ABSTRACT
Circular RNAs (circRNAs) constitute a large class of RNA species formed by the back-splicing of co-linear exons, often within protein-coding transcripts. Despite much progress in the field, it remains elusive whether the majority of circRNAs are merely aberrant splicing by-products with unknown functions, or their production is spatially and temporally regulated to carry out specific biological functions. To date, the majority of circRNAs have been cataloged in resting cells. Here, we identify an LPS-inducible circRNA: mcircRasGEF1B, which is predominantly localized in cytoplasm, shows cell-type specific expression, and has a human homolog with similar properties, hcircRasGEF1B. We show that knockdown of the expression of mcircRasGEF1B reduces LPS-induced ICAM-1 expression. Additionally, we demonstrate that mcircRasGEF1B regulates the stability of mature ICAM-1 mRNAs. These findings expand the inventory of functionally characterized circRNAs with a novel RNA species that may play a critical role in fine-tuning immune responses and protecting cells against microbial infection.
KEYWORDS: CircRNA, ICAM-1, LPS, RasGEF1B, TLR4
Introduction
Circular RNAs (circRNAs) were first identified in the early 1990s as scrambled exons in mouse and human cells.1 Additional studies reported further new examples of circRNAs over the years,2-9 yet the extent of the prevalence of the circularization phenomenon was not clear, as the available examples remained limited. The advent of high-throughput sequencing revealed that circRNAs are a large class of abundant RNAs with complex developmental- and cell-specific expression patterns.3,10 Stimulated by these findings, several reports in recent years have demonstrated the functional significance of some of these molecules. Some circRNAs function as miRNA “sponges” by sequestering miRNAs and thus preventing their binding to their target genes. For example, the human CDR1as transcript harbors a cluster of more than 70 binding sites for miR-7,4,11 while circSry has 16 binding sites for miR-138.11 CircRNAs have also been shown to function as RNA-binding protein decoys. For instance, circMbl derived from the mbl gene in Drosophila, contains binding sites for the MBL proteins itself.12 MBL auto-regulates the production of circMbl by binding to flanking intronic sequences. On the other hand, binding of MBL to circMbl prevents further induction of circMbl production. Finally, circRNAs have been implicated in transcriptional regulation. A recent study suggested that a subclass of circRNAs, exon-intron circRNAs (EIciRNAs), interact with the U1 snRNP and promote the transcription of their parental genes.13
Despite the progress made so far, the number of functionally characterized circRNAs remains very low. Thousands of cytoplasmic circRNAs have been identified, with most of them having only 1 or 2 binding sites for a particular miRNA, which limits their potential regulatory potency as miRNA “sponges”. How many of these circRNAs are functional or merely a splicing by-product remains an open question.14 ENCODE transcriptome datasets show that circRNAs expression can be cell-type specific, suggesting that their production might be regulated.10 However, most ENCODE experiments have been carried out in cell lines under unperturbed conditions, leaving circRNAs expressed in many important biological contexts largely unexplored. One of them is the transcriptomic response of immune cells following exposure to inflammatory stimuli.
Macrophages play a central role in antimicrobial responses and their action involves the modulation of the expression of hundreds of genes. Microbial and viral products, such as lipopolysaccharide (LPS), initiate signaling cascades through the TLR pathways that lead to the activation of transcription factors such as NF-κB and to changes in the expression of hundreds of genes in antimicrobial defenses and adaptive immunity.15,16 Understanding the potential regulatory role of circRNAs in immune responses is important for the complete understanding of their regulation and is thus of significant relevance to a number of therapeutic contexts, including cancer, heart disease and autoimmunity.
In this study, we initially characterized circRNAs in mouse macrophage cells treated with or without lipid A (the active component of LPS).17 While mouse macrophages produce thousands of distinct circRNAs species at different time points after LPS stimulation, we focused on one of them, mouse circRasGEF1B (mcircRasGEF1B), which we demonstrate to function as a positive regulator of LPS response and to be conserved between mouse and human. We showed that several TLR pathways regulate the expression of mcircRasGEF1B, including TLR4, TLR9, TLR3 and TLR2/TLR1. In addition, we demonstrate that knocking down mcircRasGEF1B expression reduces LPS-induced expression of ICAM-1, an intercellular adhesion molecule, the induction of which plays a role in immune responses by facilitating the binding of leukocytes to endothelium cells and their subsequent transmigration into tissues.
Results
Identification of LPS-induced circRNAs
To determine if any circRNAs might regulate the immune response, we cataloged circRNA expressed upon LPS stimulation using publicly available RNA-seq data from mouse macrophages (see the Experimental Procedures for details).17 We successfully identified a total of 1916 circRNAs across different subcellular fractions and treatment conditions (Table S1). We then compared the performance and validity of our annotation-based pipeline with published pipeline.4 We compared the circRNAs catalog from our annotation-based pipeline with circRNAs database (circBase).18 We found that 208 (10.8%) circRNAs identified using our annotation-based pipeline overlapped with circBase (Fig. S1). We validated our predictions by carrying out RT-PCR on 5 various sizes circRNA candidates from the mEtv6 (132 nt), mLilrb3 (1935 nt), mRasGEF1B (2423 nt), mPlcl2 (4900 nt), and mUbe2d2 (7902 nt) genes. We harvested RNA from mouse macrophage RAW264.7 cells and directly measured the presence of candidate circRNAs using circRNAs-specific divergent primers and Sanger sequencing. (Fig. 1A and Fig. S2A). All candidate circRNAs were confirmed by this approach.
Figure 1.
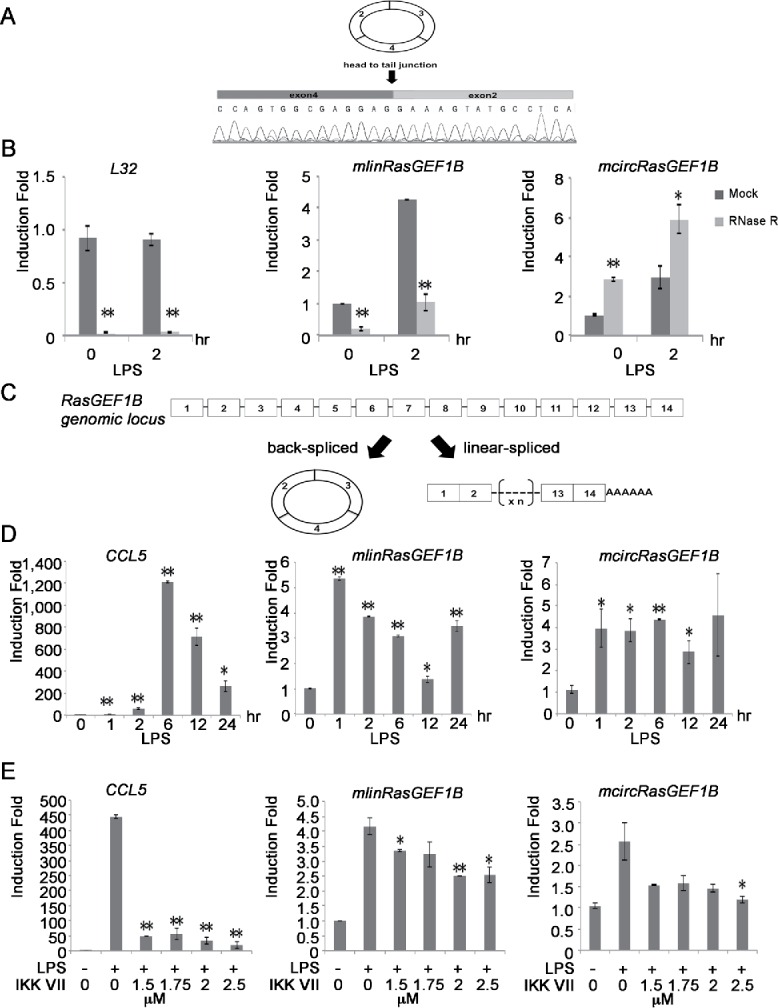
Identification of inducible and NF-κB dependent circRNAs in mouse macrophages. (A) A chromatogram of Sanger sequencing showing the sequence of the back-splice junction of mcircRasGEF1B (exon2 and 4). (B) RAW264.7 cells were induced with or without LPS for 2 hours and total RNA was treated with RNase R to confirm the circularity of mcircRasGEF1B. After RNase R treatment, levels of L32, mlinRasGEF1B and mcircRasGEF1B were measured by qRT-PCR. (C) Schematic depiction of the exon structure of linear RasGEF1B (right) and the back-splicing circular transcript (left). (D) RAW264.7 cells were treated with or without LPS for the indicated time periods. The expression levels of CCL5, mlinRasGEF1B and mcircRasGEF1B were measured by qRT-PCR. (E) RAW264.7 cells were pre-treated with the indicated doses of IKK VII for 1 hour before induction with or without LPS for 2 hours. The expression levels of CCL5, mlinRasGEF1B and mcircRasGEF1B were measured by qRT-PCR. All experiments were carried out in duplicates. (*, p < 0.05; **, p < 0.01).
To rule out the possibility that back-splicing might be the result of trans-splicing or genomic rearrangements, we used RNase R, an exonuclease that degrades linear but not circularized RNA molecules. We used the linear mRNAs of L32, a ribosomal protein, and of mRasGEF1B (mlinRasGEF1B), linear mRNA RasGEF1B, as negative controls. We found that all circRNAs except mcircLilrb3 were resistant to RNase R treatment, whereas L32 and mlinRasGEF1B were highly sensitive to it (Fig. 1B and Fig. S2B). We were thus able to verify 4 out of 5 candidates as bona fide circRNAs. To test whether these circRNAs are induced upon LPS stimulation, we treated RAW264.7 cells with or without LPS and measured the level of circRNAs using qRT-PCR. LPS induced the expression of mcircRasGEF1B, but not of mcircPlcl2, mcircEtv6, and mcircUbe2d2 (Fig. 1B and Fig. S2B). We therefore focused our subsequent efforts on the characterization of mcircRasGEF1B.
Mouse RasGEF1B contains 14 exons while mcircRasGEF1B is the result of the circularization of exons 2–4 (Fig. 1C). To study the expression dynamics of mcircRasGEF1B in a more detail manner, we stimulated RAW264.7 cells with LPS and measured its expression at various time points. Similar to linear mlinRasGEF1B, mcircRasGEF1B was induced as early as 1 hour post LPS stimulation (Fig. 1D). Furthermore, we observed stable expression of mcircRasGEF1B up to 12 hours after LPS treatment, while mlinRasGEF1B was reduced by 50% by that time. In addition, we found that mcircRasGEF1B is less abundant than mlinRasGEF1B (Fig. S3).
NF-κB-dependent expression of mcircRasGEF1B
NF-κB is one of the key transcription factors activated by LPS stimulation.19 To investigate if LPS-induced expression of mcircRasGEF1B is dependent on NF-κB, we blocked NF-κB activation by treating RAW246.7 cells with IKK inhibitor VII.20 Treatment of RAW264.7 cells with increasing doses of IKK inhibitor VII significantly inhibited LPS-induced expression of CCL5, a known NF-κB target gene, mlinRasGEF1B and mcircRasGEF1B (Fig. 1E). For example, in the presence of 1.5 μM inhibitor, induction of CCL5 was reduced by 90% while induction of mcircRasGEF1B was reduced by 42%. Increasing IKK VII inhibitor concentration to 2.5 μM led to almost complete abolishment of LPS-induced expression of mcircRasGEF1B. These results demonstrate that LPS induces the expression of mlinRasGEF1B and mcircRasGEF1B in an NF-κB-dependent manner.
TLR-mediated mcircRasGEF1B expression
A previous study by Andrade et al. suggested that mlinRasGEF1B is strongly induced upon poly I:C and LPS treatments (stimulating TLR3 and TLR4, respectively), and to a lesser extent by ODN CpG DNA and the synthetic triacylated lipopeptide Pam3CYS (stimulating TLR9 and TLR1/TLR2, respectively) treatments.21 In addition, flagellin stimulates TLR5 while imiquimod (R837) can be used to specifically activate TLR7.22 To test if mcircRasGEF1B is regulated by TLRs other than TLR4, we treated RAW264.7 cells with PAM3CSK4, ODN1826, LPS, FLA-ST (flagellin from S. typhimurium), poly I:C and R837. RAW264.7 cells responded to all of the stimulants except FLA-ST as evidenced by the induction of TNFα (Fig. 2A). Expression of mcircRasGEF1B and mlinRasGEF1B was robustly induced by LPS and ODN CpG DNA, and to a lesser extent by poly I:C and Pam3CSK4. Our results suggest that mcircRasGEF1B and mlinRasGEF1B expression is induced through several TLR pathways, including TLR4, TLR9, TLR3 and TLR1/TLR2.
Figure 2.
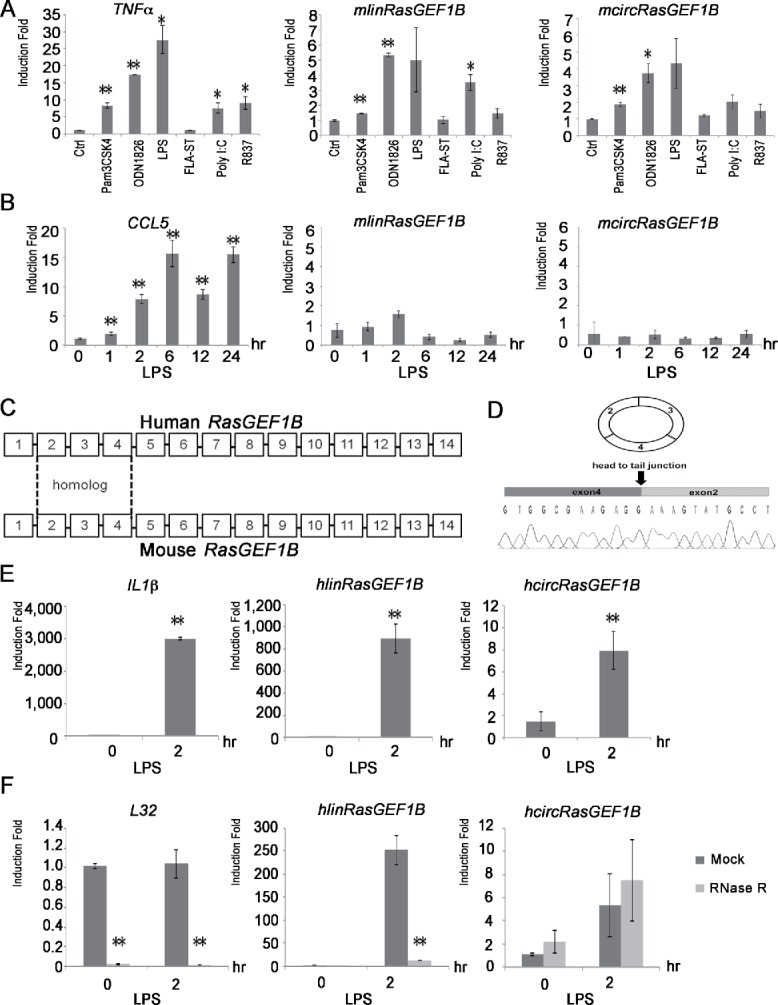
Cell type specific and evolutionary conserved mcircRasGEF1B. (A) The indicated TLR ligands were used to treat RAW264.7 cells for 2 hours. The expression levels of TNFα, mlinRasGEF1B and mcircRasGEF1B were measured by qRT-PCR. (B) MEF cells were induced with or without LPS for the indicated time periods. The expression levels of CCL5, mlinRasGEF1B and mcircRasGEF1B were measured by qRT-PCR. (C) Schematic representation of human RasGEF1B (top) and mouse RasGEF1B (bottom); sequence homology between conserved exons 2, 3, and 4 is highlighted (dashed lines). (D) A chromatogram of Sanger sequencing showing the sequence of the back-splicing junction of hcircRasGEF1B (exons 2 and 4). (F) THP-1 cells were induced with or without LPS for 2 hours and total RNA was subjected to RNase R treatment to confirm the circularity of hcircRasGEF1B. The levels of L32, hlinRasGEF1B and hcircRasGEF1B were measured by qRT-PCR. All experiments were carried out in duplicates. (*, p < 0.05; **, p < 0.01). (E) Human THP-1 cells were induced with or without LPS for 2 hours. The expression levels of IL1β, hlinRasGEF1B and hcircRasGEF1B were measured by qRT-PCR.
Cell-type specific expression of mcircRasGEF1B
An analysis of circRNA expression patterns among 15 expression cell lines by the ENCODE consortium highlighted that many circRNAs are cell-type specific.10 To examine whether the induction of mcircRasGEF1B is cell-type specific, we treated mouse embryonic fibroblast (MEF) cells with LPS and measured the expression of CCL5, mlinRasGEF1B and mcircRasGEF1B. Expression of CCL5 was induced in response to LPS stimulation in MEF cells (Fig. 2B). However, LPS failed to induce either mlinRasGEF1B or mcircRasGEF1B in MEF cells. This result implies that LPS induces the expression of circRasGEF1B in a cell-type specific manner.
Evolutionary conservation of circRasGEF1B
To assess the conservation of circRasGEF1B, we first aligned the sequences of human and mouse RasGEF1B. Both mouse and human RasGEF1B contain 14 exons and exons 2–4 share high sequence homology with 86% identity (Fig. 2C). We then designed divergent primers to detect and study the expression of hcircRasGEF1B in a human macrophage cell line, THP-1. The predicted hcircRasGEF1B is detected in these cells (Fig. 2D). Similar to what we observed in mouse, expression of hcircRasGEF1B in THP-1 cells is induced upon LPS stimulation (Fig. 2E). Furthermore, we confirmed the circularity of hcircRasGEF1B using an RNase R treatment, to which it was resistant unlike L32 and hlinRasGEF1B (Fig. 2F). Taken together, our results show that circRasGEF1B is conserved between human and mouse.
mcircRasGEF1B preferentially localizes to the cytoplasm and is not translated
As a first step toward understanding the physiological role of circRasGEF1B, we determined its subcellular localization. To this end, RAW264.7 cells were treated with LPS for 2 hours and fractionated into nuclear and cytoplasmic fractions. The cytoplasmic L32 and nuclear U6 transcripts were used as controls of the purity of cytoplasmic and nuclear fractions, respectively. As expected, L32 was predominantly enriched in the cytoplasmic fraction while U6 was enriched in the nuclear fraction (Fig. 3A). Intriguingly, mcircRasGEF1B was predominantly localized to the cytoplasm similar to mlinRasGEF1B. These results are consistent with previous reports showing that a majority of circRNAs are cytoplasmic 3,23 and suggest that mcircRasGEF1B might play a role in the post-transcriptional regulation of gene expression.
Figure 3.
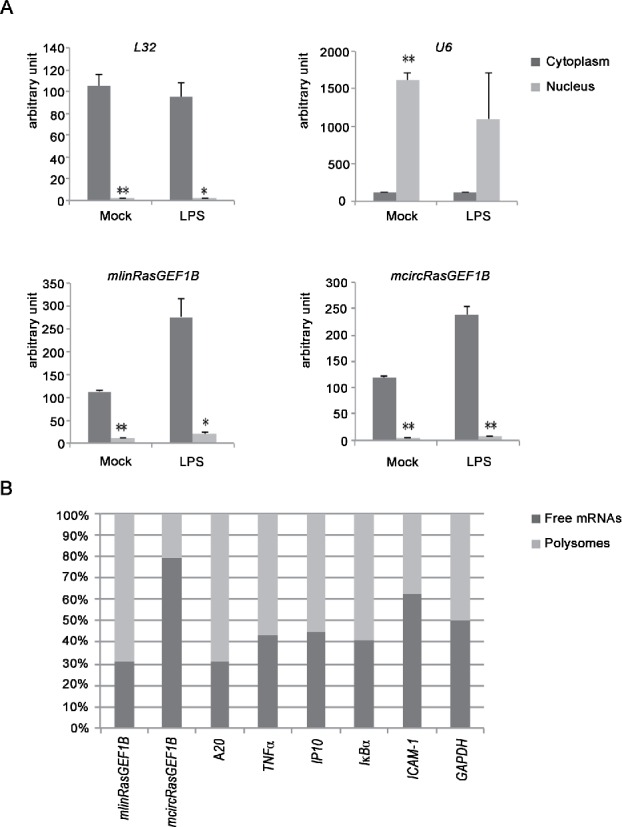
mcircRasGEF1B is predominantly located in cytoplasm and is not translated. (A) RAW264.7 cells were induced with and without LPS for 2 hours. Whole cell lysates were fractionated into cytoplasmic and nuclear fractions. The levels of L32, U6, mlinRasGEF1B and mcircRasGEF1B in these fractions were measured by qRT-PCR. All experiments were carried out in duplicates. (*, p < 0.05; **, p < 0.01). (B) RAW264.7 cells were induced with for 2 hours. Cytoplasmic supernatants were separated by sucrose gradient centrifugation. The levels of linear transcripts (mlinRasGEF1B, A20, TNFα, IP10, IκBα, ICAM-1 and GAPDH), and circular transcript mcircRasGEF1B, in free mRNA and polysome-bound fractions were measured by qRT-PCR (n = 2).
The translation start site of mlinRasGEF1B is in exon 2, which is part of mcircRasGEF1B. To test if mcircRasGEF1B is being translated, we isolated free and polysome-bound mRNAs by sucrose gradient ultracentrifugation. We ran an agarose gel to verify separation of 18S, 28S, and polysome fractions and pooled earlier fractions (fractions 1–9) as free mRNAs while remaining fractions (fractions 10–23) as polysomes (Fig. S4).24 We then measured the relative quantity of linear transcripts (mlinRasGEF1B, A20, TNFα, IP10, IκBα, ICAM-1 and GAPDH) and circular transcript (mcircRasGEF1B) by qRT-PCR (Fig. 3B). Linear products were enriched in the ribosome bound fraction for the genes assayed. Circular product, mcircRasGEF1B, however, was highly abundant in the free mRNA fraction (Fig. 3B). This is consistent with previous reports that failed to identify any polysome-bound circRNAs.3,10 Together, this result indicated that the AUG-containing mcircRasGEF1B is not translated.
mcircRasGEF1B regulates the expression of ICAM-1 in the LPS/TLR4 signaling pathway
To test if mcircRasGEF1B plays a role in regulating the LPS pathway, we knocked down the expression of mcircRasGEF1B using 2 RNase-H based antisense oligonucleotides (ASOs), ASO I and II, of which both targeting the back-splice junction of mcircRasGEF1B. A sense-strand version of ASO I was used as a control ASO (Fig. 4A). ASO I and II specifically knocked down the expression of mcircRasGEF1B but had no or little effect on mlinRasGEF1B (Fig. 4B and 4C). We then examined the effect of mcircRasGEF1B knockdown on LPS target genes and found that it resulted in reduction of ICAM-1 levels at 2 hours after LPS induction (Fig. 4B). LPS-induced ICAM-1 expression was reduced by 27% in ASO I, and 39% in ASO II. However, there were no significant differences observed in other LPS target genes, including CCL5, IP10 and A20 (Fig. S5). Thus we focused our subsequent efforts on understanding mcircRasGEF1B mediated regulation of LPS-induced ICAM1 expression. A more detailed time course using ASO I transfected cells revealed that LPS-induced ICAM-1 expression was reduced by 27% and 30% at 2 hours and 6 hours respectively in the absence of mcircRasGEF1B (Fig. 4C). To minimize the possibility that the effect we observed with mcircRasGEF1B ASO-mediated silencing was caused by an ASO off-target effect, we also constructed an inducible short hairpin RNA (shRNA) targeting the junction of exon 4 and exon 2 of mcircRasGEF1B (Fig. 4D). We knocked down mcircRasGEF1B by treating stable RAW264.7 cells carrying the inducible shRNA transgene with doxycycline for 2 days prior to LPS induction. Treating the cells with doxycycline significantly reduced the expression of mcircRasGEF1B but not the linear mlinRasGEF1B (Fig. 4E). More importantly, we observed a 30% reduction of LPS-induced expression of ICAM-1 in the absence of mcircRasGEF1B (Fig. 4E). To further confirm the effect on ICAM-1 at the protein level, we conducted western blotting in mcircRasGEF1B deficient cells. We knocked down mcircRasGEF1B with ASO I and ASO II and treated with LPS for 6, 9, and 12 hours. The reduction of ICAM-1 protein was detected across every time point, suggesting that mcircRasGEF1B effect was confirmed in both ICAM-1 mRNA and protein levels (Fig. 4F). Together, these data indicate that mcircRasGEF1B positively regulates the expression of ICAM-1 in the LPS/TLR4 signaling pathway.
Figure 4.
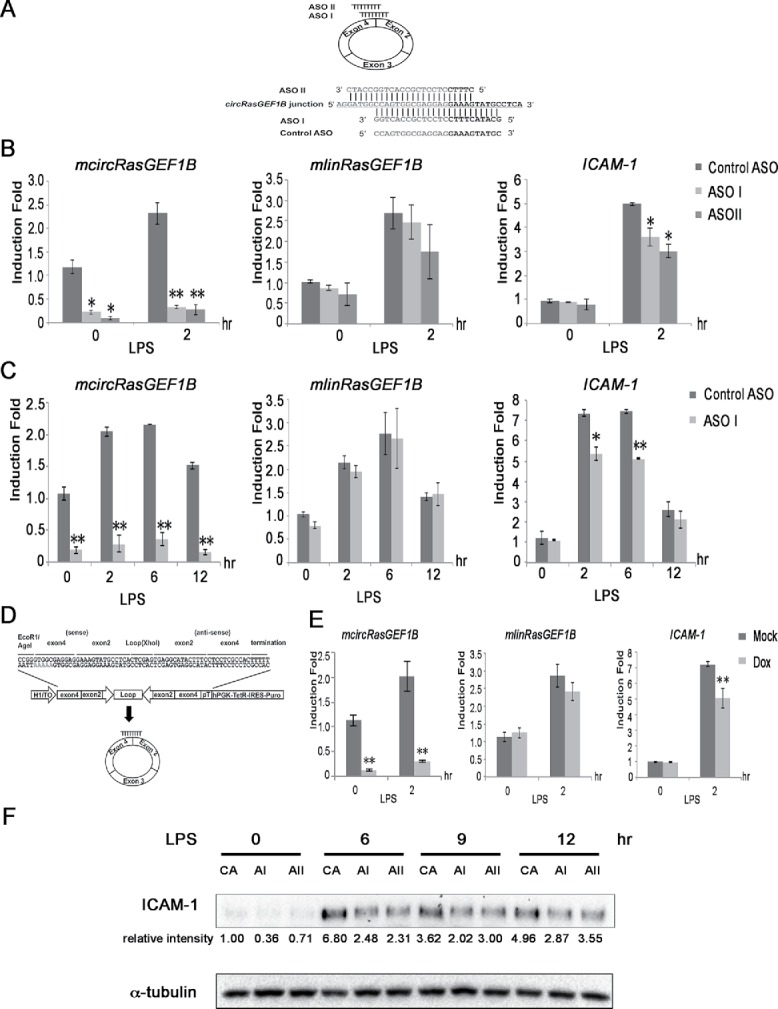
mcircRasGEF1B positively regulates the LPS-induced expression of ICAM-1. (A) ASO I and II targeting mcircRasGEF1B at the junction of exons 4 and 2. The control ASO is in the sense orientation but with the same coordinates as ASO I. (B) RAW264.7 cells were transfected with ASO I, ASO II and control ASO, and induced with LPS for 2 hours. The expression levels of ICAM-1, mlinRasGEF1B and mcircRasGEF1B were measured by qRT-PCR. (C) RAW264.7 cells were knocked down with ASO I or control ASO and induced with LPS for the indicated time periods. The expression levels of ICAM-1, mlinRasGEF1B and mcircRasGEF1B were measured by qRT-PCR. (D) Schematic depiction of the inducible shRNA construct targeting the back-splice junction of mcircRasGEF1B. (E) A stable RAW264.7 clone carrying the shRNA construct was induced with 2.5 μg of Doxycycline for 2 days before treatment with or without LPS. The expression levels of ICAM-1, mlinRasGEF1B and mcircRasGEF1B were measured by qRT-PCR. (*, p < 0.05; **, p < 0.01). Experiments were carried out in duplicates, n = 2 (B,C) and triplicates, n = 3 (E). (F) RAW264.7 cells were knocked down with control ASO, ASO I, and ASO II and then induced with or without LPS for the indicated time periods. Whole cell extracts were immunoblotted with the indicated antibodies. Intensity of bands was quantified using Image Lab (Biorad) software normalized to α-tubulin and shown in relative to 0 minute control ASO. (CA: control ASO, AI: ASO I, AII: ASO II). This is a representative data from 3 independent time course experiments.
mcircRasGEF1B does not affect upstream signal transduction of TLR4 pathway
Decreased mRNA levels of ICAM-1 could be due to a variety of mechanisms. We considered 2 possibilities in which either mcircRasGEF1B reduces transcription of ICAM-1 or it reduces stability of ICAM-1 mRNA. The reduction of the transcription of ICAM-1 could result from blocking of the TLR4 signaling or direct inhibition of transcription by mcircRasGEF1B. First, we tested if knockdown of the expression of mcircRasGEF1B affects the TLR4 signaling. We transfected RAW264.7 cells with control or mcircRasGEF1B-specific ASO I and fractionated cell lysates into cytoplasmic and nuclear fractions. Since LPS induces the activation of NF-κB and IRF3, we examined the IκBα degradation and the nuclear translocation of p65 and IRF3, of which are biochemical hallmarks of NF-κB and IRF3 activation respectively. We found that mcircRasGEF1B knockdown led to no obvious differences in the degradation of IκBα, nuclear translocation of p65, or IRF3 activation (Fig. 5A). These results imply that mcircRasGEF1B does not regulate the signal transduction of LPS pathway.
Figure 5.

mcircRasGEF1B regulates the stability of ICAM-1 mRNA. (A) RAW264.7 cells were knocked down with ASO I or control ASO, and then induced with or without LPS for the indicated time periods. Whole cell extracts were fractionated and the fractions immunoblotted with the indicated antibodies. (B) RAW264.7 cells were transfected with ASO I or control ASO, and then treated with LPS for 2 hours followed by treatment with 1 μg/ml of ActD for the indicated time periods. The expression levels of mlinRasGEF1B and mcircRasGEF1B were measured by qRT-PCR. (C) Relative levels of ICAM-1 pre-mRNA and mature mRNA were measured relative to the levels of L32's mRNA. (D) The stability of ICAM-1 pre-mRNA and mature mRNA measured relative to L32. All experiments were carried out in quadruplicates, n = 4. (*, p < 0.05; **, p < 0.01).
mcircRasGEF1B regulates the stability of ICAM-1 transcripts
Given that mcircRasGEF1B is enriched in the cytoplasm, it is unlikely that it directly regulates transcription in the nucleus. Therefore, we investigated whether mcircRasGEF1B affects the stability of ICAM-1 transcripts. We assessed the stability of ICAM-1 pre-mRNA and mature mRNA by quantitative RT-PCR measurements after blocking transcription with actinomycin D (ActD) for 1, 2, and 4 hours in the presence and absence of ASO I. We measured mRNA stability after 2 hours of LPS induction and normalized to that of the relatively stable L32 mRNA. These assays reveal that mcircRasGEF1B is more stable than mlinRasGEF1B while, as observed before, ASO I specifically reduced the expression of mcircRasGEF1B but not mlinRasGEF1B (Fig. 5B). Interestingly, in mcircRasGEF1B-deficient cells, there was a reduction of the levels of ICAM-1 mature mRNA but not of its pre-mRNA (Fig. 5C). More importantly, we also observed small but reproducible decreases in the stability of mature ICAM-1 mRNA (13% at 1 hour, 23% at 2 hours, and 12% at 4 hours post ActD treatment) in mcircRasGEF1B-depleted cells (Fig. 5D). That mcircRasGEF1B does not affect the transcription of ICAM-1 is supported by the fact that LPS-induced levels of ICAM-1 pre-mRNA are similar between control and mcircRasGEF1B-depleted cells. Taken together, our results suggest that mcircRasGEF1B controls LPS-induced ICAM-1 expression through regulating the stability of its mature mRNA.
Discussion
In this study, we report a novel LPS-inducible cytoplasmic circular RNA, mcircRasGEF1B that modulates the expression of ICAM-1 in response to LPS stimulation. We show that mcircRasGEF1B expression is induced by agonists of TLR4, TLR9, TLR3 and TRL2/1 in RAW264.7 cells but not in MEF cells. These treatments induces transcription of RasGEF1B gene, which results in both mlinRasGEF1B and mcircRasGEF1B expression (Fig. 2A). Biogenesis study also shows that circRNA are generated co-transcriptionally and circRNAs can function by competing with linear splicing.12 Furthermore, circRasGEF1B is conserved between human and mouse. Silencing the expression of mcircRasGEF1B moderately reduces the mRNA expression and protein levels of ICAM-1 upon challenging the cells with LPS. Interestingly, we find that mcircRasGEF1B is required for maintaining the stability of the mature mRNA of ICAM-1 in LPS-activated cells.
ICAM-1 is an important adhesion molecule that has been studied especially on endothelial cells due to its role in leukocyte recruitment to inflamed sites. In antigen presenting cells including macrophages, ICAM-1 participates in cell-cell interactions during antigen presentation while in other cell types ICAM-1 functions in microbial pathogenesis and as a signal transduction molecule.25,26 Physiologically, ICAM-1 is expressed at a low basal level,27 however during inflammatory and immune responses, ICAM-1 level increased substantially and aberrantly at sites of inflammation contributing to a number of inflammation-related diseases and injuries such as endotoxin-induced airway disease28,29 and asthma,27,30 arthritis,31 ulcerative colitis 32 and chronic cholangiopathies.21 In cancer, ICAM-1 has been mainly implicated in local inflammatory tumor microenvironment,33 tumor progression and metastasis.34 The molecular mechanisms underlying the transcriptional regulation of ICAM-1 gene has an important implication in term of inflammatory-related diseases.
Of importance, mcircRasGEF1B-mediated regulation of ICAM-1 indicates that circRasGEF1B may have functions in innate immune response such as inflammatory-related diseases, autoimmunity and cancer. For example, depletion of mcircRasGEF1B in tumor-associated macrophage (TAM) may cause these cells to adopt the pro-metastasis M2 phenotype as ICAM-1 expression has been reported to suppress the M2 macrophage polarization in a tumor microenvironment.35 Although macrophage is used as a model system here, it is tempting to speculate that circRasGEF1B may also regulate ICAM-1 level in other cell types. In particular, ICAM-1 plays a major role in the recruitment of neutrophils and lymphocytes in many tissues via leukocyte-endothelial cell bridging, thus mcircRasGEF1B deficiency may prevent migration of leukocyte cells to inflammatory sites.36,37 In addition, down-regulation of mcircRasGEF1B in cancer cells may also affect the cytotoxic T-lymphocytes (CTL)-mediated cytotoxicity due to engagement of LFA-1 on CTL by ICAM-1 on target cells is essential for T-cell activation and for directing the released of cytolytic granules into the tumor cells.38
With the discovery of circRNAs as miRNA sponges, it is tempting to speculate that mcircRasGEF1B could sequester miRNAs targeting ICAM-1. The expression of ICAM-1 has been shown to be regulated by several miRNAs, including miR-223,39 miR-141,40 and miR-296-3p.41 However, sequence analysis of mcircRasGEF1B does not reveal any enrichment of multiple (≥3) binding sites for any known miRNAs within mcircRasGEF1B, and it harbors no binding sites for miR-223, miR-141 and miR-296-3p (data not shown). Thus, it is unlikely that mcircRasGEF1B plays a major role in RAW264.7 cells as a miRNA sponge. These observations are consistent with previous reports by Guo et al. and Conn et al. that the majority of circRNAs do not act as miRNA sponges.14,42 We estimated that for every 2580 molecules of ICAM-1, there is 1 molecule of mcircRasGEF1B (Fig. S3). Despite the low mcircRasGEF1B: ICAM-1 ratio, the dynamic expression of mcircRasGEF1B in macrophages suggests that this small population of mcircRasGEF1B might exert its function through direct- or indirect binding to ICAM-1 mRNAs. It has been noted that, for the cis effect, the abundance of individual circRNAs do not need to be high to exert an effect. For example, low abundance of ElciRNAs is shown to regulate the transcription of more abundance parental genes.13
The biochemical fractionation analysis of cellular RNAs indicates that mcircRasGEF1B is predominantly found in the cytoplasm. This result prompted us to analyze if mcircRasGEF1B might regulate the upstream signaling cascade of TLR4 pathway. However, activation of NF-κB and IRF3 is normal in mcircRasGEF1B-deficient cells upon LPS stimulation (Fig. 5A). Furthermore, measurements of ICAM-1 pre- and mature mRNA levels in control and ASO-transfected cells show that LPS-induced transcription of ICAM-1 pre-mRNA is not affected by mcircRasGEF1B (Fig. 5C). Taken together these results suggest that mcircRasGEF1B regulates ICAM-1 at the post-transcriptional level.
The LPS-induced expression of the mature ICAM-1 mRNA is reduced in mcircRasGEF1B-deficient cells. A reduction of a mature mRNA could due to less efficient mRNA splicing or to a decrease in mRNA stability. We favor the latter possibility for the following reasons. First, mRNA splicing takes place in the nucleus while mRNA degradation occurs both in the cytoplasm and the nucleus. However, mcircRasGEF1B is enriched in the cytoplasm (Fig. 3). Nonetheless, we cannot rule out the possibility that a small amount of mcircRasGEF1B is present in the nucleus and may affect mRNA splicing there. Second, if splicing of ICAM-1 is blocked in mcircRasGEF1B-deficient cells, ICAM-1 pre-mRNA should accumulate over time, which is not the case (Fig. 5C). Third, treating cells with ActD blocks RNA synthesis but not pre-mRNA splicing. The turnover rate of ICAM-1 pre-mRNA is comparable between control and mcircRasGEF1B–depleted cells when treated with ActD, suggesting that mRNA splicing is unaffected (Fig. 5C). Finally, we observed a reproducible reduction of the stability of mature mRNA of ICAM-1 in mcircRasGEF1B-deficient cells (Fig. 5C). Thus, our data suggest that mcircRasGEF1B positively regulates the expression of ICAM-1 through modulating the stability of mature mRNA of ICAM-1. Given that is unlikely to function as a classic miRNA sponge, mcircRasGEF1B might exert its effects on ICAM-1 expression through a novel, previously unreported mechanism, elucidating which should be an exciting subject for future study.
Materials and methods
Identification of circular splice junctions
Except where explicitly stated otherwise, all RNA-seq analyses were carried out using custom-written python scripts.
Total RNA-seq sequencing reads for subcellular fractions from LPS-stimulated macrophages were downloaded from GEO series GSE32916.17 The sequences of all possible circular splice junctions within the same gene based on annotated exons (the ENSEMBL63 annotation and the mm9 version of the mouse genome were used) were compiled, retaining RL15 bp on each side of the junctions (equivalent to requiring at minimal length of 15bp for spliced alignment overhangs) where RL is the read length. The circular junction sequences were then combined with the sequences of the full-length annotated transcripts and a Bowtie index was created, which was used to align reads that do not map to the whole genome sequence.43 Candidate circular RNAs were then identified based on reads mapping to circular junctions. Candidate circRNAs present at at least 1 RPM (Reads Per Million mapped reads) in any library are listed in Table S1.
Cell culture and reagents
RAW264.7, mouse embryonic fibroblast (MEF), and THP-1 cells were cultured in Rosewell Park Memorial Institute medium (RPMI) supplemented with 10% FBS, 100 U/ml penicillin and streptomycin (100 µg/ml) (GIBCO). LPS (Sigma), actinomycin D (Sigma), PAM3CSK4 (Invivogen), ODN1826 (Invivogen), FLA-ST (Invivogen), R837 (Merck), doxycycline (Fisher Scientific), IKK inhibitor (Merck), and poly I:C (Tocris) were purchased from the respective sources. Anti-HSP90 (Santa Cruz, sc-8262), anti-SNF2H (Santa Cruz, sc-13054 X), anti-IRF3 (Santa Cruz, sc-15991), anti-p65 (Santa Cruz, sc-372), anti-IκBα (Santa Cruz, sc-203), anti-α-tubulin (Santa Cruz, sc-8035), and anti-ICAM-1 (Santa Cruz, sc-1511) were obtained from the respective companies.
Quantitative RT-PCR and RNase R treatment
Total RNA was isolated with the Thermo Scientific GeneJET RNA Purification Kit. cDNAs were synthesized and quantitative RT-PCR was performed with 2X SYBR Green PCR Master mix (Thermo Scientific) and run on a Bio-Rad Connect Real-Time PCR System. Expression levels of circular RNAs described in this study were measured by qPCR using gene specific divergent primers (Table S2). The relative expression levels of circular versus linear isoforms were normalized to L32.
RNAse R experiments were carried out by incubating purified total RNA (35 μg) at 37°C for 30 minutes with or without 15U of RNase R (Epicentre Biotechnologies). RNA was subsequently purified with the Thermo Scientific GeneJET RNA Purification Kit.
Polysome analysis
Twenty million RAW264.7 cells were grown and treated with LPS for 2 hours. The cells were then treated with 200 μM cycloheximide to stabilize polysome complexes. Cytoplasmic supernatant was loaded onto a continuous sucrose gradients 10% to 50% containing 400 mM KOAc (pH 7.5), 25 mM HEPES, 15 mM Mg(OAc)2, 200 μM cycloheximide and 50 units/mL RNase Inhibitor (NEB). Sucrose gradients were centrifuged at 4°C for 3 hours at 100,000g in a SW41 rotor. Equal volume of fractions was collected and total RNA was extracted. The identity of individual fractions was confirmed by loading equal volume of eluted RNA samples in agarose gel with ethidium bromide staining to visualize the rRNAs. Free mRNAs and polysome fractions were pooled and reverse transcribed with equal input of RNA. To determine the relative abundance of free mRNA and polysomes, we used the equation:
free mRNA: [1/(input/total RNA)], polysomes: [2^(Ctpolysome-CtfreemRNA)/(input/total RNA)] and presented as 100% stacked graph.
Plasmids
An shRNA targeting the exon junction of mcircRasGEF1B containing 11 bases of exon 4 and 14 bases of exon 2 was subcloned into a PLKO-Tet-Puro vector purchased from Addgene. The plasmid was subsequently verified by automatic DNA sequencing.
ASOs transfection
ASOs (Table S2) were synthesized by IDT technologies. ASOs (20 nM) were transfected into RAW264.7 cells with the X-tremeGENE HP DNA (Roche) according to the manufacturer's protocol. To maximize knockdown efficiency, ASO transfection was repeated 24 hours after the initial transfection.
Cell fractionation
RAW264.7 cells were resuspended in a homogenization buffer containing 10 mM HEPES, 10 mM KCl, 10 mM EDTA, 10 mM EGTA, 1 mM DTT, 1 mM MgCl2, 0.5% NP-40, and 5% glycerol. Cells were incubated on ice for 20 minutes and then centrifuged at 4 °C for 10 minutes at 500g. Supernatants were collected as cytoplasmic fractions while the pellets were washed 3 times with the homogenization buffer. Total RNA from both cytoplasmic and nuclear fractions was purified with the Thermo Scientific GeneJET RNA purification kit. Arbitrary unit was calculated based on the equation:
Cytoplasm fraction: [1/(input/total RNA)], Cytoplasm+/Nucleus/Nucleus+: [2^(Ctcytoplasm-Ctcytoplasm+/nucleus/nucleus+)/(input/total RNA)].
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the University of Malaya through High Impact Research Grant (HIR) [UM.C/625/1/HIR/MOHE/CHAN/14/2]; and the Post Graduate Research Grant [PG055-2014B].
Author contributions
WL.N., ES.L., YL.L., and C-K.E. performed the experiments. GK.M., performed bioinformatics analysis. WL.N., Y-Y.L., and C-K.E. designed experiments. WL.N., Y-Y.L., and C-K.E. analyzed the data. WL.N., GK.M., Y-Y.L., and C-K.E wrote the manuscript.
References
- 1.Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell 1991; 64:607-13; PMID:1991322; http://dx.doi.org/ 10.1016/0092-8674(91)90244-S [DOI] [PubMed] [Google Scholar]
- 2.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014; 32:453-61; PMID:24811520; http://dx.doi.org/ 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19:141-57; PMID:23249747; http://dx.doi.org/ 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al.. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333-8; PMID:23446348; http://dx.doi.org/ 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 5.Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993; 73:1019-30; PMID:7684656; http://dx.doi.org/ 10.1016/0092-8674(93)90279-Y [DOI] [PubMed] [Google Scholar]
- 6.Gualandi F, Trabanelli C, Rimessi P, Calzolari E, Toffolatti L, Patarnello T, Kunz G, Muntoni F, Ferlini A. Multiple exon skipping and RNA circularisation contribute to the severe phenotypic expression of exon 5 dystrophin deletion. J Medical Genetics 2003; 40:e100; PMID:12920092; http://dx.doi.org/ 10.1136/jmg.40.8.e100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res 2006; 34:e63; PMID:16682442; http://dx.doi.org/ 10.1093/nar/gkl151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J 1993; 7:155-60; PMID:7678559 [DOI] [PubMed] [Google Scholar]
- 9.Zaphiropoulos PG. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol 1997; 17:2985-93; PMID:9154796; http://dx.doi.org/ 10.1128/MCB.17.6.2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genetics 2013; 9:e1003777; PMID:24039610; http://dx.doi.org/ 10.1371/journal.pgen.1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384-8; PMID:23446346; http://dx.doi.org/ 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 12.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014; 56:55-66; PMID:25242144; http://dx.doi.org/ 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al.. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015; 22:256-64; PMID:25664725; http://dx.doi.org/ 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- 14.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014; 15:409; PMID:25070500; http://dx.doi.org/ 10.1186/s13059-014-0409-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol 2009; 9:692-703; PMID:19859064; http://dx.doi.org/ 10.1038/nri2634 [DOI] [PubMed] [Google Scholar]
- 16.Murray PJ, Smale ST. Restraint of inflammatory signaling by interdependent strata of negative regulatory pathways. Nat Immunol 2012; 13:916-24; PMID:22990889; http://dx.doi.org/ 10.1038/ni.2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 2012; 150:279-90; PMID:22817891; http://dx.doi.org/ 10.1016/j.cell.2012.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA 2014; 20:1666-70; PMID:25234927; http://dx.doi.org/ 10.1261/rna.043687.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin H, Wilson CA, Lee SJ, Zhao X, Benveniste EN. LPS induces CD40 gene expression through the activation of NF-kappaB and STAT-1alpha in macrophages and microglia. Blood 2005; 106:3114-22; PMID:16020513; http://dx.doi.org/ 10.1182/blood-2005-02-0759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waelchli R, Bollbuck B, Bruns C, Buhl T, Eder J, Feifel R, Hersperger R, Janser P, Revesz L, Zerwes HG, et al.. Design and preparation of 2-benzamido-pyrimidines as inhibitors of IKK. Bioorganic Med Chem Lett 2006; 16:108-12; PMID:16236504; http://dx.doi.org/ 10.1016/j.bmcl.2005.09.035 [DOI] [PubMed] [Google Scholar]
- 21.Andrade WA, Silva AM, Alves VS, Salgado AP, Melo MB, Andrade HM, Dall'Orto FV, Garcia SA, Silveira TN, Gazzinelli RT. Early endosome localization and activity of RasGEF1b, a toll-like receptor-inducible Ras guanine-nucleotide exchange factor. Genes Immunity 2010; 11:447-57; PMID:20090772; http://dx.doi.org/ 10.1038/gene.2009.107 [DOI] [PubMed] [Google Scholar]
- 22.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, Horiuchi T, Tomizawa H, Takeda K, Akira S. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol 2002; 3:196-200; PMID:11812998; http://dx.doi.org/ 10.1038/ni758 [DOI] [PubMed] [Google Scholar]
- 23.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One 2012; 7:e30733; PMID:22319583; http://dx.doi.org/ 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fallot S, Ben Naya R, Hieblot C, Mondon P, Lacazette E, Bouayadi K, Kharrat A, Touriol C, Prats H. Alternative-splicing-based bicistronic vectors for ratio-controlled protein expression and application to recombinant antibody production. Nucleic Acids Res 2009; 37:e134; PMID:19729510; http://dx.doi.org/ 10.1093/nar/gkp716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staunton DE, Merluzzi VJ, Rothlein R, Barton R, Marlin SD, Springer TA. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell 1989; 56:849-53; PMID:2538244; http://dx.doi.org/ 10.1016/0092-8674(89)90689-2 [DOI] [PubMed] [Google Scholar]
- 26.Hubbard AK, Rothlein R. Intercellular adhesion molecule-1 (ICAM-1) expression and cell signaling cascades. Free Radic Biol Med 2000; 28:1379-86; PMID:10924857; http://dx.doi.org/ 10.1016/S0891-5849(00)00223-9 [DOI] [PubMed] [Google Scholar]
- 27.Mukhopadhyay S, Malik P, Arora SK, Mukherjee TK. Intercellular adhesion molecule-1 as a drug target in asthma and rhinitis. Respirol 2014; 19:508-13; PMID:2489994; http://dx.doi.org/ 10.1111/resp.12285 [DOI] [PubMed] [Google Scholar]
- 28.Moreland JG, Fuhrman RM, Pruessner JA, Schwartz DA. CD11b and intercellular adhesion molecule-1 are involved in pulmonary neutrophil recruitment in lipopolysaccharide-induced airway disease. Am J Respir Cell Mol Biol 2002; 27:474-80; PMID:12356581; http://dx.doi.org/ 10.1165/rcmb.4694 [DOI] [PubMed] [Google Scholar]
- 29.Kumasaka T, Quinlan WM, Doyle NA, Condon TP, Sligh J, Takei F, Beaudet Al, Bennett CF, Doerschuk CM. Role of the intercellular adhesion molecule-1(ICAM-1) in endotoxin-induced pneumonia evaluated using ICAM-1 antisense oligonucleotides, anti-ICAM-1 monoclonal antibodies, and ICAM-1 mutant mice. J Clin Invest 1996; 97:2362-9; PMID:8636417; http://dx.doi.org/ 10.1172/JCI118679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wegner CD, Gundel RH, Reilly P, Haynes N, Letts LG, Rothlein R. Intercellular adhesion molecule-1 (ICAM-1) in the pathogenesis of asthma. Sci 1990; 247:456-9; PMID:1967851; http://dx.doi.org/ 10.1126/science.1967851 [DOI] [PubMed] [Google Scholar]
- 31.Seidel MF, Keck R, Vetter H. ICAM-1/LFA-1 expression in acute osteodestructive joint lesions in collagen-induced arthritis in rats. J Histochem Cytochem 1997; 45:1247-53; PMID:9283612; http://dx.doi.org/ 10.1177/002215549704500908 [DOI] [PubMed] [Google Scholar]
- 32.Vainer B. Intercellular adhesion molecule-1 (ICAM-1) in ulcerative colitis: presence, visualization, and significance. APMIS Suppl 2010; 118:1-46; PMID:20653648; http://dx.doi.org/ 10.1111/j.1600-0463.2010.02647.x [DOI] [PubMed] [Google Scholar]
- 33.Liou GY, Doppler H, Necela B, Edenfield B, Zhang L, Dawson DW, Storz P. Mutant KRAS-induced expression of ICAM-1 in pancreatic acinar cells causes attraction of macrophages to expedite the formation of precancerous lesions. Cancer Discov 2015; 5:52-63; PMID:25361845; http://dx.doi.org/ 10.1158/2159-8290.CD-14-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayes SH, Seigel GM. Immunoreactivity of ICAM-1 in human tumors, metastases and normal tissues. Int J Clin Exp Pathol 2009; 2:553-60; PMID:19636402 [PMC free article] [PubMed] [Google Scholar]
- 35.Yang M, Liu J, Piao C, Shao J, Du J. ICAM-1 suppresses tumor metastasis by inhibiting macrophage M2 polarization through blockade of efferocytosis. Cell Death Dis 2015; 6:e1780; PMID:26068788; http://dx.doi.org/ 10.1038/cddis.2015.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long EO. ICAM-1: getting a grip on leukocyte adhesion. J Immunol 2011; 186:5021-3; PMID:21505213; http://dx.doi.org/ 10.4049/jimmunol.1100646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Basit A, Reutershan J, Morris MA, Solga M, Rose CE Jr, Ley K. ICAM-1 and LFA-1 play critical roles in LPS-induced neutrophil recruitment into the alveolar space. Am J Physiol Lung Cell Mol Physiol 2006; 291:L200-7; PMID:16461431; http://dx.doi.org/ 10.1152/ajplung.00346.2005 [DOI] [PubMed] [Google Scholar]
- 38.Hamai A, Meslin F, Benlalam H, Jalil A, Mehrpour M, Faure F, Lecluse Y, Vielh P, Avril MF, Robert C, et al.. ICAM-1 has a critical role in the regulation of metastatic melanoma tumor susceptibility to CTL lysis by interfering with PI3K/AKT pathway. Cancer Res 2008; 68:9854-64; PMID:19047166; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-0719 [DOI] [PubMed] [Google Scholar]
- 39.Tabet F, Vickers KC, Cuesta Torres LF, Wiese CB, Shoucri BM, Lambert G, Catherinet C, Prado-Lourenco L, Levin MG, Thacker S, et al.. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat Commun 2014; 5:3292; PMID:24576947; http://dx.doi.org/ 10.1038/ncomms4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu RR, Li J, Gong JY, Kuang F, Liu JY, Zhang YS, Ma QL, Song CJ, Truax AD, Gao F, et al.. MicroRNA-141 regulates the expression level of ICAM-1 on endothelium to decrease myocardial ischemia-reperfusion injury. Am J Physiol Heart Circulatory Physiol 2015; 309:H1303-13; PMID:26371161; http://dx.doi.org/ 10.1152/ajpheart.00290.2015 [DOI] [PubMed] [Google Scholar]
- 41.Liu X, Chen Q, Yan J, Wang Y, Zhu C, Chen C, Zhao X, Xu M, Sun Q, Deng R, et al.. MiRNA-296-3p-ICAM-1 axis promotes metastasis of prostate cancer by possible enhancing survival of natural killer cell-resistant circulating tumour cells. Cell Death Dis 2013; 4:e928; PMID:24263102; http://dx.doi.org/ 10.1038/cddis.2013.458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015; 160:1125-34; PMID:25768908; http://dx.doi.org/ 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- 43.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10:R25; PMID:19261174; http://dx.doi.org/ 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


