ABSTRACT
Long non-coding RNAs (lncRNAs) are widely expressed and play various roles in cell homeostasis. However, because of their low conservation at the sequence level, recapitulating lncRNA evolutionary history is often challenging. While performing an ultrastructural analysis of viral particles present in uterine glands of gestating opossum females, we serendipitously noticed the presence of numerous structures similar to paraspeckles, nuclear bodies which in human and mouse cells are assembled around an architectural NEAT1/MENε/β lncRNA. Here, using an opossum kidney (OK) cell line, we confirmed by immuno-electron microscopy the presence of paraspeckles in marsupials. We then identified the orthologous opossum NEAT1 gene which, although poorly conserved at the sequence level, displays NEAT1 characteristic features such as short and long isoforms expressed from a unique promoter and for the latter an RNase P cleavage site at its 3′-end. Combining tissue-specific qRT-PCR, in situ hybridization at the optical and electron microscopic levels, we show that (i) NEAT1 is paraspeckle-associated in opossum (ii) NEAT1 expression is strongly induced in late gestation in uterine/placental extracts (iii) NEAT1 induction occurs in the uterine gland nuclei in which paraspeckles were detected. Finally, treatment of OK cells with proteasome inhibitors induces paraspeckle assembly, as previously observed in human cells. Altogether, these results demonstrate that paraspeckles are tissue-specific, stress-responding nuclear bodies in marsupials, illustrating their structural and functional continuity over 200 My of evolution throughout the mammalian lineage. In contrast, the rapid evolution of the NEAT1 transcripts highlights the relaxed constraint that, despite functional conservation, is exerted on this lncRNA.
KEYWORDS: Immuno-electron microscopy, lncRNAs, NEAT1, nuclear bodies, opossum, paraspeckles, pregnancy, ultrastructure, uterine glands
Abbreviations
- lncRNA
long non-coding RNA
- NEAT1 /MENε/β
nuclear-enriched abundant transcript 1/Multiple Endocrine Neoplasia ε/β
- OK
opossum kidney cell line
- MALAT1
metastasis associated in lung adenocarcinoma transcript 1
- DBHS
(Drosophila Behavior Human Splicing) protein family
- TD
Tasmanian devil
- PAS
polyadenylation signal
- I-EM
immuno-electron microscopy
- EM-ISH
in situ hybridization at the electron microscopic level
Introduction
The diversity and frequency of nuclear bodies in various cell types is a landmark, at the ultrastructural level, of the dynamic organization of the cell nucleus.1-6 Nuclear bodies such as the Cajal (Coiled) bodies are ubiquitous in animal and plant cells1 whereas some others, such as the paraspeckles, appear to be cell-type specific.7 In addition, some nuclear bodies are evolutionarily recent with components and functions that might be restricted to certain clades, such as that of mammals. For instance, there is no known homolog of the PML gene in Caenorhabditis elegans, Drosophila, Xenopus or the newt genomes and there is no evidence for PML bodies in these organisms.2,8 Similarly, paraspeckles are belatedly recognized nuclear bodies,9,10 that have been detected only in human and mouse cells, as distinct nuclear structures flanking reservoirs of splicing factors organized in clusters of interchromatin granules (IG) under the electron microscope (EM) and as speckles (SC35 splicing domains) by immunofluorescence (IF) studies. In addition, paraspeckle assembly occurs on NEAT1 (or MEN e/β) lncRNA nascent transcripts expressed by a gene with obvious orthologs found only in eutherian mammals, such as human, mouse or horse.11,12 In human and mouse cells, the NEAT1 gene is transcribed into 2 unspliced transcripts from a unique promoter.12,13 NEAT1_1, the shortest isoform (3.7 and 3.2-kb in human and mouse cells, respectively) is polyadenylated,14,15 while the 3′-end of the long NEAT1_2 isoform (22.7 and 20.7-kb in human and mouse cells respectively) is generated by RNase P cleavage.12 The cleavage site is located immediately upstream of a tRNA-like stem-loop structure and downstream of conserved motifs (U-rich Motif 1, U-rich Motif 2 and an A-rich tract) that were shown to form a triple helix structure critical for stabilization of the human and mouse NEAT1_2 transcripts.12,16-18 Intriguingly, the MALAT1 lncRNA which, in the syntenic region of the eutherian genomes, is encoded by a gene immediately adjacent to NEAT1 is also terminated by this RNase P-driven unorthodox 3′-end processing.17
Competing binding of hnRNPK and of the CPSF6-NUDT21 (CFIm) complex, in the vicinity of the NEAT1_1 polyadenylation signal (PAS), determines the relative abundance of the NEAT1 isoforms in human cells.19 In this model, CFIm binding to UGUA sequences upstream of PAS promotes cleavage and the ensuing polyadenylation of NEAT1_1 whereas hnRNPK binding to a closely-associated polypyrimidine UCCCCUU sequence prevents cleavage and favors NEAT1_2 synthesis. Both NEAT1_1 and NEAT1_2 isoforms are nuclear-retained and were shown to be located within paraspeckles, though with a different pattern. The central part of NEAT1_2 is located within the fibrillar core region of the paraspeckles, whereas its 5′- and 3′-end regions are only found at the edge of the nuclear bodies, together with NEAT1_1.20 The architectural role of each NEAT1 isoform is also distinct: NEAT1_2 is essential for assembly of the paraspeckle components while NEAT1_1 plays an accessory role, enhancing paraspeckle formation only when overexpressed in cells expressing NEAT1_2.13,19 Accordingly, while NEAT1_1 is widely expressed in adult mice tissues, paraspeckle formation is restricted to a small population of cells, such as within the epithelium of the gastric glands, where NEAT1_2 is also detected.7
Among the many RNA- and/or DNA-binding proteins characterized as paraspeckle proteins (PSPs), including the PSPC1, NONO and SFPQ paraspeckle markers of the Drosophila Behavior/Human Splicing (DBHS) family,9,21,22 some are essential for paraspeckle integrity. As shown by RNAi experiments, these PSPs are either required for the synthesis and stability of the architectural NEAT1 lncRNA or indispensable for paraspeckle assembly even in the presence of a steady-state NEAT1 level.19 Moreover, PSPs are significantly enriched in intrinsically-disordered Prion-like domains (PLDs) and the PLD of either RBM14 or FUS have been shown to be required for paraspeckle integrity.23,24
Two molecular functions have been assigned to paraspeckles: first, as a retention site for adenosine-to-inosine hyper-edited mRNAs, modulating their nuclear export and consequently their translation;22,25,26 secondly, as “molecular sponges” that sequestrate and inhibit regulatory factors such as the transcriptional regulators SFPQ and NONO.27,28 However, NEAT1-knock out mice developed normally and did not show any apparent phenotype when compared to their wild-type littermates,7 suggesting that NEAT1 and paraspeckles are accessory stress-responding elements. Along this line, NEAT1 expression and paraspeckle formation were shown to be enhanced in vitro by various stress conditions such as proteasome inhibition, hypoxia, cell differentiation and viral infection.24 In addition, upregulation of NEAT1 in some types of human tumors have been reported recently with, in some cases, a poor prognostic associated with high levels of NEAT1 expression.29-31
It is only recently that obvious NEAT1 induction and paraspeckle formation were observed in vivo and shown to play a role under physiological conditions. NEAT1 KO mice are affected in their reproductive capacity through 2 distinct mechanisms: i) Morphogenesis of the mammary glands is aberrant in NEAT1 KO females, leading to lactation defects and a reduction of pup viability 32 ii) Corpus luteum formation, progesterone secretion and establishment of pregnancy are stochastically impaired in a sub-population of NEAT1 KO mice.33
The evolutionary origin of NEAT1 and paraspeckles remains uncertain. Despite a conserved structural organization of the NEAT1 gene, with 2 transcripts of widely different sizes processed in a different way at their 3′-ends, only the repeat-free NEAT1_1 sequence is highly-conserved between human and mouse. In contrast, NEAT1_2 shows little conservation, except for the 5′-end region shared with NEAT1_1, and it is scattered with organism-specific repeats such as B1 and B2 elements in rodents and Alu sequences in humans. Moreover, neither NEAT1 isoforms was detected by homology search in opossum, chicken or stickleback genomes.12
However, in a study devoted to the role of endogenous retroviral gene expression in opossum placenta formation,34 we serendipitously detected under the EM numerous paraspeckle-like nuclear bodies in the opossum uterine glands at days 12–13 post-coitum (dpc). In the opossum (Monodelphis domestica), a transient placenta is formed shortly before parturition, between 12 and 15 dpc of a 15 day-long gestation, with the marsupial fetus being rapidly released and further developing via lactation. Formation of the transitory placenta takes place in close contact with a uterine epithelium that covers secretory uterine glands. These glands degenerate during late gestation, concomitant with the development of the placenta, suggesting a shift of the embryo's nutrition from histotrophic (uptake of uterine secretions) to hemotrophic (exchange of material and gases between fetal and maternal blood vessels within the placenta) in early and late gestation, respectively.35 Here we show that isoforms of the lncRNA NEAT1 are specifically expressed in the opossum uterine glands during late gestation and that NEAT1 gene organization and paraspeckle formation predates the divergence of marsupials and eutherian mammals that took place about 190 Mya.
Results
Opossum uterine glands in late gestation contain paraspeckle-like structures
We previously reported that, during pregnancy (12–13 dpc), opossum uterine glands contain retroviral particles which were budding from - or trapped within - microvilli of the lumen.34 Unexpectedly, over the time course of these experiments, small dense structures were frequently observed under the EM, within the nuclei of the peripheral cell layer of the uterine glands (Fig. 1A and Fig. S1). By their aspect, size and distribution, these structures are reminiscent of the paraspeckle nuclear bodies previously characterized by conventional electron microscopy in human and mouse cell lines.10,20,36,37 Notably, they were fibrillar, closely associated with clusters of IG (the ultrastructural counterpart of the IF speckles) and occasionally found as a group of 2 to 5 elements (Fig. 1B). Finally, they were cylindrical in shape with a constrained diameter of about 300 nm as measured on EM micrographs (Fig. 1C), 2 properties that, among known nuclear structures, are unique to paraspeckles.20,27 These structures were absent in nuclei from adjacent tissues such as the underlying uterine muscle or the placental epithelium and therefore appeared to be cell-type specific.
Figure 1.

Paraspeckle-like structures as observed in opossum uterine glands by conventional EM. (A) Left frame. Hematoxylin eosin saffron (HES)-stained section showing cellular organization of placental and uterine tissues during late gestation (12dpc). The placental syncytiotrophoblast layer (St) is in direct contact with the maternal uterine epithelium (Ue). Underneath the uterine epithelium, uterine glands (Ug) form rings, with heavily-stained peripheral cell nuclei. L: lumen. Right frame. EM micrograph showing dense paraspeckle-like nuclear structures (arrows) as observed in ultra-thin sections of opossum uterine glands at the same stage. Notice the compact fibrillar structure (inset). Ch: peripheral heterochromatin. Scale bars: 20, 2 and 0.2 µm respectively. (B) Micrographs showing paraspeckle-like fibrillar structures that are (i) tightly-associated with Interchromatin Granules (IG) (ii) cylindrical (iii) in clusters. Scale bars: 0.2 µm. (C) Values of the short (Sx) and long axis (Lx) of 31 paraspeckle-like structures (as measured in B, right frame), when plotted by increasing Lx, illustrate variable length and constant diameter (mean Sx 295 +/− 35 nm). Among nuclear bodies in human cells, this elongation profile is distinctive of paraspeckles.20,27
Characterization of opossum paraspeckles by immunogold electron microscopy
The tissue samples analyzed under the EM had been post-fixed with osmic acid and embedded in Epoxy resin (Epon). As a consequence, they were not suitable for an unequivocal characterization of bona fide paraspeckles by immunogold electron microscopy (I-EM). To further assess the presence of paraspeckles in marsupial, an opossum kidney cell line (OK cells) was grown for both IF and, following Lowicryl K4M embedding, immuno-gold electron microscopic (I-EM) studies with antibodies directed against the human DBHS proteins. IF labeling of OK cells with an anti-NONO antibody revealed the presence of distinct but rare nuclear dots (Fig. 2). Treatment of OK cells with 5 µM MG132 (a proteasome inhibitor) for 6 h increased the frequency of NONO-positive foci (Fig. 2A), as previously shown in human and mouse cell lines upon proteasome inhibition.27 Further analysis by I-EM confirmed that OK cells contain genuine paraspeckles, characterized by their ultrastructure, their frequent association with IG and their high content in PSPC1, SFPQ and NONO paraspeckle markers (Fig. 2B). Consistently, under the EM, paraspeckles were seen sporadically on thin-sections of untreated OK cells and were more frequent following proteasome inhibition. Finally, using a biotinylated DNA probe from the 5′ region of the opossum NEAT1 gene (see below), we show by electron microscopic in situ hybridization (EM-ISH) that it hybridized to NEAT1 RNA sequences located exclusively at the edge of the paraspeckles (Fig. 2C). This is consistent with our previous demonstration in both human and mouse cells that the 5′-ends of the NEAT1 isoforms are strictly located at the periphery of the paraspeckles.20,27 Altogether, these results show that paraspeckle assembly, in strict association with NEAT1 lncRNA and under control of the proteasome, is an ancient and conserved attribute of mammalian cells.
Figure 2.

OK cells contain NEAT1-associated paraspeckle nuclear bodies that are induced by proteasome inhibition. (A) IF of control and MG132-treated (5 µM, 6h) OK cells (left and right frames respectively) stained with an anti-NONO antibody (green) and counterstained with DAPI (DNA, blue) reveals scattered fluorescent foci in nuclei of untreated OK cells and their increased frequency upon proteasome inhibition (arrow-heads). Scale bars: 10 µm. (B) Ultrastructural characterization of opossum paraspeckles by I-EM with antibodies against human DBHS proteins. Ultrathin sections of Lowicryl-embedded MG132-treated OK cells were incubated with the indicated primary antibodies and secondary antibodies conjugated to 10 nm gold particles. Labeled paraspeckles are shown. IG: Interchromatin granules. Scale bars: 100 nm. (C) A biotinylated opossum NEAT1 5′-end DNA probe (see below) hybridized onto ultrathin sections of Lowicryl-embedded MG132-treated OK cells detects NEAT1 RNA 5′-end at the paraspeckle periphery. DNA/RNA hybrids were detected with an anti-biotin antibody conjugated to 10 nm gold particles. Scale bars: 100 nm.
Mapping of the opossum NEAT1 gene
To characterize the NEAT1 gene in the opossum genome, we first analyzed its genetic structure at the syntenic locus, which in eutherian mammals, contains the FRMD8, NEAT1, MALAT1 and SCYL1 genes in this order and in the same transcriptional orientation. Comparison of the opossum, human and mouse genomic sequences revealed significant conservation of the FRMD8, MALAT1 and SCYL1 gene regions but, as previously reported,12 no sequence similarity in the NEAT1 region (Fig. 3A). For instance, while an RNase P consensus sequence was easily recognizable at the 3′-end of the opossum MALAT1 gene, no such sequence was found within the putative opossum NEAT1 region extracted from the UCSC genomic database.12,18 Because of the ancient divergence around 190 Mya between marsupials and eutherian mammals, a homology-based search of non-coding genes with low sequence conservation is challenging. In order to reduce the evolutionary distance, we then compared the opossum and Tasmanian devil (TD) genomic sequences (Fig. 3B). As expected, conservation was increased for the FRMD8, MALAT1 and SCYL1 genes. Interestingly, it also revealed an approximately 5-kb long repeat-free region, downstream of the FRMD8 gene, with significant homologies between the 2 marsupial genomes. This region resembled, in size and positioning, the homologous repeat-free region shared by the human and mouse NEAT1_1 loci.12,13 In addition, in a region yet only partially assembled in the opossum genome, a downstream RNase P consensus sequence was found in the corresponding region of the TD genome, suggesting conservation of the 3′-end cleavage site of the long NEAT1_2 isoform.
Figure 3.

Homology search of NEAT1 in the opossum genome. (A) Alignment of syntenic FRMD8-SCYL1 region in human, mouse and opossum genomes shows significant DNA sequence conservation at the protein coding FRMD8 and SCYL1 and non coding MALAT1 genes but not within the presumptive NEAT1 region. RNase P cleavage sites (P) at the 3′-end of the human NEAT1 and MALAT1 genes and at the 3′-end of the opossum MALAT1 gene are underlined (arrow-head). (B) Comparison of opossum and Tasmanian Devil (TD) marsupial genomes highlights an homologous repeat-free DNA sequence downstream of FRMD8 (star) and, within TD but not opossum DNA, an RNase P site corresponding to the NEAT1_2 3′-end. (C) Mapping of the RNase P processed 3′-end of the opossum NEAT1 gene by sequencing of un-annotated regions. The location on chromosome 8 of the opossum genome (UCSC database) of 3 gaps that were PCR-amplified and sequenced is shown as red boxes. The triple-helix forming motifs (green) and modeled structure of the downstream tRNA-like element found in gap 3 are depicted. The putative RNase P and RNase Z cleavage sites at the 3′-end of the opossum NEAT1_2 isoform are inferred by comparison with the tRNA-like structure in the human locus.
To reconstitute a full-length NEAT1 opossum sequence, 3 gaps of undefined nucleotides (stretches of N's within the UCSC genomic database) of 87, 532 and 951 nt were PCR-amplified and sequenced (Fig. 3C), resulting in a continuous 31-kb long stretch of genomic sequence starting at nt 13 631 789 of the opossum chromosome 8. Within the reconstituted sequence corresponding to the longest gap, a typical RNase P consensus cleavage site was uncovered. Composed of a downstream 52 bp tRNA-like stem-loop structure and of 3 upstream canonical motifs, a U-rich Motif 1, a U-rich Motif 2 and an A-rich tract (Fig. 3C), this sequence, clearly, delineated a conserved 3′-end of the opossum NEAT1_2 isoform.12,16-18
Evidence for 2 opossum NEAT1_1 isoforms
Using this information and in the absence of reported ESTs in opossum or TD databases, we determined the sequence of the 5′- and 3′-ends of the short opossum NEAT1_1 isoform by RACE experiments. By 5′-RACE with cDNAs from uterine/placental tissues at 13 dpc, a single transcription initiation site was found 30 bp downstream of a presumptive TATA box. Interestingly, the nucleotide sequence of both the TATA box and the orthologous transcription initiation site were conserved in both TD and wallaby (Macropus eugenii) NEAT1 loci (Fig. 4A).
Figure 4.
Delineating the opossum NEAT1_1 transcription unit by RACE. (A) A single NEAT1 transcription initiation site was mapped by 5′-RACE on cDNA from total RNA extracted from a mixed uterine/placental sample at 13 dpc. Conservation of proximal DNA sequence in marsupials is illustrated. TATA box is underlined by a red square. Positions of qPCR, riboprobes and DNA probe primers are depicted. (B) Two polyadenylated NEAT1_1 3′-ends downstream of canonical (AATAAA) PAS were uncovered by 3′-RACE from the same RNA sample. The last nucleotide preceding the poly-A tail is indicated for both 3′-ends. Different DNA sequence conservation of the NEAT1_1a and NEAT1_1b PAS within the marsupial and eutherian genomes is shown in red boxes. (C). Relative abundance of the 2 NEAT1_1 opossum isoforms within oligo-dT-primed 3′-RACE cDNA from a uterine/placental sample at 13 dpc. Positions of qPCR primers specific of the NEAT1 1_1b or 1_1a + 1_1b isoforms are depicted in (B). Relative abundance (relative to the amount of the housekeeping RPLP19 gene) +/- SEM was determined from duplicate samples in 2 independent qPCR experiments.
Two different 3′-ends for NEAT1_1 were obtained by 3′-RACE. Both corresponding transcripts were poly-adenylated and cleaved 14 and 12 nt downstream of canonical (AATAAA) PAS. As the 2 3′-ends were 478 bp apart, they defined 2 opossum NEAT1_1 isoforms, NEAT1_1a and NEAT1_1b, of 2 716 and 3 194 nt respectively (Fig. 4B), contrasting with the single NEAT1_1 isoform found in human and mouse cell lines. 12-14 To determine 3′-end usage of both NEAT1_1 isoforms, we measured their relative abundance by qPCR using the oligodT-primed cDNAs derived from uterine/placental tissues at 13 dpc. In these tissues, NEAT1_1a and 1_1b were found to be approximately equally represented at this developmental stage (Fig. 4C).
Homology search indicated that the DNA sequence of the NEAT1_1a extremity was specific to the marsupial genomes whereas the DNA sequence of the further downstream NEAT1_1b 3′-end was well-conserved in the genomes of eutherian mammals, including human and mouse (Fig. 4B). Notably, the NEAT1_1b 3′-end of the marsupial genomes contains the closely-spaced TGTA repeats and the poly-pyrimidine stretch immediately upstream of the PAS (Fig. S2), that have been shown to regulate the balance between the short and long NEAT1 isoforms in human cells. 19 In addition, the opossum NEAT1_1b isoform and the mouse NEAT1_1 transcript are very similar in size (3 177 and 3 194 nt, respectively). Finally, the mapping of the NEAT1 transcription initiation site and of the RNase P cleavage consensus sequence in the opossum genome defines an 18 607 nt long NEAT1_2 isoform, as compared to the 20 771 and 22 743 nt long mouse and human transcripts. Collectively, these data demonstrate a global structural conservation of the NEAT1 gene in mammals despite an overall lack of sequence similarity between the marsupial and eutherian orthologs.
NEAT1_2 is specifically expressed in uterine gland nuclei that contain paraspeckles
To determine whether a high frequency of paraspeckle-like bodies in opossum uterine glands in late gestation correlates with high NEAT1 expression, we examined the transcript levels of the short (NEAT1_1a+b) and long (NEAT1_2) isoforms in a panel of opossum tissues. At 12, 13 and 14 dpc, the uterus and the attached placenta are tightly interpenetrated and were collected and analyzed as a whole, while embryonic tissues were collected separately. Quantitative RT-PCR (qRT-PCR) analyses were performed using primers designed to amplify either the NEAT1_1 and NEAT1_2 transcripts or only the NEAT1_2 transcript (as shown in Fig. 4A). Although detectable in all tissues (Fig. 5), NEAT1 expression is particularly high in uterus/placenta samples taken at late gestation stages (12–14 dpc). Interestingly, NEAT1_2 is highly expressed only in uterus/placenta samples and its expression, particularly low in non-pregnant uterus, increases during gestation. In contrast, the NEAT1 isoforms are poorly expressed in the embryos at the same developmental stages.
Figure 5.
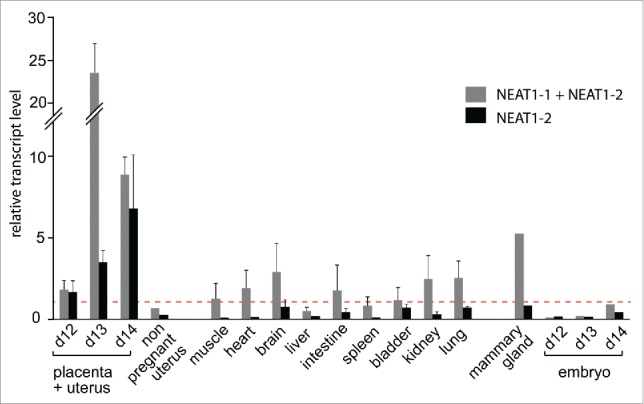
Real-time RT-qPCR analysis of the opossum NEAT1 transcripts in various tissues.Transcript levels are expressed as the ratio of their expression level to that of the RPL0 control gene (see Methods). Due to the high interpenetration of maternal and fetal tissues, placental and uterine tissues are analyzed as a whole at 3 gestational times (12, 13 and 14 dpc). Values are the means of duplicates from 3 samples ± SEM, except for the non pregnant uterus, the mammary gland and embryo samples, where only one sample was analyzed. Dotted line indicates the ratio value equal to 1. Location of primers with respect to NEAT1 isoforms is indicated in Fig. 4A.
In mouse cells, the NEAT1_2 isoform is essential for paraspeckle assembly whereas NEAT1_1 plays an accessory role. 24 To further characterize the expression pattern of NEAT1_2, we performed in situ hybridization (ISH) experiments on paraffin sections of opossum uterine/placental samples. Specific digoxigenin-labeled antisense riboprobes were synthesized for the detection of NEAT1_2 and the corresponding sense riboprobes were made in parallel to be used as negative controls (see Fig. 4A). As illustrated in Fig 6, specific labeling was observed only with the antisense, but not the sense, probes. Specific staining with the antisense probes was observed at the level of the uterine glands, and not in the adjacent uterine epithelial cells (Fig. 6). Remarkably, labeled foci can be seen within the nuclei of the peripheral cells of the endometrial uterine glands, precisely overlapping the nuclei in which paraspeckles-like structures were originally detected under the EM. In conclusion, NEAT1-driven strong and transient induction of paraspeckle assembly occurs specifically within the opossum uterine glands at the end of gestation. This physiological process precedes only shortly parturition and degeneration of the uterine glands.
Figure 6.
NEAT1_2 expression in opossum pregnant uterine glands. Serial sections of opossum pregnant uterus stained with (A) HES, (B-D) using digoxigenin-labeled NEAT1_2 sense (B) or antisense (C) riboprobes revealed with an alkaline phosphatase-conjugated antidigoxigenin antibody. (D) Enlargement of the antisense riboprobe staining. Specific staining is observed at the level of the nuclei of the uterine gland cells, while the uterine epithelium is unlabelled. Location of riboprobes is indicated in Fig. 4A. (A-C): scale bar: 50 μm. (D): scale bar: 10µm.
Discussion
Given that the description of paraspeckle nuclear bodies was so far limited to human and mouse cell nuclei, that they were dispensable for normal development in mice, and that there was not yet evidence of a clear ortholog of the NEAT1 gene in the opossum genome, the hypothesis of a recent origin of paraspeckles, with recently acquired non-essential functions, could not be ruled out. In contrast, the present results, providing evidence of their existence in the opossum uterine glands, demonstrate that these nuclear bodies are an ancient attribute of nuclear organization in the entire placental mammal lineage. The structural conservation of the paraspeckles despite distinct relaxed constraint on the NEAT1 isoforms is illustrated in Fig. 7.
Figure 7.

Paraspeckle formation predates divergence of marsupial and eutherian mammals. (A) Paraspeckle emergence at least 200 Mya in a common ancestor to eutherian and marsupial mammals is suggested by their structural conservation in human, mouse and opossum cell nuclei. Micrograph scale bars: 100 nm. (B) Level of NEAT1 identity between species was defined by measuring the % of sequence aligned (cov) and the % of identity within aligned segments (id), using BLASTN. These values are reported on the graph with the identity color code shown on the left scale to underline the higher conservation of NEAT1_1 as compared to NEAT1_2 and the overall low conservation of both NEAT1 isoforms between marsupial and eutherian mammals.
Conservation of the NEAT1 gene structure in marsupials
We provide evidence here that the main structural features of the NEAT1 locus, with a single initiation site for short and long non coding transcript isoforms that are differentially processed at their 3′-ends, are conserved in opossum. Our study, however, further illustrates the high level of variability of the NEAT1 primary DNA sequence. The overall low level of sequence homology of the long NEAT1_2 isoforms, as seen when comparing human and mouse DNA, is also found when comparing opossum and TD DNA. As, in both cases, these organisms diverged about 60–80 Mya, this observation attests the high degree of evolvability of this locus at work in all mammalian genomes. Along these lines, the higher level of sequence conservation restricted to the NEAT1_1 region in mouse and human cells was also found when comparing the opossum and TD transcripts. However, when comparing marsupial and eutherian NEAT1_1 DNA sequences by BLASTN, again no significant homology was found, indicating, this time on a larger scale (190 Mya), a slower, yet substantial instability. This is likely reflecting different constraints linked to different functions: so far NEAT1_2 has been only implicated in paraspeckle assembly whereas NEAT1_1 is also associated with - and regulates - a number of promoters.31,38 In this context, despite overall genomic variability, we show that some key NEAT1 sequence elements are highly conserved. This is the case for the tRNA-like and triplex-forming sequences found at the 3′-end of the opossum NEAT1_2 transcript, which are required respectively for RNase P recognition and cleavage and for stabilization of transcripts lacking a poly(A) tail. 16-18 This is also the case for the TGTA repeats and polypyrimidine stretch immediately upstream of the NEAT1_1 3′-end that is conserved in mammals (NEAT1_1b 3′-end in marsupials). The specific conservation of these regulatory elements in the marsupial genomes strengthens and extends the model postulating that CFIm binding to TGTA/UGUA repeats results in NEAT1_1 cleavage and polyadenylation whereas alternative HNRNPK binding to a TCCCCT/UCCCCU sequence arrests CFIm-dependent NEAT1_1 polyadenylation and promotes NEAT1_2 synthesis in human cells. 19 However, the absence of these conserved sequences upstream of the opossum NEAT1_1a 3′-end suggests a different mechanism regulating this marsupial specific isoform.
Specificity of NEAT1 gene expression in opossum
A singularity of the opossum NEAT1 gene is the expression of 2 poly-adenylated NEAT1_1 isoforms with canonical PAS (AATAAA) immediately upstream of the poly(A). Quantitation by qRT-PCR of RNA extracted from a mixed uterus/placenta sample in late gestation demonstrate similar abundance for both NEAT1_1 isoforms in opossum, indicating that neither of these 2 PAS corresponds to a cryptic 3′-end. Moreover, both PAS DNA sequences and their spacing (478, 557 and 580 bp in the opossum, TD and wallaby genomes, respectively) are conserved in marsupial genomes. Further comparisons of the NEAT1_1 3′-end usage in marsupials was impeded, however, by a lack of annotated transcripts, except for a limited repertoire of ESTs in wallaby. Examination of reported 3′-end-primed wallaby cDNAs reveals only one occurrence of NEAT1_1a in a hypothalamus-derived EST collection (FY 596756) whereas the NEAT1_1b 3′-end was found 7 times in the hypothalamus (e.g. FY590610) and 10 times in gravid uterus (e.g., FY 539726) ESTs. Thus, the 3′-end that appears most frequently used in wallaby is the one that is conserved and currently in use in human and mouse cells. Intriguingly, a 3.9-kb non coding transcript (XR_485095) originating downstream of the FRMD8 gene in platypus (Ornithorhynchus anatinus) has been reported to be a NEAT1_1 homolog.39 Although not detectable by homology with the marsupial NEAT1_1 isoforms, this transcript also ends at the NEAT1_1 3′-end that is common to all mammals. However, it does not contain any upstream PAS corresponding to the NEAT1_1a marsupial isoform. Therefore, it remains uncertain whether a shorter NEAT1_1a isoform pre-existing in early mammals was lost in eutherians or whether it evolved specifically in the marsupial lineage.
Conservation trends in paraspeckle ultrastructure and functions
Contrasting with the low sequence conservation of the NEAT1 primary sequence, the opossum proteins of the DBHS family show a high level of amino-acid identity (in the range of 90%) with their human orthologs. This explains why antibodies directed against the human proteins were efficient opossum paraspeckle markers in this study. This is also likely important for the ultrastructural constancy of the paraspeckles, resulting in their fortuitous detection in this study. Under the EM, paraspeckles were highly frequent in the opossum uterine glands in late gestation while they were not detected in adjacent embryonic and maternal tissues or in opossum kidney (not shown). Such a high rate of occurrence was unexpected because paraspeckles are rarely seen in our routine EM observations of various mouse tissues, such as heart, liver, kidney or when detected in these tissues by RNA-FISH and IF. 7 However, in vitro, enhanced NEAT1 expression and paraspeckle accumulation were shown to occur during human ES cell differentiation and murine myoblast differentiation,12,25 suggesting links with specific transcriptional programs. We also previously showed that paraspeckles become very abundant following proteasome inhibition in HeLa cells. In this setting, paraspeckles act as molecular sponges, recruiting and sequestrating transcriptional regulators such as SFPQ and NONO from the nucleoplasm, including by displacement from gene promoters.27 Their abundance in opossum uterine glands, as assessed by their presence in adjacent nuclear profiles under the EM, was as high as in HeLa cells after a 17h long MG132 treatment. It therefore suggests that the paraspeckles naturally induced in late gestation also modulate gene expression by sequestrating opossum regulatory factors.
Of note, this is the third report of paraspeckle induction in vivo. As was the case for mammary glands and corpus luteum development in mice,32,33 this event occurs in a gland with functions related to female reproduction. Although further insights into paraspeckle functions in opossum would likely require generating NEAT1 KO animals, we further noticed that in vivo dual NEAT1 up-regulation and paraspeckle assembly is taking place in transient tissues shortly before they undergo degeneration.32,33 During opossum gestation, the embryo first relies on uterine secretions and the development of the short-lived placenta between 11 and 15 dpc is associated with a drastic reduction in thickness and density of the uterine gland pad, from 3 mm at 9 dpc to 0.2–0.4 mm at 14 dpc.35 As observed in cancer cells29 and upon proteasome-inhibition in MEFs,27 NEAT1 enhanced expression (and the likely ensuing paraspeckle assembly) is protecting cells against stress-induced apoptosis. Hence, high NEAT1 expression and paraspeckle formation in degenerating tissues might be specifically involved in slowing-down cell dismantlement under harsh conditions. This would also be compatible with their notable presence in the epithelium of gastric glands in mice, independently of their gender.7 Alternatively, NEAT1 induction and paraspeckle enlargement have previously been reported in response to viral infections.28,40,41 Here, the formation of NEAT1-driven paraspeckles is observed in a tissue secreting viral particles, albeit at a low level. Hence, formation of paraspeckles in the opossum uterine glands could also be a response to this viral load, most probably resulting from the expression during pregnancy of an endogenous retrovirus.34
Collectively, the above studies indicate that enhanced paraspeckle assembly and concurrent NEAT1 upregulation in vivo in transient tissues linked to pregnancy or in vitro upon proteasome inhibition are nuclear mechanisms that have been conserved over the last 200 million years, suggesting pivotal roles in mammalian biology.
Material and methods
Database screening and sequence analyses
Genomic sequences from the FRMD8-SCYL1 locus were extracted from the opossum (6.8X coverage assembly of the Monodelphis domestica genome, Broad monDom5, Oct. 2006), Tasmanian devil (Sarcophilus harrisii, UCSC/NCBI, sarHar1, Feb. 2011), human (Homo sapiens, UCSC GRCh37/hg19, 2009) and mouse (Mus musculus, UCSC GRCm38, 2011) genomes from the UCSC website. Analyses of syntenic conserved sequences were performed using the MultiPipMaker alignment tool (http://pipmaker.bx.psu.edu) using either the human or the opossum genome sequences as a reference. Multiple alignments of the NEAT1 transcripts were performed using the EMBL-EBI web_server and the Clustal Omega package (http://www.ebi.ac.uk/Tools/msa/clustalo/). tRNA-like structures were predicted with Quickfold (http://unafold.rna.albany.edu/?q=DINAMelt/Quickfold).
Cells, tissues and reagents
Maintenance of and experiments with animals were performed under registration of the responsible State Office of Health and Social Affairs Berlin (LaGeSo), Germany. Post-coïtum tissue sampling and dissection was carried out as previously reported.34 The opossum OK cells (ATCC CRL-1840) were grown in DMEM supplemented with 10% (vol/vol) FCS (Invitrogen), 100 mg/mL streptomycin, and 100 U/mL penicillin at 37°C with 5% CO2. MG132 (Sigma) was prepared as a 20 mM stock solution in DMSO and diluted in culture growth medium to a final concentration of 5 µM. Antibodies for immuno-fluorescent (IF) and immuno-electron microscopic (I-EM) studies were as follows: anti-SFPQ (mouse, Sigma P2860, 1:50 in PBS for I-EM); anti-NONO: (mouse monoclonal antibody, diluted 1:25 in PBS for I-EM, 1:200 for IF) and anti-PSPC1 (rabbit, 1:25 in PBS for I-EM) as in.20
Immunofluorescent staining of OK cells
Cells were fixed with 4% formalin/H2O for 20 min at RT, permeabilized in 0.2% Triton-X-100/PBS, and incubated 1h at room temperature with primary anti-NONO antibody and then with secondary Alexa Fluor™ 488 anti-mouse antibodies, diluted at 1:500 (Life technologies, Carlsbad, CA). Antibodies were diluted in PBST/BSA (PBS/Tween20 0.5%/BSA 3). Cells were stained with 4,6-diamidino-2-phenylindole (DAPI), mounted in Vectashield (Vector Laboratories, Burlingame, CA), and imaged with an Olympus microscope (Nikon, Tokyo, Japan) using a 40/0.75 objective.
Optical In situ hybridization
Freshly collected opossum (Monodelphis domestica) placentae were fixed in 4% paraformaldehyde at 4°C, embedded in paraffin, and serial sections (7 μm) were either stained with hematoxylin and eosin or used for in situ hybridization. Three distinct NEAT1_2 specific PCR-amplified fragments of around 400 bp (primers listed in Table S1 and mapped on Fig 4A) were cloned into pGEM-T Easy (Promega) for in vitro synthesis of the antisense and sense riboprobes, generated with SP6 RNA polymerase and digoxigenin 11-UTP (Roche Applied Science) after cDNA template amplification. Sections were processed, hybridized at 42°C overnight with the pooled riboprobes and incubated further at room temperature for 2 h with alkaline phosphatase-conjugated anti-digoxigenin antibody Fab fragments (Roche Applied Science). Staining was performed with nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indoyl phosphate (BCIP) phosphatase alkaline substrate, as indicated by the manufacturer (Roche Applied Science).
Electron microscopic analysis of cells and tissues
Conventional ultrastructural microscopy after Epon embedding was as in.42 Freshly grown OK cells and tissue fragments stored in 4% formaldehyde were fixed in 2% glutaraldehyde in 0.1 M phosphate buffer for 1 h. Cells were collected by scraping and centrifuged. After rinsing in phosphate buffer, tissue fragments or cell pellets were post-fixed in 2 % osmium tetroxide, dehydrated in ethanol and embedded in Epon™ 812. Ultrathin sections were stained with standard uranyl acetate and lead citrate solutions prior to observation with a Tecnai 12 electron microscope (FEI, Hillsboro, OR). Digital images were taken with a SIS MegaviewIII charge-coupled device camera (Olympus, Tokyo, Japan).
Immuno-electron microscopy (I-EM)
Protein and nucleic acid localization were performed on thin sections of cells embedded in Lowicryl K4M (Polysciences Inc., PA, USA) as in.5 OK cells fixed in situ for 1h at 4°C with 4% formaldehyde (freshly prepared from paraformaldehyde in 0.1M Sörensen phosphate buffer pH 7.3) were scraped-off and centrifuged. After rinsing in phosphate buffer, cell pellets were equilibrated in 30% methanol and deposited in a Leica EM AFS2/FSP automatic reagent handling apparatus (Leica Microsystems). Lowicryl polymerization under UV was for 48h at −20°C followed by 40h at +20°C. Ultra-thin sections were incubated at room temperature for 1 h with the primary antibody and for 30 min with the secondary anti-mouse or anti-rabbit antibody coupled to 10 nm gold particles (BBI international, Cardiff, UK). To better visualize gold particles on electron dense structures such as the paraspeckles, thin-sections were briefly contrasted with uranyl acetate but lead citrate staining was omitted.
Electron microscopic in situ hybridization (EM-ISH)
A 1.674-kb long DNA fragment PCR-amplified from opossum genomic DNA was biotinylated by nick-translation with biotin16-dUTP (Roche Applied Science). This opossum NEAT1 5′-end DNA probe (nt 1025-2699) hybridizes to an RNA region that is common to all opossum NEAT1 isoforms. Hybridization conditions and detection of RNA/DNA hybrids with goat anti-biotin antibody conjugated to 10 nm gold particles (BBI International) was as previously described in.5,20
RT-PCR
5′ and 3′ RACE were performed with 100 ng of DNase-treated RNA, using the SMARTer RACE cDNA Amplification Kit (Clontech). In order to detect additional 5′- or 3′-ends within the opossum NEAT1_1 genomic sequence, interspersed 5′ and 3′ RACE primers were used. Only proficient 5′ and 3′ RACE primers are listed in Table S1.
NEAT1 mRNA expression was determined by qRT-PCR. Reverse transcription was performed with 100 ng of DNase-treated RNA. PCR was carried out with 5 μl of diluted (1:20) cDNA in a final volume of 25 μl using the FastSYBR Green PCR Master Mix (Qiagen) in an ABI PRISM 7000 sequence detection system. Transcript levels were normalized relative to the amount of the housekeeping RPL0 gene (ribosomal protein L0). Samples (3 samples per organ unless otherwise stated) were assayed in duplicate. Primers are listed in Table S1 and mapped on Fig 4A. NEAT1 genomic and transcripts sequences have been submitted to the GenBank database under accession KX036207 and KX036208-210 respectively.
Pairwise comparison of NEAT1 orthologs
BLASTN default values were used except for “open gap” and “extension gap” values that were minimized so as to favor global alignment. A percentage of coverage was defined as the fraction of the 2 sequences that were aligned and a mean percentage of identity within the aligned sequences was further calculated. The product of these 2 values was used to generate a color-coded homology score in Fig. 7.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Professor Ulrich Zeller (Humbolt University, Berlin) for providing opossum tissue samples at precisely defined time points during gestation; A. Leskowicz and V. Marquis, UMR5503, for the gift of OK cells; Anne Dupressoir, Christian Lavialle and Dominique Weil for discussion and critical reading of the manuscript.
Funding
This work was supported by general funding from the CNRS and by grants from Foundation ARC to G.P and from Ligue Nationale Contre Le Cancer (Equipe Labellisee) and Agence Nationale de la Recherche (Retro-Placenta) to T.H.
References
- 1.Gall JG. The centennial of the Cajal body. Nat Rev Mol Cell Biol 2003; 4:975-80; PMID:14685175; http://dx.doi.org/ 10.1038/nrm1262 [DOI] [PubMed] [Google Scholar]
- 2.Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol 2010; 2:a000661; PMID:20452955; http://dx.doi.org/ 10.1101/cshperspect.a000661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet 2011; 27:295-306; PMID:21680045; http://dx.doi.org/ 10.1016/j.tig.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sleeman JE, Trinkle-Mulcahy L. Nuclear bodies: new insights into assembly/dynamics and disease relevance. Curr Opin Cell Biol 2014; 28:76-83; PMID:24704702; http://dx.doi.org/ 10.1016/j.ceb.2014.03.004 [DOI] [PubMed] [Google Scholar]
- 5.Souquere S, Weil D, Pierron G. Comparative ultrastructure of CRM1-Nucleolar bodies (CNoBs), Intranucleolar bodies (INBs) and hybrid PML/p62 bodies uncovers new facets of nuclear body dynamic and diversity. Nucleus 2015; 6:326-38; PMID:26275159; http://dx.doi.org/ 10.1080/19491034.2015.1082695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spector DL. SnapShot: Cellular bodies. Cell 2006; 127:1071; PMID:17129789; http://dx.doi.org/ 10.1016/j.cell.2006.11.026 [DOI] [PubMed] [Google Scholar]
- 7.Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol 2011; 193:31-9; PMID:21444682; http://dx.doi.org/ 10.1083/jcb.201011110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borden KL. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol Cell Biol 2002; 22:5259-69; PMID:12101223; http://dx.doi.org/ 10.1128/MCB.22.15.5259-5269.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. Paraspeckles: a novel nuclear domain. Curr Biol 2002; 12:13-25; PMID:11790299; http://dx.doi.org/ 10.1016/S0960-9822(01)00632-7 [DOI] [PubMed] [Google Scholar]
- 10.Visa N, Puvion-Dutilleul F, Bachellerie JP, Puvion E. Intranuclear distribution of U1 and U2 snRNAs visualized by high resolution in situ hybridization: revelation of a novel compartment containing U1 but not U2 snRNA in HeLa cells. Eur J Cell Biol 1993; 60:308-21; PMID:8330629 [PubMed] [Google Scholar]
- 11.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 2011; 13:95-101; PMID:21170033; http://dx.doi.org/ 10.1038/ncb2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 2009; 19:347-59; PMID:19106332; http://dx.doi.org/ 10.1101/gr.087775.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A 2009; 106:2525-30; PMID:19188602; http://dx.doi.org/ 10.1073/pnas.0807899106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009; 33:717-26; PMID:19217333; http://dx.doi.org/ 10.1016/j.molcel.2009.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 2007; 8:39; PMID:17270048; http://dx.doi.org/ 10.1186/1471-2164-8-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc Natl Acad Sci U S A 2012; 109:19202-7; PMID:23129630; http://dx.doi.org/ 10.1073/pnas.1217338109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 2008; 135:919-32; PMID:19041754; http://dx.doi.org/ 10.1016/j.cell.2008.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev 2015; 26:2392-407; PMID:23073843; http://dx.doi.org/ 10.1101/gad.204438.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. Embo J 2012; 31:4020-34; PMID:22960638; http://dx.doi.org/ 10.1038/emboj.2012.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Souquere S, Beauclair G, Harper F, Fox A, Pierron G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell 2010; 21:4020-7; PMID:20881053; http://dx.doi.org/ 10.1091/mbc.E10-08-0690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell 2005; 16:5304-15; PMID:16148043; http://dx.doi.org/ 10.1091/mbc.E05-06-0587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell 2005; 123:249-63; PMID:16239143; http://dx.doi.org/ 10.1016/j.cell.2005.08.033 [DOI] [PubMed] [Google Scholar]
- 23.Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A, Knott GJ, Iyer KS, Ho D, Newcombe EA, et al.. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J Cell Biol 2015; 210:529-39; PMID:26283796; http://dx.doi.org/ 10.1083/jcb.201504117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamazaki T, Hirose T. The building process of the functional paraspeckle with long non-coding RNAs. Front Biosci (Elite Ed) 2015; 7:1-41; PMID:25553361; http:/dx.doi.org/ 10.2741/e715 [DOI] [PubMed] [Google Scholar]
- 25.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell 2009; 35:467-78; PMID:19716791; http://dx.doi.org/ 10.1016/j.molcel.2009.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elbarbary RA, Maquat LE. CARMing down the SINEs of anarchy: two paths to freedom from paraspeckle detention. Genes Dev 2015; 29:687-9; PMID:25838539; http://dx.doi.org/ 10.1101/gad.261438.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Benard M, Fox AH, et al.. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell 2014; 25:169-83; PMID:24173718; http://dx.doi.org/ 10.1091/mbc.E13-09-0558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, et al.. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell 2014; 53:393-406; PMID:24507715; http://dx.doi.org/ 10.1016/j.molcel.2014.01.009 [DOI] [PubMed] [Google Scholar]
- 29.Choudhry H, Albukhari A, Morotti M, Haider S, Moralli D, Smythies J, Schodel J, Green CM, Camps C, Buffa F, et al.. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene 2015; 34:4546; PMID:26289678; http://dx.doi.org/ 10.1038/onc.2014.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Li Y, Chen W, He F, Tan Z, Zheng J, Wang W, Zhao Q, Li J. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget 2015; 6:27641-50; PMID:26314847; http://dx.doi.org/ 10.18632/oncotarget.4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, et al.. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun 2014; 5:5383; PMID:25415230; http://dx.doi.org/ 10.1038/ncomms6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Standaert L, Adriaens C, Radaelli E, Van Keymeulen A, Blanpain C, Hirose T, Nakagawa S, Marine JC. The long noncoding RNA Neat1 is required for mammary gland development and lactation. Rna 2014; 20:1844-9; PMID:25316907; http://dx.doi.org/ 10.1261/rna.047332.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakagawa S, Shimada M, Yanaka K, Mito M, Arai T, Takahashi E, Fujita Y, Fujimori T, Standaert L, Marine JC, et al.. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 2014; 141:4618-27; PMID:25359727; http://dx.doi.org/ 10.1242/dev.110544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cornelis G, Vernochet C, Carradec Q, Souquere S, Mulot B, Catzeflis F, Nilsson MA, Menzies BR, Renfree MB, Pierron G, et al.. Retroviral envelope gene captures and syncytin exaptation for placentation in marsupials. Proc Natl Acad Sci U S A 2015; 112:E487-96; PMID:25605903; http://dx.doi.org/ 10.1073/pnas.1417000112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeller U, Freyer C. Early ontogeny and placentation of the grey short-tailed opossum, Monodelphis domestica (Didelphidae: Marsupialia): contribution to the reconstruction of the marsupial morphotype. J Zool Syst Evol Research 2001; 39:137-58; http://dx.doi.org/ 10.1046/j.1439-0469.2001.00167.x [DOI] [Google Scholar]
- 36.Cmarko D, Verschure PJ, Martin TE, Dahmus ME, Krause S, Fu XD, van Driel R, Fakan S. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol Biol Cell 1999; 10:211-23; PMID:9880337; http://dx.doi.org/ 10.1091/mbc.10.1.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malatesta M, Fakan S, Fischer U. The Sm core domain mediates targeting of U1 snRNP to subnuclear compartments involved in transcription and splicing. Exp Cell Res 1999; 249:189-98; PMID:10366418; http://dx.doi.org/ 10.1006/excr.1999.4468 [DOI] [PubMed] [Google Scholar]
- 38.West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY, Kingston RE. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell 2014; 55:791-802; PMID:25155612; http://dx.doi.org/ 10.1016/j.molcel.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stadler P. Evolution of the Long Non-coding RNAs MALAT1 and MENβ/ε. Adv Bioinformatics Comput Biol 2010;1-12; http://dx.doi.org/ 10.1007/978-3-642-15060-9_1 [DOI] [Google Scholar]
- 40.Saha S, Murthy S, Rangarajan PN. Identification and characterization of a virus-inducible non-coding RNA in mouse brain. J Gen Virol 2006; 87:1991-5; PMID:16760401; http://dx.doi.org/ 10.1099/vir.0.81768-0 [DOI] [PubMed] [Google Scholar]
- 41.Zhang Q, Chen CY, Yedavalli VS, Jeang KT. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio 2013; 4:e00596-12; PMID:23362321; http://dx.doi.org/ 10.1128/mBio.00596-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Souquere S, Mollet S, Kress M, Dautry F, Pierron G, Weil D. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J Cell Sci 2009; 122:3619-26; PMID:19812307; http://dx.doi.org/ 10.1242/jcs.054437 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




