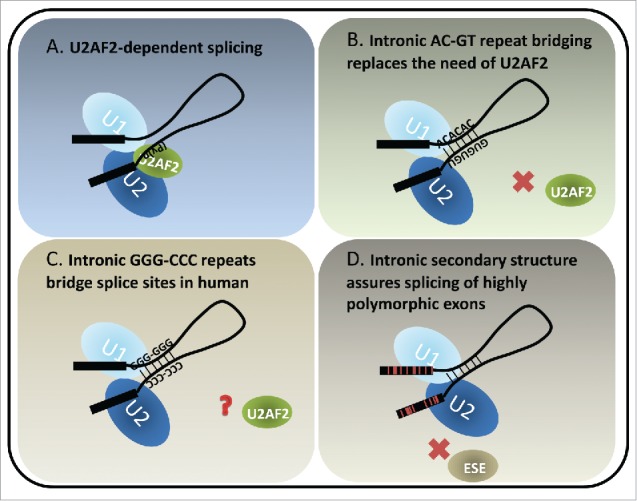
Models of U2AF2-dependent and structure-dependent splicing. (A) U2AF2 recognizes the polypyrimidine tract upstream of the 3ss. It dimerizes with U2AF1 (not shown) which recognizes the 3ss AG to facilitate U2 docking on the 3ss. The interactions between U1 and U2 snRNPs bring the 5ss and 3ss in close proximity, forming a pre-splicing complex (A complex) toward the first step of splicing. (B) A special class of introns identified in fish and lamprey contains complementary runs of AC's and GU's across introns. The complementarity bridges the introns so that the 5ss and 3ss are positioned for splicing. (C) Intronic bridging by complementary repeat sequences were also suggested in humans. A variety of G, C and GC-rich repeats could play structural roles in pairing splice sites across introns. (D) Highly polymorphic genes, such as human HLAs exhibit rapid and frequent sequence changes in the coding exons to accommodate the need of peptide variety. Therefore the exonic splicing elements, for example exonic splicing enhancers (ESE's), are not preserved. Introns flanking the highly polymorphic exons are GC-rich and have lower folding energy compared to introns within the same length range. We hypothesize that the secondary structures of such introns bridge the splice sites, similarly to the fish AC- and GT-repeat containing introns, which compensate for the potential loss of exon definition. Red lines in the exons indicate single nucleotide polymorphisms.
Structure-mediated splicing
Two key features of RNA molecules are the ability to form secondary structures and the transient nature of RNA. RNA is bound by protein partners and forms structure soon after or even during transcription. Complexes are recycled as RNA degrades in the cell. RNA structures can alter the physical spacing among sequence elements and may provide binding platforms for RNA-binding proteins.24 Proteins recognizing seemingly the same primary sequences may prefer distinct secondary structures or may differ in their ability to unwind structures.25 In this regard, RNA structures of the transcripts could play important roles of regulating protein binding and splice site interactions but, until recently, specific examples of RNA structure and splicing remained anecdotal.
It is hypothesized that pre-mRNA splicing evolved from the autocatalytic splicing observed in group II introns. Group II introns are found in rRNA, tRNA, and mRNA of organelles (chloroplasts and mitochondria) in fungi, plants, and protists, and also in mRNA in bacteria. This hypothesis is due to the remarkable structural similarity between domain V of group II introns and the active catalytic site formed by U2 and U6 small RNA pairing.26 Both group II introns and spliceosomal introns utilize 2 transesterification steps and generate an intronic lariat byproduct with an adenine branchpoint.27,28 For splicing to occur, 2 splice sites must be brought to proximity.29 In group II introns, the correct identification and pairing of splice sites is performed exclusively through RNA secondary structure. For spliceosomal introns, the self-catalytic features of group II introns are largely replaced by spliceosomal RNPs and RNA-binding proteins. For example, heterogeneous nuclear ribonucleoproteins (hnRNPs) at 2 ends of an intron could interact with each other to aid in the looping out of introns.30 Additionally, interactions between U1 hnRNP at the 5ss and the exon-binding serine/arginine-rich (SR) proteins,31 and interactions between U1 and U2AFs at the 3ss32,33 are known to enhance splicing (Fig. 1A). The correct pairing of splice sites in nuclear introns could also be achieved by RNA-RNA interaction similar to the mechanism observed in group II introns.
Secondary structures can either inhibit or enhance splicing. Studies have shown that stem structures at splice sites weaken exon inclusion34 in a dosage-dependent manner.35,36 Sequence variations that disrupt secondary structures of the splice sites or introns can direct expression of disease-specific or tissue-specific mRNA isoforms.37,38 Also, splicing enhancers and silencers residing in a double-stranded region have less effect on splicing because most of the RNA-binding proteins bind to single-stranded sequences.39 Interestingly, regional variation of GC composition can favor the formation of stronger or weaker secondary structure. Systematic analysis of splice sites show that high GC content of splice sites is associated with alternatively spliced exons,40 suggesting that regulation of openness or accessibility of splicing elements could be a means of alternative splicing regulation. Positive examples of secondary structure include the well-studied structure-mediated splicing of Drosophila Dscam (Down syndrome cell-adhesion molecule) gene. Dscam gene can potentially produce 38,016 different mRNA isoforms (in D. melanogaster) by selecting mutually exclusive exons from multiple exon clusters. Selection of exons relies on competitive basepairing between the “docking” element and the complementary “selector” element of each exon cluster to bring the splice site to proximity for splicing.41-48 In yeast S. cerevisae, secondary structures between 5ss and branch site have been shown to facilitate splicing by minimizing physical distance between the U1 and U2 complex.49-51 Similarly, stem-loop structures between the branch site and 3ss also facilitate splicing for distally located branch sites.52,53 A very interesting example in S. cerevisiae shows that the stem structure between the branch site and 3ss could serve as a thermosensor for alternative 3ss selection, depending on which 3ss was accessible by the spliceosome under the temperature-modulated structure.54 In addition, in mouse, long-range RNA-RNA interactions that position Rbfox binding sites close to its regulated exon facilitate the exon inclusion.55 These reports all together indicate that secondary structure plays a critical role throughout evolution in regulating the accessibility and the functional outcome of primary sequence elements for pre-mRNA splicing.
RNA structure replaces the need for essential factors in splicing
In our recent report published in Genome Research,56 we identified evolutionarily conserved long complementary runs of AC and GT repeats co-occurring at both ends of introns, bringing the 5ss and 3ss in close proximity, thereby enhancing splicing (Fig. 1B). Introns containing these repeats comprise ∼2% of total introns and are found in ∼10% of zebrafish genes. This analysis introduced thousands of new examples of structured introns to the field. These repeat elements are enriched in fish and lamprey but lost in mammals, suggesting it may be an ancient splicing mechanism that preceded the divergence of tetrapods from fish. Interestingly, the sizes of AC and GT repeat-containing introns across species regress toward a mean. In other words, they tend to be larger than average in fish with short introns and shorter than average in fish species with larger introns, arguing that they evolved toward an optimal range of length due to structural constraints. Importantly, The AC and GT repeat-containing introns contain weaker polypyrimidine tracts and are insensitive to U2AF2 antibody blockage or knockdown. Systematic search of the intronic conserved complementary “boxes” in Drosophila has shown the optimal stem structures are 50 and 100 nt away from the 5ss and 3ss, respectively, potentially to avoid disrupting essential splicing elements, such as polypyrimidine tracts and splice sites; otherwise the boxes would have inhibitory effect for exon inclusion.57 Therefore, the proximity of AC and GT runs to the splice sites are unusual in that the secondary structures reduce the need of essential splicing signals encoded closely to the splice sites.
It has not yet been proved that a similar mechanism of secondary structure replacing the need for essential splicing factors exists for human pre-mRNAs. However, the simultaneous pairing of G triplets at the 5′ end of introns with C triplets at the 3′ end of introns are enriched in human introns.56 Moreover, complementary C-rich and G-rich intronic sequences occurring across conditionally skipped exons may loop out the exon to support exon skipping.58 Systematic search of human intronic complementary “boxes” suggest that long range RNA-RNA interactions could facilitate splicing of introns with suboptimal splice sites.59 These observations suggest that complementary sequences and intronic secondary structures could potentially bring splice sites to proximity and regulate splicing in mammalian genomes (Fig. 1C).
While no enriched gene ontology was identified from zebrafish genes containing AC-GT repeat introns, one property of structured introns is a reduced dependence on trans-acting factors (i.e. the activators and repressors of splicing). A typical exon encodes 4-5 exonic splicing enhancers in addition to the codons required to specify the amino acids in the gene's product. Certain genes (e.g. human leukocyte antigen, HLA) are under unusual selective pressure that drives those loci to increased levels of polymorphism. In a case where splice site recognition is dependent on exonic splicing enhancers (ESE's) in exonic sequence, increased polymorphism may result in a loss of processing. The HLA locus is a potentially instructive example of this dilemma. HLA encodes proteins present peptide fragments on the cell surface. Foreign (e.g., viral) protein fragments that are successfully loaded intracellularly into the HLA, effectively target the infected cell for clearance by the immune system. This mechanism creates selective pressure on the pathogen to evolve away from the specificity of the HLA receptor. Pathogens that escape recognition create selective pressure on the host to generate a new allele of HLA loci to recognize the novel pathogen and the ensuing “arms race” drives diversity at the HLA loci. One way to compensate for the potential loss of splicing enhancer and/or generation of splicing silencer in a rapidly evolving exon is to utilize an alternate method of enforcing the correct splice site pairing that is independent of variations in the exon (Fig. 1D). Extensive regions of secondary structure that pair the 5ss to the 3ss are likely to be resistant to point mutations. Intron 2 in HLA gene family is flanked by exons that encode the recognition cleft of the HLA receptor, the most variable exons in the transcript. This second intron is by far the most structured intron in the transcript but also ranks highly in the genome: ΔG of intron 2 of HLA-A, HLA-B and HLA-C are: −124.61 (stability of structure: 96th percentile in genome), −132.46 (98th percentile) and −135.70 kcal/mol (98th percentile), respectively (Fig. 2A). Expanding the search to all genes, exons that neighbor structured introns are significantly enriched for SNPs (Fig. 2B). These results suggest that intron bridging structure can bypass the need for auxiliary splicing elements such as ESE's in the primary sequence. Structured introns may also be less vulnerable to variants that create negative silencer elements in the exon, making this region unaffected by high levels of exonic variation.
Figure 2.
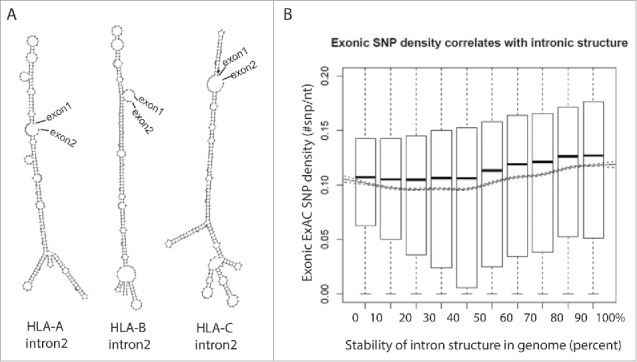
Highly polymorphic exons are flanked by structured introns. (A) Structures of intron 2 of the HLA family genes are predicted using an RNA structure prediction program, RNAfold.68 (B) Introns are ranked in decile by their folding energy calculated by RNAfold. SNP density of their flanking exons is calculated using all SNPs from the Exome Aggregation Consortium (ExAC) data and presented by boxplot for each bin. Trend of means (solid line) is depicted with 95% confidence interval (dashed lines). The significance of the positive correlation is determined by trend test, P value < 2 × 10−16.
RNA structures in evolution
Mutation and selection drive evolution and are major determinants of sequence conservation observed between orthologous loci in different species. Mutations can alter primary sequence information as well as residues involved in secondary structure formation. However, stable structures can be more resistant to mutations and it is why mutations occur at higher frequencies in many structured introns. Indeed, human structured introns are significantly less conserved than average (PhyloP100wayAll score −0.2 vs. 0.0, P-value < 0.0001). The dimer repeats used to bridge introns in fish are one of the fastest-evolving motifs presumably due to the high rate of mutation through replication slippage.60,61 The expansions and contractions of zebrafish AC repeats occur at a frequency of 1.5 × 10−4 per locus per gamete per generation, about 10 thousand times higher than other types of mutations.62,63 Although the mutation rate of microsatellites (i.e., AC repeats) in fish corresponds to that in mammals, the mutation in fish often causes gain or loss of multiple repeats whereas it rarely does in mammals.62,64, 65 Given this bias toward creating short blocks of AC or GT repeats, it is possible to see how a hairpin that bridges introns could arise or be destroyed by a single mutational event. However, we have shown that the bridging efficiency plateaus at 6 repeats, which may explain how its function is preserved even with high mutation rates.56
Concluding remarks
The recently discovered class of structured introns shares characteristics of both spliceosomal and group II introns. In this regard, these introns, which predate the divergence of tetrapods from teleosts, could represent a potential intermediate in the origin of modern nuclear intron.
Probing RNA secondary structure is not a simple task. The computational calculation often has length limitations and bias toward to a close-end structure,66,67 in which the lowest energy structure often places the ends of the RNA folding window in close proximity. Additionally, the biochemical approach is limited to a few number of candidates and is best established in vitro without taking the dynamic in vivo environment into account. However, as no RNAs are unstructured, secondary structure must be considered for all aspects of RNA biology, including but not limited to intermolecular interactions, spacing of sequence elements and catalytic functions. Cis-regulatory elements could be buried deep in the intron for splicing regulation. Moving forward, a major focus of RNA biology will be increasing the understanding of how secondary structures are controlled under different stimuli and various physiological/pathological conditions.
Disclosure of potential confllicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Yen-Tsung Huang, Dr. Rachel Soemedi and Kamil Cygan for their help on the SNP analysis. Part of this research was conducted using computational resources and services at the Center for Computation and Visualization at Brown University. This work was supported by National Institutes of Health under award number R01GM105681.
References
- 1.Matlin AJ, Moore MJ. Spliceosome assembly and composition. Adv Exp Med Biol 2007; 623:14-35; PMID:18380338; http://dx.doi.org/ 10.1007/978-0-387-77374-2_2 [DOI] [PubMed] [Google Scholar]
- 2.Hoskins AA, Moore MJ. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem Sci 2012; 37:179-88; PMID:22480731; http://dx.doi.org/ 10.1016/j.tibs.2012.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell 2003; 12:5-14; PMID:12887888; http://dx.doi.org/ 10.1016/S1097-2765(03)00270-3 [DOI] [PubMed] [Google Scholar]
- 4.Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, Munoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol 2013; 14:153-65; PMID:23385723; http://dx.doi.org/ 10.1038/nrm3525 [DOI] [PubMed] [Google Scholar]
- 5.Reed R. Mechanisms of fidelity in pre-mRNA splicing. Curr Opin Cell Biol 2000; 12:340-5; PMID:10801464; http://dx.doi.org/ 10.1016/S0955-0674(00)00097-1 [DOI] [PubMed] [Google Scholar]
- 6.Chen C, Zhao X, Kierzek R, Yu YT. A flexible RNA backbone within the polypyrimidine tract is required for U2AF65 binding and pre-mRNA splicing in vivo. Mol Cell Biol 2010; 30:4108-19; PMID:20606010; http://dx.doi.org/ 10.1128/MCB.00531-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al.. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011; 478:64-9; PMID:21909114; http://dx.doi.org/ 10.1038/nature10496 [DOI] [PubMed] [Google Scholar]
- 8.Padgett RA. New connections between splicing and human disease. Trends Genet 2012; 28:147-54; PMID:22397991; http://dx.doi.org/ 10.1016/j.tig.2012.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al.. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012; 150:1107-20; PMID:22980975; http://dx.doi.org/ 10.1016/j.cell.2012.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet 2013; 45:133-5; PMID:23313955; http://dx.doi.org/ 10.1038/ng.2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D, Bailey SL, Bhavsar EB, Chan B, Colla S, et al.. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3′ Splice Site Selection through Use of a Different Branch Point. Cell Rep 2015; 13:1033-45; PMID:26565915; http://dx.doi.org/ 10.1016/j.celrep.2015.09.053 [DOI] [PubMed] [Google Scholar]
- 12.Valcarcel J, Gaur RK, Singh R, Green MR. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA [corrected]. Science 1996; 273:1706-9; PMID:8781232; http://dx.doi.org/ 10.1126/science.273.5282.1706 [DOI] [PubMed] [Google Scholar]
- 13.Zhang M, Zamore PD, Carmo-Fonseca M, Lamond AI, Green MR. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc Natl Acad Sci U S A 1992; 89:8769-73; PMID:1388271; http://dx.doi.org/ 10.1073/pnas.89.18.8769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zamore PD, Green MR. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J 1991; 10:207-14; PMID:1824937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanaar R, Roche SE, Beall EL, Green MR, Rio DC. The conserved pre-mRNA splicing factor U2AF from Drosophila: requirement for viability. Science 1993; 262:569-73; PMID:7692602; http://dx.doi.org/ 10.1126/science.7692602 [DOI] [PubMed] [Google Scholar]
- 16.Romfo CM, Lakhe-Reddy S, Wise JA. Molecular genetic analysis of U2AF59 in Schizosaccharomyces pombe: differential sensitivity of introns to mutational inactivation. RNA 1999; 5:49-65; PMID:9917066; http://dx.doi.org/ 10.1017/S1355838299981323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudner DZ, Kanaar R, Breger KS, Rio DC. Mutations in the small subunit of the Drosophila U2AF splicing factor cause lethality and developmental defects. Proc Natl Acad Sci U S A 1996; 93:10333-7; PMID:8816800; http://dx.doi.org/ 10.1073/pnas.93.19.10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zorio DA, Blumenthal T. U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. RNA 1999; 5:487-94; PMID:10199565; http://dx.doi.org/ 10.1017/S1355838299982225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Potashkin J, Naik K, Wentz-Hunter K. U2AF homolog required for splicing in vivo. Science 1993; 262:573-5; PMID:8211184; http://dx.doi.org/ 10.1126/science.8211184 [DOI] [PubMed] [Google Scholar]
- 20.Wu T, Fu XD. Genomic functions of U2AF in constitutive and regulated splicing. RNA Biol 2015; 12:479-85; PMID:25901584; http://dx.doi.org/ 10.1080/15476286.2015.1020272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pacheco TR, Coelho MB, Desterro JM, Mollet I, Carmo-Fonseca M. In vivo requirement of the small subunit of U2AF for recognition of a weak 3′ splice site. Mol Cell Biol 2006; 26:8183-90; PMID:16940179; http://dx.doi.org/ 10.1128/MCB.00350-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho S, Moon H, Loh TJ, Jang HN, Liu Y, Zhou J, Ohn T, Zheng X, Shen H. Splicing inhibition of U2AF65 leads to alternative exon skipping. Proc Natl Acad Sci U S A 2015; 112:9926-31; PMID:26216990; http://dx.doi.org/ 10.1073/pnas.1500639112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sridharan V, Heimiller J, Singh R. Genomic mRNA profiling reveals compensatory mechanisms for the requirement of the essential splicing factor U2AF. Mol Cell Biol 2011; 31:652-61; PMID:21149581; http://dx.doi.org/ 10.1128/MCB.01000-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol 2004; 24:10505-14; PMID:15572659; http://dx.doi.org/ 10.1128/MCB.24.24.10505-10514.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warf MB, Diegel JV, von Hippel PH, Berglund JA. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc Natl Acad Sci U S A 2009; 106:9203-8; PMID:19470458; http://dx.doi.org/ 10.1073/pnas.0900342106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y, Rio DC. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Ann Rev Biochem 2015; 84:291-323; PMID:25784052; http://dx.doi.org/ 10.1146/annurev-biochem-060614-034316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hang J, Wan R, Yan C, Shi Y. Structural basis of pre-mRNA splicing. Science 2015; 349:1191-8; PMID:26292705; http://dx.doi.org/ 10.1126/science.aac8159 [DOI] [PubMed] [Google Scholar]
- 28.Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 2015; 349:1182-91; PMID:26292707; http://dx.doi.org/ 10.1126/science.aac7629 [DOI] [PubMed] [Google Scholar]
- 29.Robinson R. Looping out introns to help splicing. PLoS Biol 2006; 4:e41; PMID:20076527; http://dx.doi.org/ 10.1371/journal.pbio.0040041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Contreras R, Fisette JF, Nasim FU, Madden R, Cordeau M, Chabot B. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol 2006; 4:e21; PMID:16396608; http://dx.doi.org/ 10.1371/journal.pbio.0040021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graveley BR. Sorting out the complexity of SR protein functions. RNA 2000; 6:1197-211; PMID:10999598; http://dx.doi.org/ 10.1017/S1355838200000960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Gen Dev 1996; 10:1356-68; PMID:8647433; http://dx.doi.org/ 10.1101/gad.10.11.1356 [DOI] [PubMed] [Google Scholar]
- 33.Abovich N, Liao XC, Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Gen Dev 1994; 8:843-54; PMID:7926772; http://dx.doi.org/ 10.1101/gad.8.7.843 [DOI] [PubMed] [Google Scholar]
- 34.Solnick D. Alternative splicing caused by RNA secondary structure. Cell 1985; 43:667-76; PMID:4075405; http://dx.doi.org/ 10.1016/0092-8674(85)90239-9 [DOI] [PubMed] [Google Scholar]
- 35.Goguel V, Wang Y, Rosbash M. Short artificial hairpins sequester splicing signals and inhibit yeast pre-mRNA splicing. Mol Cell Biol 1993; 13:6841-8; PMID:8413277; http://dx.doi.org/ 10.1128/MCB.13.11.6841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plass M, Codony-Servat C, Ferreira PG, Vilardell J, Eyras E. RNA secondary structure mediates alternative 3′ss selection in Saccharomyces cerevisiae. RNA 2012; 18:1103-15; PMID:22539526; http://dx.doi.org/ 10.1261/rna.030767.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, et al.. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 1998; 393:702-5; PMID:9641683; http://dx.doi.org/ 10.1038/31508 [DOI] [PubMed] [Google Scholar]
- 38.Clouet d'Orval B, d'Aubenton Carafa Y, Sirand-Pugnet P, Gallego M, Brody E, Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science 1991; 252:1823-8; PMID:2063195; http://dx.doi.org/ 10.1126/science.2063195 [DOI] [PubMed] [Google Scholar]
- 39.Hiller M, Zhang Z, Backofen R, Stamm S. Pre-mRNA secondary structures influence exon recognition. PLoS Genet 2007; 3:e204; PMID:18020710; http://dx.doi.org/ 10.1371/journal.pgen.0030204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Kuo CC, Chen L. GC content around splice sites affects splicing through pre-mRNA secondary structures. BMC genomics 2011; 12:90; PMID:21281513; http://dx.doi.org/ 10.1186/1471-2164-12-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y, Zhan L, Zhang W, Sun F, Wang W, Tian N, Bi J, Wang H, Shi D, Jiang Y, et al.. RNA secondary structure in mutually exclusive splicing. Nat Struct Mol Biol 2011; 18:159-68; PMID:21217700; http://dx.doi.org/ 10.1038/nsmb.1959 [DOI] [PubMed] [Google Scholar]
- 42.McManus CJ, Graveley BR. RNA structure and the mechanisms of alternative splicing. Curr Opin Genet Dev 2011; 21:373-9; PMID:21530232; http://dx.doi.org/ 10.1016/j.gde.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin Y, Yang Y, Zhang P. New insights into RNA secondary structure in the alternative splicing of pre-mRNAs. RNA Biol 2011; 8:450-7. the Drosophila Dscam pre-mRNA is both temporally and spatially regulated. Genetics 2001; 159: 599-608; PMID:11606537; http://dx.doi.org/ 10.4161/rna.8.3.15388 [DOI] [PubMed] [Google Scholar]
- 44.Graveley BR, Kaur A, Gunning D, Zipursky SL, Rowen L, Clemens JC. The organization and evolution of the dipteran and hymenopteran Down syndrome cell adhesion molecule (Dscam) genes. RNA 2004; 10:1499-506; PMID:15383675; http://dx.doi.org/ 10.1261/rna.7105504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graveley BR. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell 2005; 123:65-73; PMID:16213213; http://dx.doi.org/ 10.1016/j.cell.2005.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kreahling JM, Graveley BR. The iStem, a long-range RNA secondary structure element required for efficient exon inclusion in the Drosophila Dscam pre-mRNA. Mol Cell Biol 2005; 25:10251-60; PMID:16287842; http://dx.doi.org/ 10.1128/MCB.25.23.10251-10260.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.May GE, Olson S, McManus CJ, Graveley BR. Competing RNA secondary structures are required for mutually exclusive splicing of the Dscam exon 6 cluster. RNA 2011; 17:222-9; PMID:21159795; http://dx.doi.org/ 10.1261/rna.2521311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goguel V, Rosbash M. Splice site choice and splicing efficiency are positively influenced by pre-mRNA intramolecular base pairing in yeast. Cell 1993; 72:893-901; PMID:8458083; http://dx.doi.org/ 10.1016/0092-8674(93)90578-E [DOI] [PubMed] [Google Scholar]
- 49.Libri D, Stutz F, McCarthy T, Rosbash M. RNA structural patterns and splicing: molecular basis for an RNA-based enhancer. RNA 1995; 1:425-36; PMID:7493320 [PMC free article] [PubMed] [Google Scholar]
- 50.Howe KJ, Ares M Jr. Intron self-complementarity enforces exon inclusion in a yeast pre-mRNA. Proc Natl Acad Sci U S A 1997; 94:12467-72; PMID:9356473; http://dx.doi.org/ 10.1073/pnas.94.23.12467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gahura O, Hammann C, Valentova A, Puta F, Folk P. Secondary structure is required for 3′ splice site recognition in yeast. Nucleic Acids Res 2011; 39:9759-67; PMID:21893588; http://dx.doi.org/ 10.1093/nar/gkr662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rogic S, Montpetit B, Hoos HH, Mackworth AK, Ouellette BF, Hieter P. Correlation between the secondary structure of pre-mRNA introns and the efficiency of splicing in Saccharomyces cerevisiae. BMC genomics 2008; 9:355; PMID:18664289; http://dx.doi.org/ 10.1186/1471-2164-9-355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyer M, Plass M, Perez-Valle J, Eyras E, Vilardell J. Deciphering 3′ss selection in the yeast genome reveals an RNA thermosensor that mediates alternative splicing. Mol Cell 2011; 43:1033-9; PMID:21925391; http://dx.doi.org/ 10.1016/j.molcel.2011.07.030 [DOI] [PubMed] [Google Scholar]
- 54.Lovci MT, Ghanem D, Marr H, Arnold J, Gee S, Parra M, Liang TY, Stark TJ, Gehman LT, Hoon S, et al.. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol 2013; 20:1434-42; PMID:24213538; http://dx.doi.org/ 10.1038/nsmb.2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin CL, Taggart AJ, Lim KH, Cygan KJ, Ferraris L, Creton R, Huang YT, Fairbrother WG. RNA structure replaces the need for U2AF2 in splicing. Genome research 2016; 26:12-23; PMID:26566657; http://dx.doi.org/ 10.1101/gr.181008.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raker VA, Mironov AA, Gelfand MS, Pervouchine DD. Modulation of alternative splicing by long-range RNA structures in Drosophila. Nucleic Acids Res 2009; 37:4533-44; PMID:19465384; http://dx.doi.org/ 10.1093/nar/gkp407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miriami E, Margalit H, Sperling R. Conserved sequence elements associated with exon skipping. Nucleic Acids Res 2003; 31:1974-83; PMID:12655015; http://dx.doi.org/ 10.1093/nar/gkg279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pervouchine DD, Khrameeva EE, Pichugina MY, Nikolaienko OV, Gelfand MS, Rubtsov PM, Mironov AA. Evidence for widespread association of mammalian splicing and conserved long-range RNA structures. RNA 2012; 18:1-15; PMID:22128342; http://dx.doi.org/ 10.1261/rna.029249.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leclercq S, Rivals E, Jarne P. DNA slippage occurs at microsatellite loci without minimal threshold length in humans: a comparative genomic approach. Genome biology and evolution 2010; 2:325-35; PMID:20624737; http://dx.doi.org/ 10.1093/gbe/evq023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.da Silva EF, Reha-Krantz LJ. Dinucleotide repeat expansion catalyzed by bacteriophage T4 DNA polymerase in vitro. J Biol Chem 2000; 275:31528-35; PMID:10924513; http://dx.doi.org/ 10.1074/jbc.M004594200 [DOI] [PubMed] [Google Scholar]
- 61.Shimoda N, Knapik EW, Ziniti J, Sim C, Yamada E, Kaplan S, Jackson D, de Sauvage F, Jacob H, Fishman MC. Zebrafish genetic map with 2000 microsatellite markers. Genomics 1999; 58:219-32; PMID:10373319; http://dx.doi.org/ 10.1006/geno.1999.5824 [DOI] [PubMed] [Google Scholar]
- 62.Conrad DF, Keebler JE, DePristo MA, Lindsay SJ, Zhang Y, Casals F, Idaghdour Y, Hartl CL, Torroja C, Garimella KV, et al.. Variation in genome-wide mutation rates within and between human families. Nat Genet 2011; 43:712-4; PMID:21666693; http://dx.doi.org/ 10.1038/ng.862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weber JL, Wong C. Mutation of human short tandem repeats. Hum Mol Genet 1993; 2:1123-8; PMID:8401493; http://dx.doi.org/ 10.1093/hmg/2.8.1123 [DOI] [PubMed] [Google Scholar]
- 64.Crawford AM, Cuthbertson RP. Mutations in sheep microsatellites. Genome Res 1996; 6:876-9; PMID:8889555; http://dx.doi.org/ 10.1101/gr.6.9.876 [DOI] [PubMed] [Google Scholar]
- 65.Yoffe AM, Prinsen P, Gelbart WM, Ben-Shaul A. The ends of a large RNA molecule are necessarily close. Nucleic Acids Res 2011; 39:292-9; PMID:20810537; http://dx.doi.org/ 10.1093/nar/gkq642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boniecki MJ, Lach G, Dawson WK, Tomala K, Lukasz P, Soltysinski T, Rother KM, Bujnicki JM. SimRNA: a coarse-grained method for RNA folding simulations and 3D structure prediction. Nucleic Acids Res 2015; 44(7):e63; PMID:26687716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res 1981; 9:133-48; PMID:6163133; http://dx.doi.org/ 10.1093/nar/9.1.133 [DOI] [PMC free article] [PubMed] [Google Scholar]


