Abstract
While the evolutionary mechanisms driving eukaryote genome size evolution are still debated, repeated element content appears to be crucial. Here, we reconstructed the phylogeny and identified repeats in the genome of 26 Drosophila exhibiting a twofold variation in genome size. The content in transposable elements (TEs) is highly correlated to genome size evolution among these closely related species. We detected a strong phylogenetic signal on the evolution of both genome size and TE content, and a genome contraction in the Drosophila melanogaster subgroup.
Keywords: genome size, phylogenetic inertia, transposable elements, flies
1. Introduction
One striking outcome of genome evolution is illustrated by the dramatic 200 000-fold variation in genome size across eukaryotes. Generally, eukaryote genome size reflects the genomic content in repeated sequences, especially in transposable elements (TEs, [1,2]). Although McClintock described TEs in the 1950s [3], it was only with the first whole genome sequencing projects that the community realized the extent of the repeatome (more than 45% of the human genome). Except for some rare cases of beneficial domestication events, TEs are seen as selfish parasitic elements that are mainly neutral or deleterious for their host [4].
Lynch and Conery proposed that genome size results from non-adaptive forces such as genetic drift and mutation. Their model predicts an accumulation of TEs—and therefore larger genomes—in species of small effective population size Ne [5]. In such species, selection to remove TEs may not be efficient compared with drift. A very intense debate on the relative role of Ne on genome size evolution among distantly related taxa followed [6–10], inter alia because the original model was not robust to phylogenetic control [8]. Indeed, a greater resemblance in inherited traits is expected among closely related species compared with distant ones, independently of selection or drift on these traits. Recently, a few studies focused on closely related species (reducing the number of potential confounding factors) to test for an accumulation of TEs in the genome of species with an expected reduced Ne followed by recent life-history trait changes, and reported contrasting results [11–15].
To date, quantifying the importance of phylogenetic inertia in TE content distribution remains a key question as the dynamic of TE accumulation is still poorly understood. Here, we analysed the evolution of genome size and genomic TE content in 26 Drosophila, using a phylogenetic framework. We estimated genomic TE content using a de novo TE assembly approach, tested the correlation between TE content and genome size among closely related species and finally estimated the phylogenetic inertia.
2. Material and methods
(a). Genome size
Genome sizes were estimated using flow cytometry on 24 species. DNA content estimates were collected from the Animal Genome Size database for 23 species (http://www.genomesize.com). Cytometry measures for D. suzukii were performed in the laboratory on fresh samples of 4 day old females, with 10 replicates, from an isofemale line collected in France (P. Gibert).
(b). Sequencing data
Public datasets of short read sequences were downloaded from the Search Read Archive (SRA) database, except for D. yakuba (provided by K. Thornton). Runs were selected with paired-end data when possible (all except D. santomea), sequenced from females. Run identification numbers and more details about the data are provided in the electronic supplementary material.
(c). Repeat content
The genomic content in repeated elements was estimated for each species from de novo assembly and annotation using dnaPipeTE [16]. We filtered raw reads using unsupervised quality trimming [17] and used a random sample corresponding to 0.25× coverage. Simple repeats, satellites and low complexity elements were pooled in the ‘simple repeats' category. To test the effect of datasets' heterogeneity on TE content estimates—as clustering efficiency might vary according to read length—we simulated 0.25× datasets of varying length (40–120 bp) from the reference genome of D. melanogaster using ART [18].
(d). Phylogeny reconstruction
Cytochrome c oxidase subunit I (coI) sequences were recovered for each of the 26 species using the following methodology. Homologous sequences to the D. melanogaster reference protein (Uniprot P00399) were identified using ncbi-tblastn. A consensus was built from the 10 best hits to account for intraspecific diversity. In parallel, sequences homologous to the reference coI sequence were identified by blast among dnaPipeTE contigs (mitochondrion is identified as repeated element owing to its higher coverage compared with nuclear genome). A consensus sequence was finally built between the two sequences. We obtained a 1536 bp long alignment, in respect of the protein sequences. A similar methodology was used to recover fill mitochondria sequences. Best-fit model of nucleotide substitution was selected using jModelTest v. 2.1.10 [19]. According to Bayes information criteria (BIC), a GTR + I + G model was used to reconstruct the species phylogeny by maximum-likelihood (100 bootstraps) using PhyML [20].
(e). Phylogenetic analyses
Comparative analyses were performed using ape [21], nlme [22] and phytools [23] packages in R. Ancestral trait reconstruction of genome size was calculated using phylogenetic independent contrasts. We tested the phylogenetic signal using Pagel's λ [24]. Best-fitting model to the trait evolution and its covariance structure was tested among (i) absence of phylogenetic signal, (ii) neutral Brownian motion and (iii) constrained evolution Ornstein–Uhlenbeck (OU) models using generalized least squares (GLS) and selected according to minimum Akaike information criterion (AIC). We then estimated OU model parameters by maximum-likelihood.
3. Results
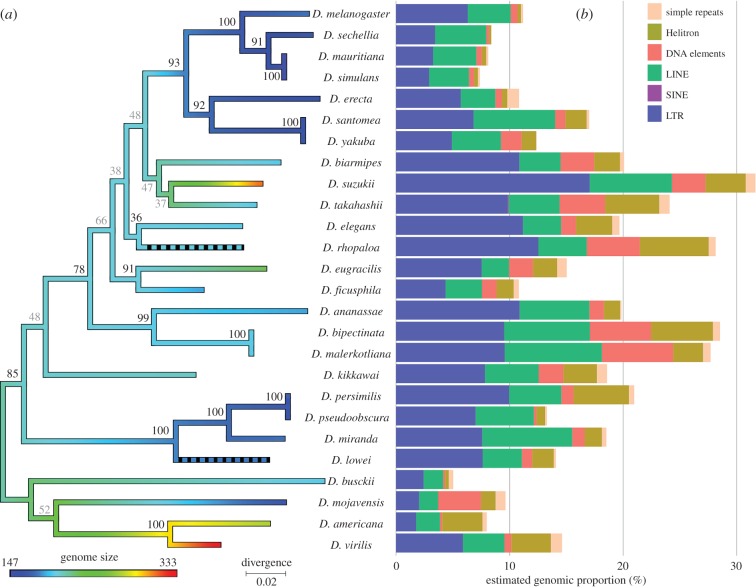
We analysed genome size and TE content evolution among 26 flies. Overall, a twofold variation in genome size was detected, ranging from 147 (D. mauritiana and D. simulans) to 333 Mb (D. virilis). The smallest genomes (less than 180 Mb) essentially clustered into the melanogaster (n = 7) and the pseudoobscura (n = 3) subgroups (figure 1a). After trimming, average read length varied from 47 (D. persimilis) to 121 bp (D. ficusphila and D. kikkawai).
Figure 1.
Phylogenetic tree representing genome size evolution for 26 Drosophila species (a) and their genomic content in repeated elements (b). Bootstrap support of each node is specified on the tree (values <70 in grey indicate less robust nodes). Colours of the branches represent genome size estimates (black dashed branches are used for lineages with unknown genome size).
The genomic content of repeats ranged from 4.65% in D. busckii to 30.80% in D. suzukii (figure 1b). TEs are major components of the repeatome, essentially with LTR and LINE elements, compared with simple repeats (less than or equal to 1%). Some species exhibit a large proportion of DNA elements (6.3% in D. malerkotliana) and Helitron (6% in D. rhopaloa). Global TE content is significantly correlated with the genome size (Spearman's ρ = 0.43, p = 0.04).
We detected a significant effect of read length on the estimated TE content using simulated D. melanogaster data (χ2 = 1780, d.f. = 16, p < 2.2 × 10−16). The repeatome tends to be underestimated using reads shorter than 80 bp (electronic supplementary material). Removing five species with reads less than 80 bp did not affect the correlation coefficient between genome size and TE content, but the relationship became non-significant as a result of the reduced test's power (ρ = 0.44, p = 0.06).
We reconstructed the phylogeny from coI (figure 1a): 15 out of 23 nodes are robust (bootstrap values more than 70) and congruent with a phylogeny reconstructed from the full mitochondria sequences (electronic supplementary material) and with previous studies, except for two branches (D. eugracilis and D. kikkawai). The clade ancestral genome size was much larger than D. melanogaster's, whose subgroup ancestor had a serious genome compaction. The phylogeny fully explains both genome size and TE content variation among the 26 flies (λ = 0.98, p = 1.45 × 10−4 and λ = 0.88, p = 2.19 × 10−3, respectively). Strong phylogenetic signal is confirmed by GLS analysis: the OU model (AIC = 262) better fits the genome size evolution than the non-phylogenetic (AIC = 270) or the Brownian (AIC = 328) model. Similar results were found for TE content (AIC of 174, 177 and 280, respectively). We detected a significantly different optimal genome size for the melanogaster subgroup (deviance = 257.7, p = 0.03).
4. Discussion
The evolution of eukaryote genome size remains mysterious. While the respective roles of neutral and selective forces are debated [6–10], TE accumulation emerges as a major factor of genome size variation. In this study, our estimates of the genomic TE content in 26 Drosophila support this claim among closely related species. We detected a strong phylogenetic signal on the evolution of both genome size and TE content, and genome contraction in the D. melanogaster subgroup.
So far, detailed analyses of genomic content in TEs have been restricted to model-species, because specific amplification methods (targeting one type of TE at a time) are time-consuming and fairly expensive, and whole-genome sequencing methods met technical limitations owing to the challenging assembly of repeat-rich regions. New methods allow this obstacle to be overcome by using the repeated nature of TEs to perform de novo identification from raw reads. Here, we detected a greater proportion of TEs in the genomes than previous estimations done on flies by means of genome assemblies (in which TE-rich regions are underrepresented owing to assembly difficulties). However, our estimates of TE content are congruent with previous ones ([25], R2 = 0.64, p = 0.04, n = 10). Although their genomic content remains limited in flies (15.8% on average) compared with other eukaryotes [26], TEs appear to be driving genome size in flies, like in plants [2] or in eukaryotes [1].
The best-fit OU model suggests some stabilizing selection on genome size evolution, with a significantly different optimal genome size for the melanogaster subgroup. The model detects the apparent genome contraction in this subgroup. While this result holds without D. kikkawai and D. eugracilis for which the position in the reconstructed phylogeny was not consistent with [27], it has to be considered with caution because a Pagel's modification of the basic Brownian model had a similar fit to the OU. It is necessary to test the constrained versus neutral evolution of the genome size on a more phylogenetically balanced sample to conclude on this point. The phylogeny fully explains the distribution of both genome size and TE content among the sampled species, while a previous study indicated that genome size varied with some life history traits (development time, body size and sperm length) in this genus (with reference to Gregory and Johnston [28]). Similarly, a very strong phylogenetic signal was found on genome size variation in liverworts [29] and evening primroses [13], independently of expected variation in Ne. In those species, variation in Ne was expected as a result of changes in some life-history trait, as determining long-term Ne is very challenging and requires, for example, polymorphism estimates. Although there is evidence of some life-history traits promoting the accumulation of TEs (e.g. mating system in Daphnia [30] or parasitism in Amanita fungi [12]) owing to their impact on Ne, empirical studies of specific clades accounting for phylogenetic signal are not unanimous.
Here, we have performed, we believe, the first phylogenetic analysis of genome size and genomic repeated content in a large set of Drosophila species. Our results suggest that the effect of life-history changes (and resulting variations of Ne) on TE spread may not be detected in a short evolutionary scale owing to the major role of phylogenetic inertia. To further test the role of drift in this clade, exhaustive estimates of Ne and unbiased sampling of the phylogeny are now required.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank D. deVienne, L. Duret and C. Vieira and two anonymous reviewers for useful comments on the manuscript.
Data accessibility
Phylogenetic data, including alignments: TreeBASE accession number 19296. (http://purl.org/phylo/treebase/phylows/study/TB2:S19296).
Authors' contributions
N.B. performed flow cytometry experiments. C.S. collected data and performed analyses. A.H. designed the study, performed data analysis and interpretation, and wrote the final manuscript. All authors gave final approval for publication and agree to be held accountable for the content herein.
Competing interests
We have no competing interests.
Funding
CNRS-APEGE to A.H.
References
- 1.Biémont C, Vieira C. 2006. Junk DNA as an evolutionary force. Nature 443, 521–524. ( 10.1038/443521a) [DOI] [PubMed] [Google Scholar]
- 2.Tenaillon MI, Hollister JD, Gaut BS. 2010. A triptych of the evolution of plant transposable elements. Trends Plant Sci. 15, 471–478. ( 10.1016/j.tplants.2010.05.003) [DOI] [PubMed] [Google Scholar]
- 3.McClintock B. 1950. The origin and behavior of mutable loci in maize. Proc. Natl Acad. Sci. USA 36, 344–355. ( 10.1073/pnas.36.6.344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackay TFC. 1986. A quantitative genetic analysis of fitness and its components in Drosophila melanogaster. Genet. Res. 47, 59–70. ( 10.1017/S0016672300024526) [DOI] [Google Scholar]
- 5.Lynch M, Conery JS. 2003. The origins of genome complexity. Science 302, 1401–1404. ( 10.1126/science.1089370) [DOI] [PubMed] [Google Scholar]
- 6.Charlesworth B, Barton N. 2004. Genome size: does bigger mean worse? Curr. Biol. 14, R233–R235. ( 10.1016/j.cub.2004.02.054) [DOI] [PubMed] [Google Scholar]
- 7.Daubin V, Moran NA. 2004. Comment on ‘The origins of genome complexity’. Science 306, 978 ( 10.1126/science.1098469) [DOI] [PubMed] [Google Scholar]
- 8.Whitney KD, Garland T. 2010. Did genetic drift drive increases in genome complexity? PLoS Genet. 6, e1001080 ( 10.1371/journal.pgen.1001080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch M, Bobay L-M, Catania F, Gout J-F, Rho M. 2011. The repatterning of eukaryotic genomes by random genetic drift. Annu. Rev. Genomics Hum. Genet. 12, 347–366. ( 10.1146/annurev-genom-082410-101412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitney KD, Boussau B, Baack EJ, Garland T. 2011. Drift and genome complexity revisited. PLoS Genet. 7, e1002092 ( 10.1371/journal.pgen.1002092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelkar YD, Ochman H. 2012. Causes and consequences of genome expansion in fungi. Genome Biol. Evol. 4, 13–23. ( 10.1093/gbe/evr124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hess J, Skrede I, Wolfe BE, LaButti K, Ohm RA, Grigoriev IV, Pringle A. 2014. Transposable element dynamics among asymbiotic and ectomycorrhizal Amanita fungi. Genome Biol. Evol. 6, 1564–1578. ( 10.1093/gbe/evu121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ågren JA, Greiner S, Johnson MTJ, Wright SI. 2015. No evidence that sex and transposable elements drive genome size variation in evening primroses. Evolution 69, 1053–1062. ( 10.1111/evo.12627) [DOI] [PubMed] [Google Scholar]
- 14.Fierst JL, Willis JH, Thomas CG, Wang W, Reynolds RM, Ahearne TE, Cutter AD, Phillips PC. 2015. Reproductive mode and the evolution of genome size and structure in Caenorhabditis nematodes. PLOS Genet. 11, e1005323 ( 10.1371/journal.pgen.1005323) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bast J, Schaefer I, Schwander T, Maraun M, Scheu S, Kraaijeveld K. 2016. No accumulation of transposable elements in asexual arthropods. Mol. Biol. Evol. 33, 697–706. ( 10.1093/molbev/msv261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goubert C, Modolo L, Vieira C, ValienteMoro C, Mavingui P, Boulesteix M. 2015. De novo assembly and annotation of the Asian tiger mosquito (Aedes albopictus) repeatome with dnaPipeTE from raw genomic reads and comparative analysis with the yellow fever mosquito (Aedes aegypti). Genome Biol. Evol. 7, 1192–1205. ( 10.1093/gbe/evv050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Modolo L, Lerat E. 2015. UrQt: an efficient software for the unsupervised quality trimming of NGS data. BMC Bioinformatics 16, 137 ( 10.1186/s12859-015-0546-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang W, Li L, Myers JR, Marth GT. 2012. ART: a next-generation sequencing read simulator. Bioinformatics 28, 593–594. ( 10.1093/bioinformatics/btr708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772 ( 10.1038/nmeth.2109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. ( 10.1080/10635150390235520) [DOI] [PubMed] [Google Scholar]
- 21.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. ( 10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 22.Pinheiro JC, Bates D, DebRoy S, Sarkar D, R-core Team 2016. nlme: linear and nonlinear mixed effects models. In R package version 3.1–127. https://cran.r-project.org/web/packages/nlme/index.html.
- 23.Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 24.Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401, 877–884. ( 10.1038/44766) [DOI] [PubMed] [Google Scholar]
- 25.Clark AG, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450, 203–218. ( 10.1038/nature06341) [DOI] [PubMed] [Google Scholar]
- 26.Biemont C. 2010. A brief history of the status of transposable elements: from junk DNA to major players in evolution. Genetics 186, 1085–1093. ( 10.1534/genetics.110.124180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Linde K, Houle D, Spicer GS, Steppan SJ. 2010. A supermatrix-based molecular phylogeny of the family Drosophilidae. Genet. Res. 92, 25–38. ( 10.1017/S001667231000008X) [DOI] [PubMed] [Google Scholar]
- 28.Gregory TR, Johnston JS. 2008. Genome size diversity in the family Drosophilidae. Heredity (Edinb) 101, 228–238. ( 10.1038/hdy.2008.49) [DOI] [PubMed] [Google Scholar]
- 29.Bainard JD, Forrest LL, Goffinet B, Newmaster SG. 2013. Nuclear DNA content variation and evolution in liverworts. Mol. Phylogenet. Evol. 68, 619–627. ( 10.1016/j.ympev.2013.04.008) [DOI] [PubMed] [Google Scholar]
- 30.Schaack S, Choi E, Lynch M, Pritham EJ. 2010. DNA transposons and the role of recombination in mutation accumulation in Daphnia pulex. Genome Biol. 11, R46 ( 10.1186/gb-2010-11-4-r46) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Phylogenetic data, including alignments: TreeBASE accession number 19296. (http://purl.org/phylo/treebase/phylows/study/TB2:S19296).